Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 18
Plasma Proteomics Study Between the Frequent Exacerbation and Infrequent Exacerbation Phenotypes of Chronic Obstructive Pulmonary Disease
Authors Yang C , Yang L, Yang L, Li S, Ye L , Ye J, Chen C, Zeng Y, Zhu M, Lin X, Peng Q, Wang Y, Jin M
Received 21 February 2023
Accepted for publication 9 July 2023
Published 9 August 2023 Volume 2023:18 Pages 1713—1728
DOI https://doi.org/10.2147/COPD.S408361
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Min Zhang
Chengyu Yang,1,2,* Li Yang,3,4,* Lei Yang,5 Shuiming Li,5 Ling Ye,1,6 Jinfeng Ye,5 Chengshui Chen,3,4,7 Yiming Zeng,8 Mengchan Zhu,1 Xiaoping Lin,8 Qing Peng,9 Yun Wang,5,10 Meiling Jin1,6
1Department of Pulmonary and Critical Care Medicine, Zhongshan Hospital, Fudan University, Shanghai, 200032, People’s Republic of China; 2Department of Pulmonary and Critical Care Medicine, Huadong Hospital, Fudan University, Shanghai, 200040, People’s Republic of China; 3Department of Pulmonary and Critical Care Medicine, the First Affiliated Hospital of Wenzhou Medical University, Wenzhou, 325015, People’s Republic of China; 4Key Laboratory of Interventional Pulmonology of Zhejiang Province, the First Affiliated Hospital of Wenzhou Medical University, Wenzhou, 325015, People’s Republic of China; 5Longhua Innovation Institute for Biotechnology, Shenzhen University, Shenzhen, Guangdong, 518055, People’s Republic of China; 6Department of Allergy, Zhongshan Hospital, Fudan University, Shanghai, 200032, People’s Republic of China; 7Department of Pulmonary and Critical Care Medicine, the Quzhou Affiliated Hospital of Wenzhou Medical University, Quzhou People’s Hospital, Zhejiang, 324000, People’s Republic of China; 8Department of Pulmonary and Critical Care Medicine, the Second Affiliated Hospital of Fujian Medical University, Respiratory Medicine Center of Fujian Province, Quanzhou, Fujian, 362000, People’s Republic of China; 9Department of Pulmonary and Critical Care Medicine, Minhang Hospital, Fudan University, Shanghai, 201199, People’s Republic of China; 10Guangdong Provincial Key Laboratory of Infectious Diseases and Molecular Immunopathology, Shantou University Medical College, Shantou, Guangdong, 515041, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Meiling Jin, Department of Pulmonary and Critical Care Medicine, Zhongshan Hospital, Fudan University, Shanghai, 200032, People’s Republic of China, Email [email protected] Yun Wang, Longhua Innovation Institute for Biotechnology, Shenzhen University, Shenzhen, Guangdong, 518055, People’s Republic of China, Email [email protected]
Background: Frequent exacerbation (FE) and infrequent exacerbation (IE) are two phenotypes of chronic obstructive pulmonary disease (COPD), of which FE is associated with a higher incidence of exacerbation and a serious threat to human health. Because the pathogenesis mechanisms of FE are unclear, this study aims to identify FE-related proteins in the plasma via proteomics for use as predictive, diagnostic, and therapeutic biomarkers of COPD.
Methods: A cross-sectional study was conducted in which plasma protein profiles were analyzed in COPD patients at stable stage, and differentially expressed proteins (DEPs) were screened out between the FE and IE patients. FE-related DEPs were identified using data-independent acquisition-based proteomics and bioinformatics analyses. In addition, FE-related candidates were verified by enzyme-linked immunosorbent assay.
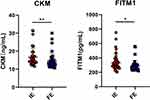
Results: In this study, 47 DEPs were screened out between the FE and IE groups, including 20 upregulated and 27 downregulated proteins. Key biological functions (eg, neutrophil degranulation, extracellular exosome, protein homodimerization activity) and signaling pathways (eg, arginine and proline metabolism) were enriched in association with the FE phenotype. Receiver operating characteristic (ROC) analysis of the 11 combined DEPs revealed an area under the curve of 0.985 (p < 0.05) for discriminating FE from IE. Moreover, correlation and ROC curve analyses indicated that creatine kinase, M-type (CKM) and fat storage-inducing transmembrane protein 1 (FITM1) might be clinically significant in patients with the FE phenotype. In addition, plasma expression levels of CKM and FITM1 were validated to be significantly decreased in the FE group compared with the IE group (CKM: p < 0.01; FITM1: p < 0.05).
Conclusion: In this study, novel insights into COPD pathogenesis were provided by investigating and comparing plasma protein profiles between the FE and IE patients. CKM, FITM1, and a combinative biomarker panel may serve as useful tools for assisting in the precision diagnosis and effective treatment of the FE phenotype of COPD.
