Back to Journals » OncoTargets and Therapy » Volume 13
Phosphoglycerate Mutase 1: Its Glycolytic and Non-Glycolytic Roles in Tumor Malignant Behaviors and Potential Therapeutic Significance
Received 16 November 2019
Accepted for publication 4 February 2020
Published 27 February 2020 Volume 2020:13 Pages 1787—1795
DOI https://doi.org/10.2147/OTT.S238920
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Na Li,1 Xinlu Liu2
1 1st Department of Gastroenterology, First Affiliated Hospital of Dalian Medical University, Dalian 116011, People’s Republic of China; 2 1st Department of General Surgery, First Affiliated Hospital of Dalian Medical University, Dalian 116011, People’s Republic of China
Correspondence: Xinlu Liu
1st Department of General Surgery, First Affiliated Hospital of Dalian Medical University, Dalian 116011, People’s Republic of China
Tel +86-411-83635963-2073
Fax +86-411-83635432
Email [email protected]
Abstract: Phosphoglycerate mutase 1 (PGAM1) is an important enzyme that catalyzes the reversible conversion of 3-phosphoglycerate and 2-phosphoglycerate during the process of glycolysis. Increasing evidence suggests that PGAM1 is widely overexpressed in various cancer tissues and plays a significant role in promoting cancer progression and metastasis. Although PGAM1 is a potential target in cancer therapy, the specific mechanisms of action remain unknown. This review introduces the basic structure and functions of PGAM1 and its family members and summarizes recent advances in the role of PGAM1 and various inhibitors of cancer cell proliferation and metastasis from a glycolytic and non-glycolytic perspective. Recent studies have highlighted a correlation between PGAM1 and clinical features and prognosis of cancer as well as the development of target drugs for PGAM1. The integrated information in this review will help better understand the specific roles of PGAM1 in cancer progression. Furthermore, the information highlights the non-glycolytic functions of PGAM1 in tumor metastasis, providing an innovative basis and direction for clinical drug research.
Keywords: PGAM1, glycolysis, non-glycolytic, proliferation, metastasis, cancer therapy
Introduction
Even in aerobic environments, most cancer cells rely mainly on glycolysis to generate energy, unlike normal cells, which mainly rely on mitochondrial oxidative phosphorylation to generate energy. This phenomenon was discovered by Warburg in 1924 and was named the “Warburg effect”1 Glycolysis is not an effective process for generating adenosine triphosphate (ATP) and the preference of cancer cells for this type of metabolic pattern has aroused intense interest and has been thought to be a hallmark of cancer therapy in past decades.2,3 Following the discovery of the Warburg effect, many glycolytic proteins were subsequently found to be involved in cancer progression, including lactate dehydrogenase A (LDHA),4,5 phosphoglycerate dehydrogenase (PHGDH),6,7 hexokinase 2 (HK2),8,9 and glucose transporter 1 (GLUT1).10 Among these proteins, phosphoglycerate mutase 1 (PGAM1), a key enzyme in the glycolytic pathway that catalyzes the reversible conversion of 3-phosphoglycerate (3-PG) into 2-phosphoglycerate (2-PG), has also received increasing attention.11 PGAM1 is overexpressed in colorectal cancer,12,13 hepatocellular carcinoma (HCC),14 non-small cell lung cancer (NSCLC),15 pancreatic ductal adenocarcinoma (PDAC),16 oral squamous cell carcinoma (OSCC),17 prostate cancer (PCa),18 urothelial carcinoma (UBC),19 glioma,20 and breast cancer.21–23 Furthermore, it plays an important role in tumor proliferation and tumor metastasis in some of these cancer types. The expression of PGAM1 was higher in tumor tissues than in adjacent normal tissues.24–27 Altogether, these findings indicate that PGAM1 could be a potential target for cancer therapy. Until recently, several factors of PGAM1 biology were still unknown such as how it affected tumor proliferation and metastasis through the regulation of glycolysis, whether its non-glycolytic effect participated in the malignant behavior of cancer and whether it is a clinically relevant therapeutic target or biomarker for cancer. In this review, we summarized the current knowledge of the role of PGAM1 and its inhibitors in the regulation of tumor malignant behaviors, as well as current developments on target drugs for PGAM1. Such information will provide novel concepts for future investigation of PGAM1 as a potential target for cancer therapy.
