Back to Journals » Journal of Inflammation Research » Volume 14
PLAUR as a Potential Biomarker Associated with Immune Infiltration in Bladder Urothelial Carcinoma
Authors Liu M , Chen S, Zhang A, Zheng Q, Fu J
Received 25 June 2021
Accepted for publication 20 August 2021
Published 15 September 2021 Volume 2021:14 Pages 4629—4641
DOI https://doi.org/10.2147/JIR.S326559
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Mulin Liu,1 Siyi Chen,2 Aihui Zhang,2 Qin Zheng,2 Juan Fu3
1Department of Clinical Laboratory, The First Affiliated Hospital of Dalian Medical University, Dalian, Liaoning Province, 116011, People’s Republic of China; 2Department of Biochemistry and Molecular Biology, College of Basic Medical Science, Dalian Medical University, Dalian, Liaoning Province, 116044, People’s Republic of China; 3Department of Obstetrics and Gynecology, The First Affiliated Hospital of Dalian Medical University, Dalian, Liaoning Province, 116011, People’s Republic of China
Correspondence: Qin Zheng
Department of Biochemistry and Molecular Biology, Liaoning Provincial Core Laboratory of Glycobiology and Glycoengineering, College of Basic Medical Science, Dalian Medical University, 9 West Section, Lvshun South Road, Dalian, Liaoning Province, 116044, People’s Republic of China
Tel/ Fax +86 411-86110308
Email [email protected]
Juan Fu
Department of Obstetrics and Gynecology, The First Affiliated Hospital of Dalian Medical University, No. 222 Zhongshan Road, Dalian, Liaoning Province, 116011, People’s Republic of China
Tel +86 411-83635963 ext. 3589
Fax +86 411-83622844
Email [email protected]
Background: Bladder urothelial carcinoma (BLCA) is one of the most lethal and aggressive malignancies of genitourinary system that affects human health. The urokinase plasminogen activator receptor (PLAUR) plays essential roles in tumorigenesis and immune modulation, and its aberrant expression is closely correlated with cancer progression. However, whether PLAUR has the potential to be one promising biomarker or immunotherapy target for BLCA is unknown.
Methodology: Various online databases were applied to assess the expression profile and prognostic value of PLAUR, as well as its correlation with immune infiltration in BLCA, including Oncomine, PrognoScan, TCGA, cBioPortal, TIMER, TISIDB, UALCAN, and MethSurv. The expression of PLAUR in BLCA was confirmed with ELISA assay for serum samples and immunohistochemistry for tissue samples.
Results: The results showed that the expression of PLAUR was elevated in BLCA, which was further confirmed by ELISA and immunohistochemistry. Patients with higher PLAUR level were predicted to have lower overall survival and disease specific survival rates, which were not impacted by the genetic alterations of PLAUR. In addition, the expression of PLAUR was positively associated with immune infiltration, and also the expression levels of gene markers of various immune cells. The negative correlation between PLAUR expression and PLAUR methylation level was observed, among which PLAUR expression was positively correlated with the abundance of 28 kinds of tumor-infiltrating lymphocytes, while PLAUR methylation level was negatively correlated with the abundance of 11 types of tumor-infiltrating lymphocytes. Moreover, the methylation level of PLAUR was closely correlated with patients’ clinicopathological features, and hypomethylation of PLAUR was associated with better outcomes of BLCA patients.
Conclusion: These findings suggested that PLAUR had the potential to serve as a valuable detection and prognostic biomarker or immunotherapeutic target for BLCA.
Keywords: bladder urothelial carcinoma, PLAUR, biomarker, prognosis, immune infiltration, DNA methylation
Introduction
Bladder urothelial carcinoma (BLCA), one of the most common type and also the malignancy of the urinary system, is the leading cause of cancer-related death in China among elderly population with increasing morbidity and mortality.1,2 Patients with BLCA have unique features, including high metastasis frequency, high recurrence rate, and poor outcomes. Most of the patients are diagnosed in the middle or advanced stage due to the lack of sensitive markers and predictive indicators.3 Even with the rapid development of the detection, treatment and surveillance technologies in recent years, the survival rate of BLCA patients is still far from satisfactory and becoming one of the major threat to human health. After progressing from non-muscle-invasive bladder cancer (NMIBC) to muscle-invasive bladder cancer (MIBC) with high metastasis rate, the 5-year survival rate of patients will drop to only 4.6%.4 Nowadays, cystoscopy and urinary cytology are still the standard diagnostic methods for BLCA, but both methods are with shortcomings. For cystoscopy, it is a high sensitivity and specificity method but an invasive means with risks of complications. For urinary cytology, it is a noninvasive, high specificity but low-sensitivity method.5–7 Thus, it is indispensable and imperative to identify novel, valuable and noninvasive serum biomarkers to ameliorate the clinical detection and prognosis evaluation of BLCA.