Keywords: chronic obstructive pulmonary disease, frequent exacerbation, data-independent acquisition, proteomics, bioinformatics analysis
Background
Chronic obstructive pulmonary disease (COPD) is a common and progressive lung disease leading to high morbidity and mortality globally.1 Acute exacerbation leads to rapid deterioration of pulmonary function and a significant decline in the quality of life of COPD patients, resulting in severe economic and social burdens.2–4 Based on the frequency of acute exacerbations per year, the COPD population can be divided into two clinical phenotypes: frequent exacerbation (FE) and infrequent exacerbation (IE). Hurst et al reported that FE is a unique phenotype of COPD that appears to be relatively stable.5 This status implies the presence of many complex underlying factors regulating the pathophysiological processes, such as genetics, biology, pathology, and behavior, which can determine susceptibility or resistance to recurrent episodes, regardless of the severity of this disease.6 Frequent exacerbators have a higher risk of hospitalization and a worse prognosis compared with infrequent exacerbators. Despite the importance of the FE phenotype, its pathogenesis has not been sufficiently studied. The identification of the FE phenotype is currently based on clinical assessment and records of previous treatments, which is complicated by the time-consuming process of establishing a unified standard. Therefore, the discovery of special, functional, and feasible biomarkers is needed to guide clinical practice and improve early interventions for patients.
The proteomics can provide additional bioinformatics information for understanding the pathophysiological processes beyond the genome and transcriptome. Proteomics is one of the rapidly developing high-throughput technologies that has gained popularity in recent years. It is used to explore the protein expression profiles of cells, tissues, and organisms during disease development. Therefore, in-depth investigation of the pathogenesis of COPD with the multi-level discovery of related protein biomarkers has become an important subject of research in modern precision medicine.
In the past, most liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) methods relied on data-dependent acquisition (DDA). Traditional DDA selects peptide ions for fragmentation based on their abundance on MS1 scans, which can be hampered by interference from chemical noise,7,8 resulting in the loss of some low-abundance precursors.9 Data-independent acquisition (DIA) has emerged to overcome this limitation. In contrast with DDA, DIA systematically parallelizes the fragmentation of all possibly generated precursor ions within a wide m/z range and quantifies peptides at the MS2 level, thereby providing better sensitivity, precise quantification, and improved reproducibility.10 The convoluted or multiplexed fragment ion spectra generated by DIA require more sophisticated processing algorithms than DDA. Therefore, by using DIA, bioinformatics, and statistics in this study, we aimed to investigate FE-related protein profiles and reveal their pathophysiological functions during the disease development to enhance the precision treatment and management of COPD.
Materials and Methods
Subject Enrollment
This research project and procedures were approved by the Ethics Committee of Zhongshan Hospital Affiliated with Fudan University and complied with the ethical guidelines and standards set by the Declaration of Helsinki (No. B2017-022R). Written informed consent was signed by every subject.
This cross-sectional study was conducted at Zhongshan Hospital Affiliated with Fudan University (Shanghai, China). Patients with COPD at stable stage (no history of acute exacerbation within 4 weeks prior to enrollment) were recruited from the outpatient department with no additional intervention from March 2017 to September 2018. All patients maintained the original prescription of therapeutic drugs when sampling.
The inclusion criteria of COPD patients were age of 40 to 80 years old, history of smoking (≥10 pack-years), and history of biofuel use (ie, frequent use of charcoal, wood, animal manure, or crops for home heating and cooking) or occupational exposure to dust and chemicals (eg, mining, quarrying, casting, grain dust, paint, chemical industry, and other occupational dust/gas smoke longer than one year).
According to the 2017 Global Initiative for Chronic Obstructive Pulmonary Disease (GOLD) guidelines,11 the key diagnostic criterion of COPD is a post-bronchodilator forced expiratory volume at 1 second (FEV1)/forced vital capacity (FVC) <0.70. The COPD patients were assigned to either the FE or IE group based on their exacerbation history. Acute exacerbation was defined as the acute deterioration of respiratory symptoms leading to the requirement for additional treatment (emergency or hospitalization with antibiotics and/or systemic glucocorticoid therapy).11 The FE subjects were defined as ≥2 acute exacerbations of COPD per year or ≥1 acute exacerbation leading to hospitalization.11
The exclusion criteria were (a) history of acute pulmonary disease (eg, pneumonia, acute lung injury) within 3 months before enrollment, (b) history of other chronic lung diseases (eg, bronchiectasis, pulmonary interstitial fibrosis), (c) history of malignant tumor, and (d) pregnancy or lactation. The research records included the patients’ demographic data, smoking history, history of biofuel use/occupational exposure, laboratory tests, and lung function.
Plasma Specimen Collection
After enrollment, 5 mL of peripheral venous blood per patient was collected into a collection tube containing ethylenediaminetetraacetic acid anticoagulant. Low-temperature centrifugation (4°C, 1000 rpm, 10 min) was immediately performed on the blood sample, and the supernatant was divided into enzyme-free Eppendorf tubes and stored at −80°C.
To detect lower concentrations of plasma proteins, high-abundance and structural proteins were removed using the Human 14 Multiple Affinity Removal Kit (Agilent, Santa Clara, CA, USA) following the manufacturer’s instructions.
Protein Digestion and Peptide Fractionation via High-pH Reverse-Phase High-Performance Liquid Chromatography
The protein concentration in the pretreated sample was measured using a Bradford assay kit (Bio-Rad, Hercules, California, USA), and 100 μg of protein from each sample was subjected to trypsin digestion based on an aided filter sample preparation protocol. Briefly, the proteins were denatured by the addition of 8 M urea and vortexed for 5 min, and the proteins were reduced with dithiothreitol at 37°C for 2 hr followed by alkylation with iodoacetamide for 30 min and incubation in the dark. The mixture was then transferred to a 10-kDa filter (Sartorius, Gottingen, Niedersachsen, Germany) and centrifuged at 14,000 ×g for 10 min until the buffer was spun down completely into the collection tube. The proteins were kept on the ultrafiltration membrane and redissolved using 50 mM ammonium bicarbonate. After centrifugation, the redissolved proteins were digested with trypsin (protein: enzyme = 40:1) overnight, and tryptic peptides were subsequently centrifuged and transferred into a fresh tube for peptide collection.