Basic Structure and Function of PGAM1 and Its Family Members
PGAM1 belongs to the phosphoglycerate mutase family, which can be subdivided into monophosphoglycerate mutases (mPGAM) and bisphosphoglycerate mutases (BPGAM). The interconversion of 3-PG and 2-PG is mainly catalyzed by mPGAM, whereas the conversion of 1,3-bisphosphoglycerate (BPG) to 2,3-BPG in the presence of 3-PG is catalyzed by BPGAM.7,11 Additionally, mPGAM can be further subdivided into two distinct categories, cofactor-dependent (dPGM) and cofactor-independent (iPGM).28 Previous studies provided evidence indicating that dPGM and BPGAM have kinetic and structural similarities and are thought to be paralog structures.29,30 For example, dPGM participates in three catalytic reactions: the reversible conversion of 3-PG to 2-PG,31,32 the phosphatase reaction transforming 2,3-BPG to PG,29,33 and the synthase reaction producing 2,3-BPG from 1,3-BPG, which is similar to BPGAM. In adult mammals, dPGM has two different subunits, BB-PGAM and MM-PGAM. In humans, BB-PGAM, another form of PGAM1, was originally isolated from the brain but has recently been found in the liver, breast and other tissues.14,21 MM-PGAM (also known as PGAM2) is a muscle-specific form mainly expressed in mature cardiac tissues and skeletal muscles.34
In humans, the cytogenetic location of PGAM1 is 10q24.1, with its cDNA encoding a 254 amino acid protein. PGAM1 is a homodimer with a molecular weight (MW) of 28,804 Da (Figure 1A). The phosphorylated HIS11 residues in the active domain are donors and acceptors of phosphate groups, with 2,3-BPG acting as an intermediate26 (Figure 1B). PGAM1 is primarily found in the cytoplasm, but has also been found on the cell membrane.35
 |
Figure 1 3D structure and the cDNA encoding of PGAM1. (A) The 3D structure of PGAM1 from SWISS-MODEL website (https://swissmodel.expasy.org/docs/terms_of_use). Reproduced from Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1), W296-W30357 and Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis. 2009;30, S162-S173.58 The active sites of PGAM1 are indicated in the form of red rods in the picture. (B) The whole protein feature view of PGAM1 from RCSB PDB website (https://www.rcsb.org). Reproduced from Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Research. 2000;28: 235-242.59 |
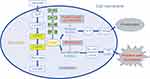
The primary role of PGAM1 is to catalyze the reversible conversion of 3-PG to 2-PG,11 a critical step in glycolysis (Figure 2). According to a study by Liu et al,36 PGAM1 is a downstream target of the PI3K/Akt/mTOR/HIF-1α pathway, which regulates cellular metabolism (Figure 2). Schrade et al found that altered expression of PGAM1 is associated with GATA4, which mostly modulates tight and adherens junction formation and extracellular matrix reorganization in mouse Sertoli cells (SCs).37
Glycolytic Role of PGAM1 in Cancer Proliferation
Glycolysis is an oxygen-independent metabolic pathway that converts glucose to ATP and combines ten enzyme-catalyzed reactions.9 The glycolytic pathway is the first step in glucose metabolism in all living cells, with multiple enzymes involved in the precise regulation of the pathway for the maintenance of homeostasis. Most normal cells generate energy through glycolysis under oxygen deficient conditions. However, the Warburg effect highlights that cancer cells mainly produce energy via glycolysis, even in an aerobic environment.38 Therefore, to provide sufficient ATP and carbon for the necessary building blocks of the cellular processes such as nucleotides, amino acids, lipids and NADPH, cancer cells require a higher glucose intake than normal cells to meet the energy requirements for rapid proliferation.38–40 Subsequently, this overactive glycolysis may play a role in promoting tumor cell proliferation.41