The urokinase plasminogen activator receptor (PLAUR), belongs to the plasminogen activation system, consists of three domains and attaches to the cell membrane by a glycolipid anchor. It has multifaceted roles in diseases, especially cancer.8–11 PLAUR involves in cell–cell and cell–extracellular matrix interactions, and participates in various cancer-specific processes, such as cell differentiation,12 proliferation,13 metastasis14 and angiogenesis,15 etc. Rysenkova et al found that uPAR was an important regulator of EGFR and ERK1/2 signaling and implicated that uPAR was closely involved in neuroblastoma cell survival and differentiation.16 Semina et al reported that uPAR silencing promoted epithelial–mesenchymal transition and increased cell migration of neuroblastoma Neuro2a cells, which indicated the existence of a functional link between the urokinase system and epithelial–mesenchymal transition program.17 Biagioni et al found that uPAR loss reduced the pro-angiogenic effects of melanoma Exos by inhibition of VE-cadherin, EGFR and ERK1/2 signaling in endothelial cells, which suggested that uPAR was required for the pro-angiogenic activity of melanoma Exos, and the identification of uPAR-expressing Exos could be a potential biomarker for assessing pro-angiogenic propensity and monitoring the response to treatment in metastatic melanoma patients.18 PLAUR was demonstrably elevated in a variety of malignant cancers and correlated with the prognosis of cancer patients.19,20 However, the role of PLAUR in BLCA remains unclear.
To identify the sufficient biomarker with diagnostic and prognostic values, as well as potential targets for BLCA, the expression level of PLAUR was explored by using Oncomine database and confirmed with ELISA assay and immunohistochemical staining. The prognostic value of PLAUR was investigated with PrognoScan, TCGA and cBioPortal databases. In addition, the correlation between PLAUR expression and immune infiltration level was analyzed in TIMER and TISIDB databases, as well as PLAUR methylation level and the abundance of tumor-infiltrating lymphocytes. The analysis of the methylation level of PLAUR was performed in UALCAN database, as well as its correlation with clinicopathological features of BLCA patients, and the impact of DNA methylation of PLAUR on the overall survival of BLAC patients was determined with MethSurv database. The findings of this study elucidated that the upregulation of PLAUR might influence the tumorigenesis and progression of BLCA, implying that PLAUR had the potential to serve as a novel detection biomarker and also an immunotherapeutic target for BLCA.
Materials and Methods
Oncomine Database Analysis
Oncomine database (https://www.oncomine.org/resource/login.html) was applied to evaluate the expression profile of PLAUR in bladder cancer by searching “PLAUR” and selected the catalog “Bladder Cancer vs Normal Analysis” under the module of “Cancer vs Normal Analysis” according to the significance threshold “P-value: 0.05”.
Ethics Statement
The study was approved by the Clinical Ethics Review Board of the First Affiliated Hospital of Dalian Medical University according to the guidelines of the Declaration of Helsinki (YJ-KY-FB-2021-17).
Clinical Samples
The serum samples from 43 patients of bladder cancer and 47 healthy controls from January 2019 to April 2021 were collected from the First Affiliated Hospital of Dalian Medical University. The clinical characteristics of bladder cancer patients is shown in Supplementary Table 1. The paraffin-embedded tissues were acquired from 39 patients diagnosed with bladder cancer from January 2019 to April 2021 at the First Affiliated Hospital of Dalian Medical University. Informed consent was obtained from each subject. The inclusion criteria for bladder cancer patients were as follows: 1. more than 18 years old; 2. patients with bladder cancer were diagnosed by at least two senior pathologists without other malignancies, and serum samples were collected before patients received any chemotherapy or radiotherapy; 3. healthy controls were age-matched subjects with cancer patients without any bladder-related diseases. Any individuals with chronic or acute infections of urinary system or with any malignancies were excluded.
Enzyme-Linked Immunosorbent Assay
The serum level of PLAUR in bladder cancer patients and healthy controls was measured with enzyme-linked immunosorbent assay (ELISA) with commercial PLAUR ELISA kit (Langton, Shanghai, China) according to the manufacturer’s instructions. The serum samples (50µL) and biotinylated PLAUR antibody (50µL) were incubated together at 37°C for 1h. After washing for five times, 50µL streptavidin-horseradish peroxidase conjugates were added to the plate, and incubated at 37°C for 1h. Then, the absorbance value at 450nm (OD450nm) was determined and recorded. The serum concentration of PLAUR in bladder cancer patients was calculated according to the standard curve with curve expert software, and statistical analysis was conducted.
Immunohistochemistry
The paraffin-embedded tissue sections were deparaffinized in xylol and rehydrated in descending concentrations of ethanol. After antigen retrieval in citrate buffer (pH6.0) for 20 min, the tissue sections were blocked with 0.3% H2O2, and non-specific binding sites were blocked by complete serum. Then, the tissue sections were blocked with primary antibody at 4°C overnight (PLAUR, 1:100, Proteintech, Wuhan, China). After washing with PBS for three times, the second antibody was incubated, and then visualized with diaminobenzidine (DAB) (ZSGB-BIO, Beijing, China). Images were captured under an inverted microscope (BX51, Olympus, Tokyo, Japan) after the tissue sections were counterstained with hematoxylin. The yellowish-brown staining represented the positive result, and the staining intensity was evaluated in comparison with normal tissue which was scored as follows: 0: no staining, 1: weak staining, 2: moderate staining, 3: strong staining.
PrognoScan Database Analysis
PrognoScan database (http://dna00.bio.kyutech.ac.jp/PrognoScan/) was used to explore the prognostic value of PLAUR for bladder cancer patients. Overall survival (OS) and disease specific survival (DSS) were assessed by entering gene identifier “PLAUR” and choosing cohort “CNUH” with probe ID “ILMN_1691508”.
cBioPortal Database Analysis
cBioPortal database (http://www.cbioportal.org/) was used to investigate the frequency of genetic alterations of PLAUR, and its effects on overall survival and disease free survival rates, as well as the correlation between the expression of PLAUR and gene markers of immune cells in bladder cancer. Bladder Tract “Bladder Cancer (TCGA, Cell 2017, 413 samples)” was chosen. The genetic alterations of PLAUR were investigated with “OncoPrint” module, and the prognostic value of PLAUR for bladder cancer was explored with “Comparison/Survival” module, and the correlation between the expression of PLAUR and gene markers of various immune cells was evaluated with “Co-expression” module.