All samples (10 µg/sample) were redissolved and fractionated using the Ultimate 3000 high-performance liquid chromatography system (Thermo Fisher Scientific, Waltham, Massachusetts, USA) coupled with an Agilent 300 Extend-C18 high pH column (2.1 × 250 mm, 3.5 μm). One sample was loaded onto a column, and the elute was collected at a flow rate of 0.3 mL/min by following gradient mobile phase B (95% acetonitrile (ACN), pH 10.0) at (1) 5% for 3 min, (2) 5–35% for 50 min, (3) 35–95% for 7 min, and (4) 100% for the last 4 min. An additional 5% mobile phase B was flowed for 6 min for equilibration. The elution peak was monitored at a wavelength of 214 nm, and the sample fractions were collected every 1 min. Finally, all components per sample were combined into a total of 25 fractions and vacuum-dried.
DDA and DIA Analyses Based on Nano-Liquid Chromatography-Mass Spectrometry
DDA and DIA analyses were performed using a Q-Exactive mass spectrometer (Thermo) coupled with an Easy-nanoLC 1200 system. The peptide digests were reconstituted in nano-LC mobile phase A (0.1% formic acid), loaded onto a NanoViper C18 trap column (3 mm, 100 Å), and separated on an analytical column (75 mm × 25 cm C18-2 mm 100 Å) via a 2-hr linear gradient from 5 to 35% mobile phase B (95% ACN with 0.1% formic acid) at a flow rate of 300 nL/min, followed by a linear increase to 95% mobile phase B for 2 min and 95% for 6 min. The Q-Exactive mass spectrometer was operated with a spray voltage of 1.9 kV and a capillary temperature of 275°C in a 350–1500 m/z scan range and 70,000 resolution, and the maximum ion injection time was 100 ms. Twenty of the most abundant precursor ions from each DDA cycle were selected for higher-energy collision dissociation fragment analysis with a maximum ion injection time of 50 ms, collision energy of 28 eV, and dynamic exclusion of 25 s. For each sample in the DIA analysis, MS1 and MS2 were set with a maximum ion injection time of 50 ms. MS1 was set as mentioned above (350–1500 m/z scan range and 70,000 resolution) and was separated into 34 acquisition windows with a mass range of 14–152 Da. The liquid conditions were consistent with the DDA model for separation. Raw DIA data were analyzed using Spectronaut Pulsar X.
Spectral Library Construction and Protein Quantification
The spectral library of plasma samples was constructed from raw MS files obtained from the Human UniProt protein database (20,336 protein sequences) and analyzed using Proteome Discoverer (V2.1.0.81). The search was conducted with a precursor tolerance of 10 ppm and a fragment ion tolerance of 0.05 Da. Identified peptides were filtered with a false discovery rate of <1% and contained at least one unique peptide segment. Protein abundance was calculated using the peak areas of the samples. Raw data from the spectral library and DIA were loaded with the Spectronaut Pulsar X software via protein relative quantification analysis. Comparative analyses of the datasets were performed after quantile normalization.
Bioinformatics Analysis
The identified proteins were screened out as differentially expressed proteins (DEPs) between the FE and IE groups. A fold change of relative expression >1.2 or <0.83 with a p-value <0.05 served as a criterion for processing the quantitative DIA data. Principal component analysis (PCA) and partial least squares discrimination analysis (PLS-DA) were performed using R package ggplot2 and mixOmics (version 4.2.0). The Metascape online website (http://metascape.org) was used for Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) signal pathway analyses.12 The protein–protein interaction (PPI) networks of DEPs were constructed using the online database of the Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org) (version 11.5) and visualized by Cytoscape (version 3.7.1).
Validation of DEPs by Enzyme-Linked Immunosorbent Assay
To validate the accuracy of proteomics results, the expression levels of the key DEPs were detected in plasma samples from an additional 40 FE and 40 IE patients by enzyme-linked immunosorbent assay (ELISA). The candidate proteins were selected based on several features: (a) their biological processes and signaling pathways related to the pathogenesis of COPD, according to previous research; (b) their expression levels related to lung function, according to the correlation analysis; and (c) their areas under the curve (AUC) >0.7, according to receiver operating characteristic (ROC) analysis. The ELISA kits were purchased from Jianglai Bio (Shanghai, China). Each sample was tested with two standard replicates at the same time. The ELISA assays were performed according to the manufacturer’s instructions.
Statistical Analysis
Statistical analysis was performed using SPSS 24.0 (IBM Corporation, Armonk, NY, USA). Continuous variables were presented as means (standard deviation) or medians (interquartile range). Independent sample t-tests and Mann–Whitney U-test were used to determine differences between two groups for continuous variables. Categorical variables were presented as percentages and compared via chi-squared test. Spearman correlation analysis was used to assess the correlation between DEP expression and lung function. The ROC curve was applied to evaluate the diagnostic value of DEPs. A p-value <0.05 was considered statistically significant.