The alternative recombinant metabolic pattern of cancer cells was considered to provide new opportunities for cancer treatment, which lead researchers to investigate the roles of metabolic enzymes during the development of cancer.42,43 Therefore, the relationship between the metabolic changes brought by PGAM1 and cancer are gradually being explored. Hitosugi et al.28,44 found that PGMI-004A, a small molecule inhibitor of PGAM1, was able to decrease the glycolytic function of PGAM1. Subsequently, a significant decrease in the pentose phosphate pathway (PPP) flux and biosynthesis as well as an attenuated cell proliferation and tumor growth were observed in the breast cancer cell line MDA-MB-231, the lung cancer cell line H1299, the acute myeloid leukemia cell line Molm14, and in the head and neck cancer cell line 212LN. In this in-depth study, several new findings were discovered. First, knocking down PGAM1 led to a significant decrease in the glycolytic rate, lactate production, lipogenesis, and RNA biosynthesis and, correspondingly, cell proliferation in H1299 cells. Second, the role of PGAM1 in promoting tumor proliferation has also been shown to be modulated by intracellular levels of 3-PG and conversely by 2-PG. Third, Y26 phosphorylation of PGAM1 was found to represent a common, short-term molecular mechanism that contributed to the upregulation of PGAM1 activity and promotion of cancer cell proliferation and tumor growth. This mechanism differs from the previously described chronic mechanism in which the upregulation of PGAM1 was thought to be caused by loss of TP53. Fourth, the crystal structure of the mechanism of Y26 phosphorylation has been revealed, showing that activation of PGAM1 is enhanced by the release of inhibitory E19 that typically blocks the active site, thereby stabilizing cofactor 2,3-BPG binding and H11 phosphorylation. In addition, Engel et al.45 also indicated that PGAM1 activity can be inhibited by exogenous polypeptides, resulting in a decrease in the glycolytic rate and cell growth arrest in a breast cancer cell line. Although there are still many unknown factors, there is a correlation between PGAM1 and cancer proliferation. Moreover, PGAM1 is thought to affect cancer cell proliferation through the regulation of glycolysis in the cell. In addition to the proliferation of tumor cells, tumor metastasis is also an important factor affecting the prognosis of cancer patients. DM et al.46 reported PGAM1 was overexpressed in the cytoplasm of capillary/artery endothelial cells, suggesting a potential correlation between PGAM1 and tumor invasion and metastasis. However, the relationship between the metabolic role of PGAM1 and tumor metastasis has been infrequently reported. It is difficult to confirm whether the mechanisms by which PGAM1 affects tumor metastasis are also achieved through glycolytic regulation.
Non-Glycolytic Role of PGAM1 in Tumor Invasion and Metastasis
Many studies have highlighted the metabolic role of PGAM1 in promoting cancer cell proliferation. However, it remains unclear whether PGAM1 can promote cancer malignant behaviors through a non-metabolic pathway. Previously, metabolites such as adenosine monophosphate (AMP), an allosteric activator for AMP-activated protein kinase which senses intracellular energy levels (ATP/AMP ratio), have been suggested to function as signaling molecules.47 Glutamine, which activates leucine uptake, leads to mTOR activation.48 The non-glycolytic role of PGAM1 has been recently uncovered. Hitosugi et al.28 found that targeting PGAM1 did not significantly influence intracellular ATP levels and showed that the decrease in ATP production caused by the attenuated glycolysis in PGAM1 knockdown cells was compensated by rescue treatment with methyl-2-PG. However, methyl-2-PG treatment only partially rescued the attenuated cell proliferation in the PGAM1 knockdown cells or cells treated with PGMI-004A, indicating that PGAM1 might contribute to cell proliferation in a 2-PG-dependent and -independent manner. The latter has been associated with the non-glycolytic function of PGAM1. The promoting role of PGAM1 on tumor metastasis has also been unveiled, but rarely related to the glycolytic functions of PGAM1. Recently, Zhang et al.23 confirmed that PGAM1 can promote tumor metastasis through a non-metabolic function. In this study, PGAM1 was found to directly interact with α-smooth muscle actin 2 (ACTA2) independent of its metabolic activity. To exclude the impact of the glycolytic pathway, numbers of glycolytic enzymes, such as HK2, PKM2, LDHA, and PDK1, were individually depleted in MDA-MB-231 cells. Following depletion of these enzymes, knocking down the expression of PGAM1 still reduced cancer cell motility. The PGAM1 metabolic inhibitor PGMI-004A28 also failed to affect cell migration in HEK 293 cells regardless of the effects of decreased PGAM1 enzymatic activity in cancer cell proliferation. This metabolism-independent role of PGAM1 in tumor invasion and metastasis has been verified through its association with ACTA2.