TIMER Database Analysis
TIMER database (https://cistrome.shinyapps.io/timer/) was used to measure the relationship between PLAUR expression and immune infiltration in BLCA, as well as the correlation between the expression of PLAUR and gene markers of immune cells. The “Gene” module was applied to explore the association between PLAUR expression and the infiltration levels of immune cells, and the “SCNA” module was used to explore the correlation between the somatic copy number alterations (SCNA) of PLAUR and the abundance of immune infiltration. The “Correlation” module was applied to draw the association between PLAUR expression and gene markers of immune cells.
TISIDB Database Analysis
TISIDB database (http://cis.hku.hk/TISIDB/index.php) was conducted to investigate the association between the expression of PLAUR, as well as the methylation level of PLAUR and the abundance of tumor-infiltrating lymphocytes. The analysis was carried out by entering “PLAUR” in “Gene Symbol” module, and the correlation was evaluated with “Lymphocyte” module.
UALCAN Database Analysis
UALCAN database (http://ualcan.path.uab.edu/analysis.html) was used to explore the correlation between the promoter methylation level of PLAUR and the clinicopathological features of BLCA patients. The analysis was conducted by entering gene symbol(s) “PLAUR” with TCGA dataset “Bladder urothelial carcinoma” and choosing the “Methylation” in “Links for analysis” module.
MethSurv Database Analysis
MethSurv database (https://biit.cs.ut.ee/methsurv/) was applied to explore the heatmap of DNA methylation of PLAUR in BLCA, as well as multivariable survival analysis of DNA methylation of PLAUR for BLCA by using TCGA dataset. For heatmap analysis, “Gene visualization” module was used by choosing “Bladder Urothelial Carcinoma [BLCA] TCGA March 2017” in “Cancer” and “PLAUR” in “Gene”. For survival analysis, “Single CpG” module was used by choosing “Bladder Urothelial Carcinoma [BLCA] TCGA March 2017” in “TCGA cancer datasets”, “PLAUR” in “Gene” and “Relation to island”, “Genomic Region” and “CpG site” modules.
Statistical Analysis
The data represented in this study were got from three independent experiments and expressed as mean ± S.E.M. The differences between two groups were analyzed with Student’s t-test by using SPSS 22.0 software (SPSS Inc., Chicago, IL). The diagnostic value of PLAUR for bladder cancer was evaluated with receiver operating characteristic curve (ROC). P<0.05 was considered as statistically significant.
Results
Increased Expression of PLAUR in BLCA Patients
To explore the role of PLAUR in BLCA, we first analyzed its expression in Oncomine database. The results showed that PLAUR expression was increased in bladder cancer tissues than in normal bladder tissues (Figure 1A). We further collected clinical serum and tissue samples to confirm the elevated expression of PLAUR. The results of ELISA assay found similar alteration of PLAUR in patients with bladder cancer (Figure 1B). ROC curve analysis was performed to evaluate the diagnostic value of PLAUR, which found that PLAUR could serve as a potential biomarker for bladder cancer detection with the area under the curve (AUC) 0.803, sensitivity 86%, and specificity 61.7% (Figure 1C). The immunohistochemical staining results indicated that PLAUR expression was upregulated in bladder cancer tissues when compared to normal bladder tissues (Figure 1D), which showed analogous changes with Oncomine database and ELISA assay results. Taken together, these data demonstrated that the expression of PLAUR was increased in bladder cancer, and might act as a potential serum biomarker for the detection of BLCA.
The Prognostic Value of PLAUR for BLCA
PrognoScan database was applied to investigate the correlation between PLAUR expression and the prognosis of BLCA patients. As a result, patients of BLCA with lower PLAUR level had better overall survival and disease specific survival rates than patients with higher PLAUR level (Figure 2A). In order to clarify whether PLAUR was a prognostic factor independent of other clinical variables including age, gender, cancer stage and pathological TNM stage, univariate and multivariate cox regression analysis were conducted. The results of univariate cox regression analysis showed that cancer stage (HR=1.7627 (1.2356–2.5148), P-value=0.0018), pathological T (HR=1.6950 (1.1516–2.4949), P-value=0.0075) and M (HR=2.0870 (0.7514–5.7967), P-value=0.0023) were independent biomarkers of overall survival, while age, gender and pathological N had no significant relationship with overall survival (Figure 2B). However, no matter age, gender, cancer stage, nor pathological TNM stage showed no significant correlation with overall survival in multivariate cox regression analysis (Figure 2C). These results suggested that PLAUR might have potential to serve as a predictive factor for the outcomes of BLCA patients. In addition, we further analyzed the effects of genetic alterations of PLAUR on the outcomes of BLCA patients via cBioPortal database. It was found that PLAUR was altered in 8% (32 out of 413) of BLCA patients, which included missense and truncating mutation, amplification and mRNA high (Figure 2D). However, the genetic alterations of PLAUR represented no significant impact on the overall survival and disease free survival rates of patients with BLCA (Figure 2E). These findings suggested that PLAUR could act as a potential prognostic indicator to evaluate the outcome of BLCA patients.