Results
Clinical Characteristics of FE and IE Subjects
The summarized workflow of the study is presented in Figure 1. A total of 76 patients with COPD were enrolled, including 34 and 42 patients in the FE and IE groups, respectively. The clinical information of the patients is summarized in Table 1. There were no differences in sex, age, body mass index, smoking history, or hemoglobin levels between the two groups. FEV1 and FEV1% predicted were significantly lower in the FE group than in the IE group (p <0.05). In addition, there was a significant difference in the GOLD classification between the two groups (p <0.05).
 |
Table 1 Clinical Characteristics of Enrolled COPD Patients |
DEPs Screened Between the FE and IE Groups
A total of 1036 proteins were identified from both the FE and IE groups. The quantile normalization of the datasets is presented in Supplementary Figure 1. Forty-seven DEPs were screened out (with a threshold of fold change >1.2 or <0.83, and p <0.05) as candidate biomarkers by comparing FE patients to IE patients, including 20 upregulated and 27 downregulated DEPs (Figure 2). The information of the DEPs is presented in Table 2.
 |
Table 2 Differentially Expressed Proteins Between FE and IE Groups of COPD patients |
Principal Component Analysis (PCA) and Partial Least Squares Discrimination Analysis (PLS-DA) of the DEPs
To identify the protein expression profiles, PCA was performed based on the DEPs via a comparison assay between the FE and IE groups. The results were visualized using a two-dimensional coordinate map (Figure 3A). The result indicated that there were only a few FE-related features in the plasma protein samples between the two groups, suggesting that strict and sensitive detection of the protein candidates was required. PLS-DA was performed to identify the differences by comparing the protein expression profiles of the FE and IE groups, and the results indicated that two groups could be distinguished (Figure 3B). Compared with the unsupervised PCA model, the supervised PLS-DA model better distinguished the FE group from the IE group, which indicated that remarkable FE-related DEPs existed in the plasma protein profiles of above two groups.
GO and KEGG Enrichment Analyses of FE-Related DEPs
To further understand the biological roles of the 47 FE-related DEPs that were screened out as candidate biomarkers by comparing the FE group with the IE group, GO and KEGG enrichment analyses were performed using Metascape (Figures 4A and B). By GO analysis, the biological processes of the DEPs were mainly enriched in the following terms: neutrophil degranulation, movement of cell or subcellular component, immune response, and lipid storage. In addition, changes in cellular components were associated with the extracellular exosome, extracellular space, and extracellular region. Moreover, changes in molecular function were relevant to protein homodimerization activity, ribonuclease activity, and endonuclease activity. GO enrichment analysis was also performed in the upregulated and downregulated DEPs, respectively (Supplementary Figure 2).
Concurrently, KEGG analysis revealed that the DEPs were involved in complement and coagulation cascades, bacterial invasion of epithelial cells, and arginine and proline metabolism.
Establishment of a Possible PPI Network of FE-Related DEPs
A total of 47 FE-related biomarkers were uploaded to the STRING database. The minimum required interaction score was set to medium confidence (0.3), and disconnected nodes were removed from the network. The PPI network graph is visualized in Figure 5 by Cytoscape to present the possible interrelationships of the DEPs and signaling pathways they involved.
Correlation Analysis of FE-Related DEPs
Correlation analyses of FEV1% and FE-related DEPs were performed to investigate the association between the expression of DEP and pulmonary function (Figure 6), individually. The results indicated that pulmonary function was correlated with the expression levels of seven proteins: actin-related protein 2/3 complex subunit 5 (ARPC5), complement C4-B (C4B), creatine kinase, M-type (CKM), Fc receptor-like 5 (FCRL5), fat storage-inducing transmembrane protein 1 (FITM1), immunoglobulin lambda constant 2 (IGLC2)/IGLC3, and ubiquitin protein ligase E3C (UBE3C). Among these proteins, expression levels of ARPC5 and C4B were negatively correlated with FEV1%, while expression levels of the other proteins were positively correlated with FEV1% (p <0.05).
ROC Analysis of FE-Related DEPs
Among the 47 FE-related DEPs, 11 proteins had AUCs >0.7 according to ROC analysis, including inter-alpha-trypsin inhibitor heavy chain 3 (ITIH3), lymphatic vessel endothelial hyaluronan receptor 1 (LYVE1), paraoxonase 1 (PON1), alanine-glyoxylate and serine-pyruvate aminotransferase (AGXT), immunoglobulin kappa variable 3D-11 (IGKV3D-11)/IGKV3-11, septin-2 (SEPTIN2), apolipoprotein F (APOF), dopamine beta-hydroxylase (DBH), CKM, extracellular serine/threonine protein kinase FAM20C (FAM20C), and FITM1 (Figure 7). The sensitivity, specificity, optimal cutoff, 95% confidence interval, and p-values are presented in Table 3. ROC analysis of the 11 combined DEPs resulted in an AUC of 0.985 (95% CI: 0.966–1.000, p <0.05) for discrimination of FE from IE (Figure 8). The max Youden index was 0.752, with a sensitivity of 90.9% and a specificity of 100%, which was superior to the results of any individual DEP, suggesting that the optimal approach was the measurement of a combined biomarker panel.
 |
Table 3 Areas Under the Curve, Sensitivities, Specificities, and Optimum Cutoff Scores of the DEPs for Predicting Frequently Exacerbated COPD |
Validation of Candidate DEPs as FE-Related Biomarkers
To confirm the results obtained from the proteomics, bioinformatics, correlation, and ROC curve analyses, two key candidate biomarkers (CKM and FITM1) were selected and individually validated in plasma samples from an additional 40 FE and 40 IE patients in a validation cohort using ELISA assays. Expression levels of both CKM and FITM1 were significantly decreased in the FE group compared with the IE group, which was consistent with the proteomics results (Figure 9).