As well as interacting with non-glycolytic proteins, the promoting role of PGAM1 in tumor invasion and metastasis was also found to correlate with other non-glycolytic pathways. Zhang et al.17 showed that reduced expression of PGAM1 in HN12 and Cal27 cells lead to a significant decrease in cell migration and in the expression levels of corresponding regulatory pathway molecules, such as focal adhesion kinase, the proto-oncogene c-SRC, and paxillin. Liu et al.36 also found that PGAM1 can promote migration and invasion of pancreatic cancer cells and may promote epithelial-to-mesenchymal transition (EMT) in pancreatic cancer cells by regulation of the Wnt/β-catenin pathway. Although the non-glycolytic function of PGAM1 was rarely described, it provided a better explanation of the mechanism by which PGAM1 modulates tumor progression, especially invasion and metastasis and led to an important new pathway for anti-cancer therapy.
Potential Clinical Value of PGAM1 as a Target for Cancer Therapy
The role of PGAM1 in cancer progression is receiving increasing attention. Recent clinical data showed a correlation between PGAM1 and the clinical features and prognosis of cancer, suggesting that PGAM1 can be a novel potential therapeutic target. Zhang et al.17 reported that PGAM1 expression was correlated with age, lymphatic metastasis, and tumor recurrence and was closely associated with poorer overall survival (OS) and disease-free survival (DFS). PGAM1 was also suggested to be an independent risk factor for OS and DFS. It also correlated with a poor differentiation status and was identified as a potential therapeutic target for urothelial cancer by Peng et al.19 who conducted a two-dimensional electrophoresis proteomic analysis of clinical tissues. Li et al.49 found that PGAM1 was highly expressed in clear cell renal cell carcinoma and that its expression was significantly associated with age, tumor size, and TNM stage. Ren et al.14 analyzed the expression of PGAM1 in 54 paired HCC samples and 21 normal liver tissues and suggested PGAM1 as a potential diagnostic biomarker, as well as an attractive therapeutic target for HCC. Finally, Liu et al.36 found that the overexpression of PGAM1 correlated with poor prognosis in PDAC patients after analyzing 54 PDAC clinical tissues. Taken together, these clinical data have emphasized the clinical research value of PGAM1 and suggest that PGAM1 is a potential therapeutic target for the treatment of cancer.