Association Between the Expression of PLAUR and the Infiltration Levels of Immune Cells
After confirming the expression profile and prognostic value of PLAUR in BLCA, we further explored the relationship between PLAUR expression and immune infiltration levels. Infiltrating immune cells were main components of the tumor microenvironment, which were closely associated with tumor progression. Among the six kinds of immune cells in TIMER database, PLAUR expression was found to be positively correlated with immune cell infiltration levels in BLCA except B cell (cor=−0.275, p=1.03e-07) (CD8+ T cell: cor=0.413, p=1.65e-16; CD4+ T cell: cor=0.226, p=1.25e-05; macrophage: cor=0.143, p= 6.09e-03; neutrophil: cor=0.549, p=5.54e-30; dendritic cell: cor=0.623, p=1.18e-40) (Figure 3A). In addition, we analyzed the relationship between the SCNA of PLAUR with immune infiltration, which showed its significant correlations with infiltrating levels of CD4+ T cell, macrophage and neutrophil (Figure 3B). Except TIMER database, we also used TISIDB database to explore the relation between PLAUR expression and the abundance of 28 kinds of tumor-infiltrating lymphocytes (Figure 3C), which corresponded to the results reported above. As shown in Table 1, we found that the expression of PLAUR was positively correlated with the abundance of tumor-infiltrating lymphocytes, including Activated CD8 T cell (rho=0.42, P-value<2.2e-16), Central memory CD8 T cell (rho=0.592, P-value<2.2e-16), Effector memory CD8 T cell (rho=0.623, P-value<2.2e-16), Activated CD4 T cell (rho=0.463, P-value<2.2e-16), Central memory CD4 T cell (rho=0.616, P-value<2.2e-16), Effector memory CD4 T cell (rho=0.209, P-value=2.11e-05), T follicular helper cell (rho=0.612, P-value<2.2e-16), Gamma delta T cell (rho=0.603, P-value<2.2e-16), Type 1 T helper cell (rho=0.639, P-value<2.2e-16), Type 2 T helper cell (rho=0.486, P-value<2.2e-16), Type 17 T helper cell (rho=0.265, P-value=6.44e-08), Regulatory T cell (rho=0.736, P-value<2.2e-16), Activated B cell (rho=0.366, P-value=3.08e-14), Immature B cell (rho=0.424, P-value<2.2e-16), Memory B cell (rho=0.348, P-value=6.79e-13), Natural killer cell (rho=0.624, P-value<2.2e-16), CD56bright natural killer cell (rho=0.195, P-value=7.35e-05), CD56dim natural killer cell (rho=0.199, P-value=5.13e-05), Myeloid derived suppressor cell (rho=0.666, P-value<2.2e-16), Natural killer T cell (rho=0.695, P-value<2.2e-16), Activated dendritic cell (rho=0.703, P-value<2.2e-16), Plasmacytoid dendritic cell (rho=0.533, P-value<2.2e-16), Immature dendritic cell (rho=0.312, P-value=1.54e-10), Macrophage (rho=0.594, P-value<2.2e-16), Eosinophil (rho=0.343, P-value=1.54e-12), Mast cell (rho=0.445, P-value<2.2e-16), Monocyte (rho=0.327, P-value=1.78e-11) and Neutrophil (rho=0.656, P-value<2.2e-16) (Table 1). Among them, the top five significant positive correlation was Regulatory T cell, Activated dendritic cell, Natural killer T cell, Myeloid derived suppressor cell and neutrophil, which played essential roles in the tumor microenvironment of cancers. Therefore, PLAUR might be involved in immune regulation of BLCA.
 |
Table 1 Association Between the Expression of PLAUR and the Abundance of Tumor-Infiltrating Lymphocytes |
After observing the positive correlation between PLAUR expression and immune infiltration in BLCA, we further detected its relationship with the expression of gene markers of diverse immune cells both in TIMER and cBioPortal databases (Table 2). Specifically, the results showed their significant positive correlation, including CD8+ T cell markers (CD8A: TIMER: cor=0.486, P-value=0; cBioPortal: cor=0.476, P-value=1.92e-24; CD8B: TIMER: cor=0.373, P-value=6.86e-15; cBioPortal: cor=0.363, P-value=4.00e-14), T cell markers (CD3D: TIMER: cor=0.424, P-value=3.02e-19; cBioPortal: cor=0.426, P-value=2.05e-19; CD3E: TIMER: cor=0.474, P-value=0; cBioPortal: cor=0.469, P-value=9.77e-24), B cell markers (CD19: TIMER: cor=0.19, P-value=1.10e-04; cBioPortal: cor=0.204, P-value=3.265e-05; CD79A: TIMER: cor=0.29, P-value=2.30e-09; cBioPortal: cor=0.297, P-value=9.56e-10), Natural killer cell markers (KIR2DL1: TIMER: cor=0.342, P-value=1.32e-12; cBioPortal: cor=0.340, P-value=1.73e-12; KIR2DL3: TIMER: cor=0.459, P-value=1.27e-22; cBioPortal: cor=0.454, P-value=3.62e-22; KIR3DL1: TIMER: cor=0.341, P-value=1.46e-12; cBioPortal: cor=0.338, P-value=2.48e-12; KIR3DL3: TIMER: cor=0.192, P-value=9.45e-05; cBioPortal: cor=0.189, P-value=1.239e-04; NCAM1: TIMER: cor=0.271, P-value=2.65e-08; cBioPortal: cor=0.259, P-value=1.13e-07), Neutrophil cell markers (ITGAM: TIMER: cor=0.66, P-value=0; cBioPortal: cor=0.641, p=1.60e-48; CD59: TIMER: cor=0.564, P-value=1.24e-35; cBioPortal: cor=0.542, P-value=1.73e-32), Type 1 T helper cell markers (TBX21: TIMER: cor=0.465, P-value=2.87e-23; cBioPortal: cor=0.455, P-value=3.38e-22; STAT4: TIMER: cor=0.598, P-value=5.30e-41; cBioPortal: cor=0.586, P-value=6.34e-39; IFNG: TIMER: cor=0.468, P-value=1.31e-23; cBioPortal: cor=0.473, P-value=4.41e-24), Type 2 T helper cell markers (STAT5A: TIMER: cor=0.308, P-value=2.62e-10; cBioPortal: cor=0.282, P-value=7.17e-09; IL13: TIMER: cor=0.231, P-value=2.36e-06; cBioPortal: cor=0.228, P-value=3.199e-06; CCR4: TIMER: cor=0.357, P-value=1.02e-13; cBioPortal: cor=0.315, P-value=7.39e-11; CCR8: TIMER: cor=0.415, P-value=1.94e-18; cBioPortal: cor=0.375, P-value=4.53e-15), Type 17 T helper cell marker (STAT3: TIMER: cor=0.474, P-value=0; cBioPortal: cor=0.465, P-value=2.56e-23), T follicular helper cell marker (IL21: TIMER: cor=0.205, P-value=3.13e-05; cBioPortal: cor=0.187, P-value=1.477e-04), Tumor associated macrophage cell markers (CCL2: TIMER: cor=0.543, P-value=0; cBioPortal: cor=0.534, P-value=2.04e-31; IL10: TIMER: cor=0.611, P-value=4.73e-43; cBioPortal: cor=0.600, P-value=3.28e-41; CD68: TIMER: cor=0.595, P-value=0; cBioPortal: cor=0.574, P-value=4.34e-37), Dendritic cell markers (CD1C: TIMER: cor=0.188, P-value=1.31e-04; cBioPortal: cor=0.168, P-value=6.631e-04; HLA-DPB1: TIMER: cor=0.543, P-value=0; cBioPortal: cor=0.531, P-value=4.62e-31; HLA-DQB1: TIMER: cor=0.506, P-value=5.76e-28; cBioPortal: cor=0.496, P-value=1.09e-26; HLA-DPA1: TIMER: cor=0.532, P-value=3.22e-31; cBioPortal: cor=0.517, P-value=2.73e-29), Regulatory T cell markers (FOXP3: TIMER: cor=0.474, P-value=0; cBioPortal: cor=0.448, P-value=1.63e-21; TGFB1: TIMER: cor=0.518, P-value=0; cBioPortal: cor=0.505, P-value=8.85e-28), Monocyte cell markers (CD86: TIMER: cor=0.671, P-value=0; cBioPortal: cor=0.664, P-value=3.51e-53; CSF1R: TIMER: cor=0.661, P-value=0; cBioPortal: cor=0.651, P-value=1.74e-50), M1 macrophage cell markers (NOS2: TIMER: cor=0.205, P-value=3.12e-05; cBioPortal: cor=0.167, P-value=7.00e-04; PTGS2: TIMER: cor=0.303, P-value=5.11e-10; cBioPortal: cor=0.257, P-value=1.42e-07), M2 macrophage cell markers (CD163: TIMER: cor=0.693, P-value=0; cBioPortal: cor=0.680, p=1.06e-56; VSIG4: TIMER: cor=0.677, P-value=0; cBioPortal: cor=0.665, P-value=1.94e-53). The results of the association between the expression of PLAUR and gene markers of immune cells further confirmed the positive correlation of PLAUR with immune infiltration in BLCA.
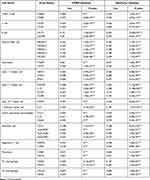 |
Table 2 Correlation Between the Expression of PLAUR and Gene Markers of Immune Cells |
These findings elucidated that PLAUR expression was positively correlated with immune infiltration, and it might serve as a potential immunotherapeutic target for BLCA treatment.
The Promoter Methylation Level of PLAUR and Its Correlation with Clinicopathological Features of BLCA Patients
To explore the potential mechanism by which PLAUR involved in the carcinogenesis of BLCA, we studied the correlation between the expression and the promoter methylation level of PLAUR with cBioPortal and UALCAN databases. It showed that the expression of PLAUR was negatively associated with the methylation level of PLAUR in BLCA (Spearman: −0.11, p=0.0305; Pearson: −0.14, p=6.042e-3) (Figure 4A). We also detected the relationships between the methylation level of PLAUR and clinicopathological characteristics of patients with BLCA. The promoter methylation level of PLAUR was downregulated in bladder cancer tissues when compared with normal bladder tissues (Figure 4B) and decreased in patients with higher cancer stage (Figure 4C) or with nodal metastasis (Figure 4D). Besides, it was negatively correlated with patients’ age (Figure 4E), weight (Figure 4F), smoking status (Figure 4G) and TP53 mutation status (Figure 4H). Thus, the decreased methylation level of PLAUR might be an underlying indicator, which reflected clinical characteristics of BLCA patients.