Discussion
In recent years, a growing number of studies have focused on the occurrence and development of COPD. However, few reports have investigated the phenotypes of COPD.13–15 The FE phenotype significantly impacts patient’s quality of life and disease prognosis. Therefore, it would be of great importance to identify patients with FE phenotype and deliver appropriate monitoring and intervention at the earliest occasion. Singh et al investigated gene expression profiles with microarray and polymerase-chain reaction analysis of sputum and blood samples from subjects in the ECLIPSE COPD cohort and screened out B3GNT, LAF4, and ARHGEF103 as biomarkers predicting FE.16 Sun et al detected changes in the lung proteome of COPD patients via tandem mass tag-labeled quantitative proteomics combined with LC-MS/MS, which revealed that major histocompatibility complex class II, DQ alpha 1 (HLA-DQA1), polymeric immunoglobulin receptor (PIGR), and biglycan (BGN) were significantly increased in FE patients compared with IE patients.17 Although the lung is the main target organ of COPD, the complex condition and severe hypoxia can cause a variety of complications in the body. Therefore, blood biomarkers may provide a better systematic assessment of the nature of COPD. The advantage of blood biomarkers is that obtaining blood samples is associated with less risk than sampling from the lung, and these biomarkers can be sampled repeatedly and dynamically to monitor disease progression. Therefore, in this study, peripheral blood samples were collected and individually tested to determine the protein expression profiles of FE and IE patients via DIA-MS.
After DIA-MS measurement and comparison analysis, 47 DEPs were screened out between the FE and IE groups, including 20 upregulated and 27 downregulated proteins. The PCA data indicated slight differences in plasma protein profiles between the two groups because of individual risk factors in each sample, further demonstrating that COPD is a heterogeneous disease. Since the two groups of patients only differed in phenotype of COPD, careful and profound analyses were needed to shed more light on these DEPs. A supervised PLS-DA model was then applied to obtain better insights into the data. FE and IE groups were relatively well separated in the PLS-DA score plot. The relationships among the DEPs were analyzed through functional and pathway enrichment analyses, which revealed that neutrophil degranulation, extracellular exosome, protein homodimerization activity, and complement and coagulation cascades were associated with FE. Hoenderdos et al reported that hypoxia augmented neutrophil degranulation and conferred enhanced injurious potential that might be relevant to neutrophilic airway inflammation in COPD.18 Lodge et al found that hypoxia promoted neutrophil degranulation and neutrophil-induced endothelial damage, which might contribute to increased cardiovascular risk in patients with COPD.19 Recently, an accumulating amount of research demonstrated the active involvement of complement and coagulation cascades pathway in COPD. Circulating C1q was found to be related to the pulmonary function of COPD and might be a biomarker to predict the risk of COPD deterioration.20 A clinical study showed that coagulation system was activated during COPD exacerbations and coagulation markers were potential predictors of COPD exacerbations.21 The results obtained from the present bioinformatics analyses were consistent with data from previous studies.
According to analyses of biological processes and signaling pathways, and results of correlation and ROC curve analyses, the downregulation of plasma CKM and FITM1 may be clinically significant in COPD patients of FE phenotype. The expression levels of CKM and FITM1 detected by ELISA were significantly decreased in FE patients compared with IE patients, which was consistent with the proteomics results. Moreover, PPI network analysis showed that there was a close interaction between the two proteins.
CKM is involved in arginine and proline metabolism pathway which may play a key role in regulating the formation and metabolism of proteins.22 Anoxic injury in patients with COPD was reported to be accompanied by the production of methylated arginine derivatives, which might be useful for recognizing acute exacerbations of COPD.23 Activation and expression of methylated arginine derivatives might also play an important role in the extrapulmonary complications of COPD.24,25 CKM is believed to play an indispensable role in maintaining energy homeostasis by providing a stable supply of creatine phosphate, which is essential for maintaining the Ca2+-ATPase channel in the sarcoplasmic reticulum.26 More than 90% of the CKM in circulation is derived from human skeletal muscle; therefore, as a biomarker of skeletal muscle protein metabolism, it can be used to measure the rate of tissue protein synthesis.27 FITM1 belongs to an evolutionarily conserved family of proteins and is involved in fat storage.28 The binding of FIT protein to triglycerides is important during FIT-mediated lipid droplet (LD) formation.29 A previous study showed that LDs could limit the level of reactive oxygen species and inhibit the oxidation of polyunsaturated fatty acids in glial cells.30 FIT proteins have varying tissue distribution, and FIT1 is mainly expressed in muscle. Yan et al proposed that FIT1 was a direct target of MyoD and participated in muscle development.31 Mormeneo et al reported that the overexpression of peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1α) could induce the expression of FITM1 in cultured human skeletal muscle cells, accompanied by the accumulation of LDs.32 There is an imbalance in the rate of muscle protein synthesis and degradation in patients with COPD. The degradation rate of skeletal muscle protein is increased in cachexic patients with COPD, which causes muscle wasting.33 Muscle wasting was reported to be associated with systemic inflammation and oxidative stress in COPD patients.34,35 Hallin et al analyzed 41 patients who were hospitalized due to exacerbation of COPD, and found that patients who lost weight during the 1-year follow-up were more likely to experience acute exacerbations than those who had no weight change or gained weight.36 In addition, it has been reported that there was an association between the occurrence of sarcopenia and deteriorating lung function (FEV1) in COPD patients.37 Therefore, CKM and FITM1 may be involved in the exacerbation and development of COPD. To date, few studies have investigated the relationship between CKM/FITM1 and COPD. Further studies are needed to explore the mechanisms of thetwo biomarkers in COPD.