Summary of PGAM1 Inhibitors and Research Directions to Explore PGAM1-Targeted Drugs
Since PGAM1 was suggested as a potential therapeutic target for multiple cancer types, several PGAM1 inhibitors have been developed for cancer therapy.21,22,28,50–53 A summary of these inhibitors and their related functions are listed in Table 1. PGAM1 inhibitors are divided into pharmacological inhibitors and genetic inhibitors. The pharmacological inhibitors are small molecular compounds, with six types of small molecules reported to inhibit PGAM1, and which are mainly associated with metabolism and cancer cell proliferation. MJE3 was the first cell-permeable, small-molecule compound inhibitor of PGAM1. It reacted specifically with lysine-100 (K100) in the PGAM1 active site and hydrolyzed in situ to produce acid products that decreased breast cancer cell proliferation.21,22 The anthraquinone derivative 3, also named PGMI-004A, is another small-molecule inhibitor of PGAM1 that inhibits the conversion of 3-PG to 2-PG in cancer cells, leading to significant inhibition of the glycolytic pathway, PPP flux and biosynthesis, subsequently decreasing cancer cell proliferation and tumor growth.28 However, this inhibitor has been reported to be ineffective for tumor invasion or metastasis.23 Epigallocatechin-3-gallate (EGCG), a natural product derived from green tea, was also identified as a PGAM1 inhibitor. EGCG was reported to inhibit PGAM1 enzymatic activity by directly impairing glycolysis and PPP flux, regardless of 3-PG competition, and further, it was shown to inhibit cancer cell proliferation by modulating the intracellular level of 2-PG.52 However, because of its multiple targets, its specificity to PGAM1 is poor.28,54 Wang et al.53 used scaffold hopping and a sulfonamide reversal strategy based on the lead compound PGMI-004A to discover a series of xanthone derivatives (12a–12s) as novel PGAM1 inhibitors. These xanthone derivatives showed stronger efficacy and better specificity than PGMI-004A in the inhibition of PGAM1, as well as an increased anti-proliferative effect in the H1299 cell line. Huang et al.51 revealed that F22, K100, and R116 of PGAM1 residues were critical for the binding of inhibitors and that compound 9i, an anthraquinone inhibitor, significantly decreased lung cancer cell proliferation in different cell lines, which is a promising inhibitor for PGAM1. Moreover, in the recent research of Wen CL et al.55 an allosteric inhibitor of PGAMI named KH3 has been explored that dramatically inhibited the proliferation of PDAC cell lines by hampering the canonical cancer metabolic pathways. In summary, inhibitors targeting PGAM1 have been developed rapidly. However, most of the PGAM1 inhibitors were glycolysis-targeted with minimal to no effect on the invasion and metastasis of cancer cells (Table 1).
 |
Table 1 Effects of Different Inhibitors of PGAM1 on Proliferation and Metastasis of Various Cancer |
Genetic inhibitors, unlike pharmacological inhibitors, interfere with RNA levels and appear to have increased inhibitory effects on cancer. Genetic inhibitors such as PGAM1-siRNA or shRNA proved to not only inhibit cancer cell proliferation, but also invasion and metastasis17,18,23,36 (Table 1). Liu et al showed that following PGAM1 inhibition in PDAC cell lines, the decrease in PDAC cell invasion occurred earlier than proliferation.16,36 This points to the presence of an active site in PGAM1 that regulates its non-glycolytic functions and has a greater correlation with cancer metastasis. Therefore, in the future, the development of PGAM1-targeted drugs should also consider the non-glycolytic pathway. Surprisingly, in the latest study by Huang et al.56 reported the first allosteric PGAM1 inhibitor HKB99, which suppresses NSCLC tumor growth through ROS-dependent activation of JNK/c-Jun and metastasis by abrogating the interaction between PGAM1 and ACTA2. This discovery provides a new understanding of the function of PGAM1’s undiscovered domain. PGAM1-targeted drugs that integrate these two functions would more likely produce a more substantial effect in tumor therapy (Figure 2).
Conclusion
Increasing evidence has indicated the vital biological roles of PGAM1 in tumor progression. On one hand, PGAM1 is thought to be involved in the glycolytic pathway to regulate tumor cells’ metabolic pattern and promote cancer cell proliferation. On the other hand, PGAM1 can promote cancer cell invasion and metastasis through a specific non-glycolytic function. Future studies should focus on the molecular pathways modulated by PGAM1 to induce cancer cell motility during invasion and metastasis and development of drugs that can target the non-glycolytic functions of PGAM1, as its metabolic changes are mainly associated with cancer cell proliferation. The latter requires a deeper understanding of the non-glycolytic functions of PGAM1. Finally, the correlation between PGAM1 and cancer prognosis has been gaining attention and further research will identify whether PGAM1 can be used as a biomarker for early cancer detection. Although there are many unsolved questions around the roles of PGAM1 in tumor malignant behaviors, increasing evidence suggests that it has become an emerging and promising target for cancer therapy and worth further investigation in the future.
Ethical Approval
This article does not include any experiments involving humans or animals.
Acknowledgment
We thank Editage for English language editing.