Association Between the Methylation Level of PLAUR and the Abundance of Tumor-Infiltrating Lymphocytes
Since the methylation level of PLAUR was closely correlated with clinicopathological features of BLCA patients, we further checked the relationship between the methylation level of PLAUR and the abundance of 28 types of tumor-infiltrating lymphocytes (Figure 5). As shown, it was significantly negatively associated with 11 kinds of tumor-infiltrating lymphocytes (Table 3), which were Effector memory CD8 T cell (rho=−0.118, P-value=0.0169), Activated CD4 T cell (rho=−0.099, P-value=0.0452), T follicular helper cell (rho=−0.11, P-value=0.0267), Type 1 T helper cell (rho=−0.103, P-value=0.0381), Regulatory T cell (rho=−0.102, P-value=0.0388), Activated B cell (rho=−0.111, P-value=0.025), Immature B cell (rho=−0.099, P-value=0.0457), Natural killer cell (rho=−0.115, P-value=0.0202), Myeloid derived suppressor cell (rho=−0.1, P-value=0.0433), Plasmacytoid dendritic cell (rho=−0.14, P-value=0.00465) and Neutrophil (rho=−0.101, P-value=0.042). These data revealed that the methylation level of PLAUR was negatively correlated, while the expression of PLAUR was positively correlated with the abundance of tumor-infiltrating lymphocytes that was opposite, which might be one of the reasons for immune infiltration, and it represented the potential to be an immunotherapeutic target for individualized treatment of BLCA patients.
 |
Table 3 Association Between the Methylation Level of PLAUR and the Abundance of Tumor-Infiltrating Lymphocytes |
 |
Figure 5 Correlation between the methylation level of PLAUR and the abundance of tumor-infiltrating lymphocytes. |
The Heatmap of DNA Methylation of PLAUR and Its Prognostic Value for BLCA
DNA methylation was a critical biological process of tumorigenesis and cancer progression, and the above results demonstrated that it was closely associated with BLCA, so we further investigated the effects of DNA methylation of PALUR on the outcomes of BLCA patients with MethSurv database. The heatmap of DNA methylation of PLAUR in BLCA was presented in Figure 6A. According to the heatmap, there were nine CpG sites in PLAUR, including cg17168630, cg25750408, cg20716515, cg06540636, cg14526204, cg19524534, cg23211714, cg02983203 and cg15459820. The influence of DNA methylation of PLAUR on the prognosis of BLCA patients were identified, which found that among these nine CpG sites, the significant prognostic values were observed in PLAUR-Body-Island-cg19524534 (HR=1.45, P-value=0.02), PLAUR-5ʹUTR;1stExon-S_Shore-cg23211714 (HR=1.457, P-value=0.014) and PLAUR-Body- Island-cg15459820 (HR=1.498, P-value=0.015). These results implied that DNA methylation of PLAUR played a crucial role in the occurrence and progression of BLCA, which was closely related with the prognosis of BLCA patients.
Discussion
Even though the diagnostic and therapeutic technologies have been improved and advanced, BLCA remains one of the most life-threatening cancer in the world. The understanding of the mechanism and the exploration of sensitive biomarkers is still not sufficient, leading to low diagnostic rate and short survival time of BLCA patients. Nowadays, serum biomarkers are widely used in clinic because of its easier accessibility and noninvasiveness for sample collection, shorter detection time, lower cost, risks and complications. Therefore, the investigation of efficient serum biomarkers for the detection, prognosis evaluation and therapy selection of BLCA is urgently needed. Thus far, a number of genes have been identified as playing key roles in BLCA, among which there is PLAUR. Liu et al found that FUT7 expression was increased in bladder cancer tissues and promoted epithelial–mesenchymal transition of bladder cancer cells, as well as immune infiltration in BLCA, which indicated its potential to be a detection biomarker or immunotherapeutic target for BLCA.21 Eich ML reported that MTHFD1L was overexpressed in bladder cancer and played a critical role in cell proliferation, colony formation and invasion, which could serve as a valuable therapeutic target for bladder cancer.22 Sidaway found that the expression of uPAR was associated with worse prognosis, staging and overall survival in patients with urothelial carcinoma of the bladder (UCB).23 Currently, there is limited literature on the potential biological influence of PLAUR in BLCA. In this study, the result of Oncomine database showed that PLAUR expression in BLCA was higher than that in normal bladder tissues, which was further confirmed with ELISA assay for serum samples and immunohistochemical staining for tissue samples (Figure 1). In addition, patients with lower PLAUR level represented a better outcome than patients with higher PLAUR level, which was not impacted by genetic alterations of PLAUR (Figure 2). Collectively, these findings reasonably suggested that PLAUR might considered as an oncogene in BLCA, and highlighted its potential role to be a serum detection and prognosis evaluation biomarker for BLCA patients.
As BLCA is a highly immunogenic malignant cancer, the abnormal regulation of immune response in the tumor microenvironment plays a decisive role in its occurrence and development.24–26 In the early stage of tumorigenesis, the immune cells will be activated to attack the tumor cells, and inhibit the progression of cancer. When the tumor progresses, the immune system will in turn to support the tumor cells and promote cancer progression mediated by immune cells.27 Understanding the tumor microenvironment may help to decipher the mechanism of cancers. Thus, it is necessary to identify useful immunotherapeutic targets that impact the immune infiltration level, which is a novel characteristic of cancer. Lv et al investigated the prognostic immune cells and potential therapeutic targets, and found that higher infiltrating levels of M0 macrophages, M2 macrophages and neutrophils were hazard factors, while CD8+ T cells and memory activated CD4+ T cells were protective factors. It shed insight on the individualized immunotherapy or potential drug targets.28 Chen et al reported that tumor derived SIGLEC family genes were related to tumor immune cell infiltration and might regulate tumor microenvironment by influencing chemokine axis, which could provide new thoughts for cancer immune therapy.29 We used TIMER and TISIDB databases to analyze the relationship between PLAUR expression and the levels of immune cell infiltration. The results showed that the expression of PLAUR was positively correlated with immune infiltration, including CD8+ T cells, CD4+ T cells, macrophage, neutrophil and dendritic cells, as well as 28 kinds of tumor-infiltrating lymphocytes (Figure 3, Table 1). In the meantime, the relationship between the expression of gene markers of immune cells and PLAUR was also investigated, which represented a significantly positive correlation (Table 2). It implied the importance of PLAUR in the regulation of tumor immune microenvironment, which might be involved in the promotion of immune infiltration in BLCA. These evidence revealed the central role of PLAUR as a novel biomarker for the functional regulation of immune infiltration and also provided novel insight into the immunotherapy of BLCA.