Previous studies have reported that frequent exacerbators tend to have a lower FEV1% predicted and a rapid decline in FEV1.38–40 Moreover, a lower FEV1 value was identified as an independent risk factor for future exacerbations.41 In this study, FEV1 and FEV1% predicted were significantly lower in the FE group than in the IE group. In addition to CKM and FITM1, ARPC5, C4B, FCRL5, IGLC2/IGLC3 and UBE3C expression levels were correlated with pulmonary function, suggesting that these proteins might have significant associations with the development of airflow obstruction and be essential in the FE phenotype. FCRL5 belongs to the immunoglobulin receptor superfamily and is expressed on both mature B cells and plasma cells. A recent study identified a novel single-nucleotide polymorphism of FCRL5 that was significantly associated with a risk of developing asthma with comorbid allergic rhinitis.42 The UBE3C gene is crucial in antigen presentation and immune invasion. Polymorphisms in this gene could affect NF-κB activation by modulating IκB ubiquitination and were involved in the decrease of lung function with airway inflammation.43
ROC analysis of the 11 selected DEPs revealed that a combinative biomarker panel was the optimal approach for discriminating FE from IE in COPD patients. LYVE-1 binds to hyaluronan on the luminal surface of lymphatic vessels. LYVE-1 expression was reported to be significantly higher in the lung tissue of COPD patients compared with non-COPD smokers and was correlated with FEV1% predicted.44 PON1, a member of the paraoxonase family of enzymes, has antioxidant properties, and is involved in redox homeostasis. Lada et al reported that patients with COPD had lower PON1 activity compared with healthy subjects,45 and they found that PON1 polymorphism could contribute to the reduction of PON1 activity in patients with COPD.46 SEPTIN2 is a member of a highly conserved GTPase family. It was revealed that SEPTIN2 fundamentally regulated airway epithelial barrier function in response to pathological luminal stimuli by altering cortical actin expression.47 The protein encoded by DBH catalyzes the conversion of dopamine to norepinephrine, which is the main neurotransmitter of the sympathetic nervous system.48 A DBH polymorphism (rs3025343) was significantly associated with smoking cessation in patients with COPD.49
Interactions between COPD and other DEPs (meaningful in the correlation and ROC analysis) have not been widely reported according to our literature retrieval. Further studies will focus on exploring the regulatory roles of those potential biomarkers for the early diagnosis and prevention of the FE phenotype of COPD.
Conclusions
In this study, two novel biomarkers (CKM and FITM1) and a comprehensive perspective on the mechanism of FE phenotype were found by detecting, analyzing, and verifying plasma samples with proteomics in this study. These findings can be applied in the precision diagnosis and treatment of COPD. Furthermore, a panel of 11 plasma biomarkers might be a useful tool for discriminating the FE patients from the IE patients. Further investigations are required to elucidate the pathophysiological function of plasma proteins involved in the FE phenotype of COPD via enlarging the sample size and performing researches in vitro and in vivo.
Abbreviations
COPD, chronic obstructive pulmonary disease; FE, frequent exacerbation; IE, infrequent exacerbation; LC-MS/MS, liquid chromatography-tandem mass spectrometry; DDA, data-dependent acquisition; DIA, data-independent acquisition; AUC, area under the curve; ROC, receiver operating characteristic; ELISA, enzyme-linked immunosorbent assay; GOLD, Global Initiative for Chronic Obstructive Lung Disease; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; DEPs, differentially expressed proteins; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein interaction; PCA, principal component analysis; PLS-DA, partial least squares discrimination analysis; ARPC5, actin-related protein 2/3 complex subunit 5; C4B, complement C4-B; CKM, creatine kinase, M-type; FCRL5, Fc receptor like 5; FITM1, fat storage-inducing transmembrane protein 1; IGLC2, immunoglobulin lambda constant 2; UBE3C, ubiquitin protein ligase E3C; ITIH3, inter-alpha-trypsin inhibitor heavy chain 3; LYVE1, lymphatic vessel endothelial hyaluronan receptor 1; PON1, paraoxonase 1; AGXT, alanine-glyoxylate and serine-pyruvate aminotransferase; IgKV3D-11, immunoglobulin kappa variable 3D-11; SEPTIN2, septin-2; APOF, apolipoprotein F; DBH, dopamine beta-hydroxylase; FAM20C, extracellular serine/threonine protein kinase FAM20C; HLA-DQA1, major histocompatibility complex class II, DQ alpha 1; PIGR, polymeric immunoglobulin receptor; BGN, biglycan; LD, lipid droplet; PGC-1α, peroxisome proliferator-activated receptor-gamma coactivator 1 alpha.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding authors upon request.
Ethical Approval
The study has been approved by the Ethics Committee of Zhongshan Hospital affiliated with Fudan University (No. B2017-022R).
Acknowledgments
The authors sincerely thank Drs. Kai Fang, Shan Zhong and Nan Niu from Shenzhen University and Dr Qingying Zhang from Shantou University for assisting data analysis of this study.
Funding
This study was supported by grants from the Shenzhen Basic Research Program of Science and Technology Innovation Commission (JCYJ20190808122413582), Shanghai Natural Science Foundation (22ZR1411100), and the National Research & Development Program (2016YFC1304000 and 2016YFC1304002).
Disclosure
The authors declare that they have no competing interests.
References
1. Celli BR, Wedzicha JA . Update on clinical aspects of chronic obstructive pulmonary disease. N Engl J Med. 2019;381(13):1257–1266. doi:10.1056/NEJMra1900500
2. Soler-Cataluna JJ, Martinez-Garcia MA, Roman Sanchez P, Salcedo E, Navarro M, Ochando R. Severe acute exacerbations and mortality in patients with chronic obstructive pulmonary disease. Thorax. 2005;60(11):925–931.
3. Thomas M, Radwan A, Stonham C, Marshall S. COPD Exacerbation Frequency, Pharmacotherapy and Resource Use: an Observational Study in UK Primary Care. COPD. 2014;11(3):300–309.
4. Ko FW, Chan KP, Hui DS, et al. Acute exacerbation of COPD. Respirology. 2016;21(7):1152–1165.
5. Hurst JR, Vestbo J, Anzueto A, et al. Susceptibility to exacerbation in chronic obstructive pulmonary disease. N Engl J Med. 2010;363(12):1128–1138.
6. Tashkin DP. Frequent exacerbations of chronic obstructive pulmonary disease--A distinct phenotype? N Engl J Med. 2010;363(12):1183–1184.
7. Fernandez-Costa C, Martinez-Bartolome S, McClatchy DB, Saviola AJ, Yu NK, Yates JR. Impact of the Identification Strategy on the Reproducibility of the DDA and DIA Results. J Proteome Res. 2020;19(8):3153–3161.
8. Willems P, Fels U, Staes A, Gevaert K, Van Damme P. Use of hybrid data-dependent and -independent acquisition spectral libraries empowers dual-proteome profiling. J Proteome Res. 2021;20(2):1165–1177.
9. Barkovits K, Pacharra S, Pfeiffer K, et al. Reproducibility, Specificity and Accuracy of Relative Quantification Using Spectral Library-based Data-independent Acquisition. Mol Cell Proteomics. 2020;19(1):181–197.
10. Bilbao A, Varesio E, Luban J, et al. Processing strategies and software solutions for data-independent acquisition in mass spectrometry. Proteomics. 2015;15(5–6):964–980.
11. Vogelmeier CF, Criner GJ, Martinez FJ, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. Gold executive summary. Am J Respir Crit Care Med. 2017;195(5):557–582.
12. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523.
13. Baralla A, Fois AG, Sotgiu E, et al. Plasma proteomic signatures in early chronic obstructive pulmonary disease. Proteomics Clin Appl. 2018;12(3):e1700088.
14. Regan EA, Hersh CP, Castaldi PJ, et al. Omics and the Search for Blood Biomarkers in Chronic Obstructive Pulmonary Disease. Insights from COPDGene. Am J Respir Cell Mol Biol. 2019;61(2):143–149.
15. Liu Y, Liu H, Li C, Ma C, Ge W. Proteome Profiling of Lung Tissues in Chronic Obstructive Pulmonary Disease (COPD): platelet and Macrophage Dysfunction Contribute to the Pathogenesis of COPD. Int J Chron Obstruct Pulmon Dis. 2020;15:973–980.
16. Singh D, Fox SM, Tal-Singer R, Bates S, Riley JH, Celli B. Altered gene expression in blood and sputum in COPD frequent exacerbators in the ECLIPSE cohort. PLoS One. 2014;9(9):e107381.
17. Sun P, Ye R, Wang C, Bai S, Zhao L. Identification of proteomic signatures associated with COPD frequent exacerbators. Life Sci. 2019;230:1–9.
18. Hoenderdos K, Lodge KM, Hirst RA, et al. Hypoxia upregulates neutrophil degranulation and potential for tissue injury. Thorax. 2016;71(11):1030–1038.
19. Lodge KM, Vassallo A, Liu B, et al. Hypoxia increases the potential for neutrophil-mediated endothelial damage in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2022;205(8):903–916.
20. Zhang K, Han K, Liu H, Zheng C. Circulating Complement C1q as a Novel Biomarker is Associated with the Occurrence and Development of COPD. Int J Chron Obstruct Pulmon Dis. 2022;17:395–404.
21. Husebo GR, Gabazza EC, D’Alessandro C, et al. Coagulation markers as predictors for clinical events in COPD. Respirology. 2021;26(4):342–351.
22. Morris SM. Arginine Metabolism Revisited. J Nutr. 2016;146(12):2579S–2586S.
23. Ruzsics I, Nagy L, Keki S, et al. L-Arginine Pathway in COPD Patients with Acute Exacerbation: a New Potential Biomarker. COPD. 2016;13(2):139–145.
24. Parmaksiz ET, Inal A, Salepci B, et al. Relationship of asymmetric dimethylarginine levels with disease severity and pulmonary hypertension in chronic obstructive pulmonary disease. Lung India. 2018;35(3):199–203.
25. van den Berg MP, Meurs H, Gosens R. Targeting arginase and nitric oxide metabolism in chronic airway diseases and their co-morbidities. CurrOpinPharmacol. 2018;40:126–133.