DNA methylation has been recognized as an essential biological process of cancer.30–32 Yu et al found that the methylation level of GALNT2/14 was negatively correlated with GALNT2/14 expression, and patients with GALNT2/14 hypomethylation had worse overall survival than patients with hypermethylation, which elucidated the role of DNA methylation of GALNT2/14 in the tumorigenesis and provided additional insight for therapy and prognosis of lung adenocarcinoma.33 Zhen et al identified 24 up-hypomethylated oncogenes and 37 down-hypermethylated tumor suppressor genes in hepatocellular carcinoma. Among them, higher expression of methylated CDC5L-cg05671347, MERTK-cg08279316, RHOA-cg05657651 and YBX1-cg16306148, and lower expression of methylated BCR-cg25410636, DFFA-cg20696875, SCUBE2-cg19000089 and TP63-cg06520450 were related with better overall survival in hepatocellular carcinoma patients. Altering the methylation status of these genes might be therapeutic targets for hepatocellular carcinoma.34 In order to elucidate the underlying mechanism of PLAUR in BLCA, we examined the relationship between PLAUR expression, the promoter methylation level and clinicopathological features of BLCA patients by using cBioPortal and UALCAN databases. The results indicated that the expression of PLAUR was negatively correlated with its promoter methylation level, which was significantly decreased in BLCA. Moreover, the promoter methylation level of PLAUR was inversely correlated with clinicopathological characteristics of BLCA patients, including cancer stage, nodal metastasis status, patients’ age, weight, smoking status and TP53 mutation status (Figure 4). Moreover, we explored the effects of methylation level of PLAUR on the survival of BLCA cases via MethSurv database (Figure 6). The prognostic values were observed in PLAUR-Body-Island-cg19524534, PLAUR-5ʹUTR;1stExon-S_Shore-cg23211714, and PLAUR-Body-Island-cg15459820. All together, the methylation status of PLAUR might be a novel direction for prognosis evaluation and treatment selection of BLCA.
Conclusion
Our study used multiple online databases to perform a comprehensive analysis between PLAUR and BLCA. We identified PLAUR as a novel diagnostic and prognostic biomarker and also a new immunotherapeutic target for BLCA. In addition, PLAUR expression was positively associated with the infiltration levels of immune cells, as well as the expression of their gene markers. We hope that our study could provide a valuable promising biomarker and therapeutic target for anti-tumor immunotherapy, as well as an additional feasible marker to evaluate the prognosis of BLCA.
Data Sharing Statement
All figures and tables adopted in this study to support the findings are included in the article.
Acknowledgments
We would like to thank Oncomine, PrognoScan, TCGA, cBioPortal, TIMER, TISIDB, UALCAN, and MethSurv databases for the availability of the data.
Funding
This study was supported by the Natural Science Foundation of Liaoning Province (grant number: 2020-MS-253, 2019-BS-075).
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Zangouei AS, Barjasteh AH, Rahimi HR, Mojarrad M, Moghbeli M. Role of tyrosine kinases in bladder cancer progression: an overview. Cell Commun Signal. 2020;18(1):127.
2. Mossanen M. The epidemiology of bladder cancer. Hematol Oncol Clin North Am. 2021;35(3):445–455.
3. Barani M, Hosseinikhah SM, Rahdar A, et al. Nanotechnology in bladder cancer: diagnosis and treatment. Cancers (Basel). 2021;13(9):2214.
4. Saginala K, Barsouk A, Aluru JS, Rawla P, Padala SA, Barsouk A. Epidemiology of bladder cancer. Med Sci (Basel). 2020;8(1):15.
5. Batista R, Vinagre N, Meireles S, et al. Biomarkers for bladder cancer diagnosis and surveillance: a comprehensive review. Diagnostics (Basel). 2020;10(1):39.
6. Sciarra A, Di Lascio G, Del Giudice F, et al. Comparison of the clinical usefulness of different urinary tests for the initial detection of bladder cancer: a systematic review. Curr Urol. 2021;15(1):22–32.
7. Georgantzoglou N, Pergaris A, Masaoutis C, Theocharis S. Extracellular vesicles as biomarkers carriers in bladder cancer: diagnosis, surveillance, and treatment. Int J Mol Sci. 2021;22(5):2744.
8. Li Santi A, Napolitano F, Montuori N, Ragno P. The urokinase receptor: a multifunctional receptor in cancer cell biology. therapeutic implications. Int J Mol Sci. 2021;22(8):4111.
9. Napolitano F, Montuori N. The role of the plasminogen activation system in angioedema: novel insights on the pathogenesis. J Clin Med. 2021;10(3):518.