26. Sprouse C, Tosi LL, Gordish-Dressman H, et al. CK-MM polymorphism is associated with physical fitness test scores in military recruits. Mil Med. 2015;180(9):1001–1005.
27. Shankaran M, King CL, Angel TE, et al. Circulating protein synthesis rates reveal skeletal muscle proteome dynamics. J Clin Invest. 2016;126(1):288–302.
28. Kadereit B, Kumar P, Wang WJ, et al. Evolutionarily conserved gene family important for fat storage. Proc Natl Acad Sci U S A. 2008;105(1):94–99.
29. Goh VJ, Silver DL. The lipid droplet as a potential therapeutic target in NAFLD. Semin Liver Dis. 2013;33(4):312–320.
30. Bailey AP, Koster G, Guillermier C, et al. Antioxidant Role for Lipid Droplets in a Stem Cell Niche of Drosophila. Cell. 2015;163(2):340–353.
31. Yan C, Xia X, He J, et al. MyoD Is a Novel Activator of Porcine FIT1 Gene by Interacting with the Canonical E-Box Element during Myogenesis. Int J Mol Sci. 2015;16(10):25014–25030.
32. Mormeneo E, Jimenez-Mallebrera C, Palomer X, et al. PGC-1alpha induces mitochondrial and myokine transcriptional programs and lipid droplet and glycogen accumulation in cultured human skeletal muscle cells. PLoS One. 2012;7(1):e29985.
33. Rutten EP, Franssen FM, Engelen MP, Wouters EF, Deutz NE, Schols AM. Greater whole-body myofibrillar protein breakdown in cachectic patients with chronic obstructive pulmonary disease. Am J Clin Nutr. 2006;83(4):829–834.
34. Ryrso CK, Thaning P, Siebenmann C, et al. Effect of endurance versus resistance training on local muscle and systemic inflammation and oxidative stress in COPD. Scand J Med Sci Sports. 2018;28(11):2339–2348.
35. Tsutsumi A, Chubachi S, Irie H, et al. Characteristics of chronic obstructive pulmonary disease patients with robust progression of emphysematous change. Sci Rep. 2021;11(1):9548.
36. Hallin R, Koivisto-Hursti UK, Lindberg E, Janson C. Nutritional status, dietary energy intake and the risk of exacerbations in patients with chronic obstructive pulmonary disease (COPD). Respir Med. 2006;100(3):561–567.
37. Martinez-Luna N, Orea-Tejeda A, Gonzalez-Islas D, et al. Association between body composition, sarcopenia and pulmonary function in chronic obstructive pulmonary disease. BMC Pulm Med. 2022;22(1):106.
38. Tomioka R, Kawayama T, Suetomo M, et al. ”Frequent exacerbator” is a phenotype of poor prognosis in Japanese patients with chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2016;11:207–216.
39. Wu YK, Su WL, Yang MC, Chen SY, Wu CW, Lan CC. Characterization Associated with the Frequent Severe Exacerbator Phenotype in COPD Patients. Int J Chron Obstruct Pulmon Dis. 2021;16:2475–2485.
40. McGarvey L, Lee AJ, Roberts J, Gruffydd-Jones K, McKnight E, Haughney J. Characterisation of the frequent exacerbator phenotype in COPD patients in a large UK primary care population. Respir Med. 2015;109(2):228–237.
41. Ozyilmaz E, Kokturk N, Teksut G, Tatlicioglu T. Unsuspected risk factors of frequent exacerbations requiring hospital admission in chronic obstructive pulmonary disease. Int J Clin Pract. 2013;67(7):691–697.
42. Gu Z, Shen Y, Tang XY, et al. Genetic risk of FCRL3 and FCRL5 polymorphisms in children with asthma and allergic rhinitis in a Chinese Han population. Int J PediatrOtorhinolaryngol. 2019;120:58–63.
43. Lee JS, Kim JH, Bae JS, et al. Association analysis of UBE3C polymorphisms in Korean aspirin-intolerant asthmatic patients. Ann Allergy Asthma Immunol. 2010;105(4):307–312.
44. Hardavella G, Tzortzaki EG, Siozopoulou V, et al. Lymphangiogenesis in COPD: another link in the pathogenesis of the disease. Respir Med. 2012;106(5):687–693.
45. Rumora L, Rajkovic MG, Kopcinovic LM, Pancirov D, Cepelak I, Grubisic TZ. Paraoxonase 1 activity in patients with chronic obstructive pulmonary disease. COPD. 2014;11(5):539–545.
46. GrdicRajkovic M, Popovic-Grle S, VukicDugac A, et al. PON1 gene polymorphisms in patients with chronic obstructive pulmonary disease. J Clin Pathol. 2018;71(11):963–970.
47. Sidhaye VK, Chau E, Breysse PN, King LS. Septin-2 mediates airway epithelial barrier function in physiologic and pathologic conditions. Am J Respir Cell Mol Biol. 2011;45(1):120–126.
48. Hirvonen K, Korhonen T, Salomaa V, Mannisto S, Kaprio J. Association of the DBH Polymorphism rs3025343 With Smoking Cessation in a Large Population-Based Sample. Nicotine Tob Res. 2017;19(9):1112–1115.
49. Siedlinski M, Cho MH, Bakke P, et al. Genome-wide association study of smoking behaviours in patients with COPD. Thorax. 2011;66(10):894–902.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.