10. Dowsett J, Ferkingstad E, Rasmussen LJH, et al. Eleven genomic loci affect plasma levels of chronic inflammation marker soluble urokinase-type plasminogen activator receptor. Commun Biol. 2021;4(1):655.
11. Hayek SS, Leaf DE, Samman Tahhan A, et al. Soluble urokinase receptor and acute kidney injury. N Engl J Med. 2020;382(5):416–426.
12. Lino N, Fiore L, Rapacioli M, et al. uPA-uPAR molecular complex is involved in cell signaling during neuronal migration and neuritogenesis. Dev Dyn. 2014;243(5):676–689.
13. Biagioni A, Chillà A, Del Rosso M, et al. CRISPR/Cas9 uPAR gene knockout results in tumor growth inhibition, EGFR downregulation and induction of stemness markers in melanoma and colon carcinoma cell lines. Front Oncol. 2021;11:663225.
14. Yang QX, Zhong S, He L, et al. PBK overexpression promotes metastasis of hepatocellular carcinoma via activating ETV4-uPAR signaling pathway. Cancer Lett. 2019;452:90–102.
15. Hu X, Mandika C, He L, et al. Construction of urokinase-type plasminogen activator receptor-targeted heterostructures for efficient photothermal chemotherapy against cervical cancer to achieve simultaneous anticancer and antiangiogenesis. ACS Appl Mater Interfaces. 2019;11(43):39688–39705.
16. Rysenkova KD, Klimovich PS, Shmakova AA, et al. Urokinase receptor deficiency results in EGFR-mediated failure to transmit signals for cell survival and neurite formation in mouse neuroblastoma cells. Cell Signal. 2020;75:109741.
17. Semina EV, Rubina KA, Shmakova AA, et al. Downregulation of uPAR promotes urokinase translocation into the nucleus and epithelial to mesenchymal transition in neuroblastoma. J Cell Physiol. 2020;235(9):6268–6286.
18. Biagioni A, Laurenzana A, Menicacci B, et al. uPAR-expressing melanoma exosomes promote angiogenesis by VE-Cadherin, EGFR and Upar overexpression and rise of ERK1,2 signaling in endothelial cells. Cell Mol Life Sci. 2021;78(6):3057–3072.
19. Bharadwaj AG, Holloway RW, Miller VA, Waisman DM. Plasmin and plasminogen system in the tumor microenvironment: implications for cancer diagnosis, prognosis, and therapy. Cancers (Basel). 2021;13(8):1838.
20. Chen X, Wei H, Qian D, et al. Predictive value of EGF and uPAR for chemoradiotherapy response and survival in patients with esophageal squamous cell carcinoma. Ann Transl Med. 2020;8(18):1152.
21. Liu M, Zheng Q, Chen S, Liu J, Li S. FUT7 promotes the epithelial-mesenchymal transition and immune infiltration in bladder urothelial carcinoma. J Inflamm Res. 2021;14:1069–1084.
22. Eich ML, Rodriguez Pena MDC, Chandrashekar DS, et al. Expression and role of methylenetetrahydrofolate dehydrogenase 1 like (MTHFD1L) in bladder cancer. Transl Oncol. 2019;12(11):1416–1424.
23. Sidaway P. Bladder cancer: uPAR expression indicates worse prognosis of urothelial carcinoma. Nat Rev Urol. 2015;12(3):120.
24. Crispen PL, Kusmartsev S. Mechanisms of immune evasion in bladder cancer. Cancer Immunol Immunother. 2020;69(1):3–14.
25. Nie Z, Chen M, Wen X, et al. Endoplasmic reticulum stress and tumor microenvironment in bladder cancer: the missing link. Front Cell Dev Biol. 2021;9:683940.
26. Annels NE, Simpson GR, Pandha H. Modifying the non-muscle invasive bladder cancer immune microenvironment for optimal therapeutic response. Front Oncol. 2020;10:175.
27. Daza J, Charap A, Wiklund PN, Sfakianos JP. Role of the innate immune system in the development, progression, and therapeutic response of bladder cancer. Eur Urol Focus. 2020;6(4):650–652.
28. Lv Y, Jin P, Chen Z, Zhang P. Characterization of hazard infiltrating immune cells and relative risk genes in bladder urothelial carcinoma. Am J Transl Res. 2020;12(11):7510–7527.
29. Chen Z, Yu M, Guo L, et al. Tumor derived SIGLEC family genes may play roles in tumor genesis, progression, and immune microenvironment regulation. Front Oncol. 2020;10:586820.
30. Peng Q, Liu Y, Kong X, et al. The novel methylation biomarker SCARA5 sensitizes cancer cells to DNA damage chemotherapy drugs in NSCLC. Front Oncol. 2021;11:666589.
31. Flausino CS, Daniel FI, Modolo F. DNA methylation in oral squamous cell carcinoma: from its role in carcinogenesis to potential inhibitor drugs. Crit Rev Oncol Hematol. 2021;164:103399.
32. Zhang J, Yin J, Luo L, et al. Integrative analysis of DNA methylation and transcriptome identifies a predictive epigenetic signature associated with immune infiltration in gliomas. Front Cell Dev Biol. 2021;9:670854.
33. Yu Y, Wang Z, Zheng Q, Li J. GALNT2/14 overexpression correlate with prognosis and methylation: potential therapeutic targets for lung adenocarcinoma. Gene. 2021;790:145689.
34. Zhen L, Ning G, Wu L, et al. Prognostic value of aberrantly expressed methylation genes in human hepatocellular carcinoma. Biosci Rep. 2020;40(10):BSR20192593.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