Author Contributions
Dr. Li was involved in the conception and design, the first draft of the article, final approval of the article, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved. Dr. Liu was involved in the conception and design, study supervision, initial drafting of the article, critical revision of the article for important intellectual content, final approval of the article, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work”.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi:10.1126/science.123.3191.309
2. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi:10.1016/j.ccr.2012.02.014
3. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
4. Shim H, Dolde C, Lewis BC, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94(13):6658–6663. doi:10.1073/pnas.94.13.6658
5. Le A, Cooper CR, Gouw AM, et al. Inhibition of lactate dehydrogenase a induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A. 2010;107(5):2037–2042. doi:10.1073/pnas.0914433107
6. Possemato R, Marks KM, Shaul YD, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi:10.1038/nature10350
7. Locasale JW, Grassian AR, Melman T, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43(9):869–874. doi:10.1038/ng.890
8. Wolf A, Agnihotri S, Micallef J, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208(2):313–326. doi:10.1084/jem.20101470
9. Mathupala SP, Rempel A, Pedersen PL. Glucose catabolism in cancer cells: identification and characterization of a marked activation response of the type II hexokinase gene to hypoxic conditions. J Biol Chem. 2001;276(46):43407–43412. doi:10.1074/jbc.M108181200
10. Szablewski L. Expression of glucose transporters in cancers. Biochim Biophys Acta. 2013;1835(2):164–169. doi:10.1016/j.bbcan.2012.12.004
11. Fothergill-Gilmore LA, Watson HC. The phosphoglycerate mutases. Adv Enzymol Relat Areas Mol Biol. 1989;62:227–313. doi:10.1002/9780470123089.ch6
12. Liu L, Wang S, Zhang Q, Ding Y. Identification of potential genes/proteins regulated by Tiam1 in colorectal cancer by microarray analysis and proteome analysis. Cell Biol Int. 2008;32(10):1215–1222. doi:10.1016/j.cellbi.2008.07.004
13. Lei Y, Huang K, Gao C, et al. Proteomics identification of ITGB3 as a key regulator in reactive oxygen species-induced migration and invasion of colorectal cancer cells. Mol Cell Proteomics. 2011;10(10):M110 005397. doi:10.1074/mcp.M110.005397
14. Ren F, Wu H, Lei Y, et al. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol Cancer. 2010;9:81. doi:10.1186/1476-4598-9-81
15. Sun Q, Li S, Wang Y, et al. Phosphoglyceric acid mutase-1 contributes to oncogenic mTOR-mediated tumor growth and confers non-small cell lung cancer patients with poor prognosis. Cell Death Differ. 2018;25(6):1160–1173. doi:10.1038/s41418-017-0034-y
16. Liu X, Weng Y, Liu P, et al. Identification of PGAM1 as a putative therapeutic target for pancreatic ductal adenocarcinoma metastasis using quantitative proteomics. Onco Targets Ther. 2018;11:3345–3357. doi:10.2147/OTT
17. Zhang D, Wu H, Zhang X, et al. Phosphoglycerate mutase 1 predicts the poor prognosis of oral squamous cell carcinoma and is associated with cell migration. J Cancer. 2017;8(11):1943–1951. doi:10.7150/jca.19278
18. Wen YA, Zhou BW, Lv DJ, et al. Phosphoglycerate mutase 1 knockdown inhibits prostate cancer cell growth, migration, and invasion. Asian J Androl. 2018;20(2):178–183. doi:10.4103/aja.aja_57_17
19. Peng XC, Gong FM, Chen Y, et al. Proteomics identification of PGAM1 as a potential therapeutic target for urothelial bladder cancer. J Proteomics. 2016;132:85–92. doi:10.1016/j.jprot.2015.11.027
20. Xu Z, Gong J, Wang C, et al. The diagnostic value and functional roles of phosphoglycerate mutase 1 in glioma. Oncol Rep. 2016;36(4):2236–2244. doi:10.3892/or.2016.5046
21. Evans MJ, Morris GM, Wu J, Olson AJ, Sorensen EJ, Cravatt BF. Mechanistic and structural requirements for active site labeling of phosphoglycerate mutase by spiroepoxides. Mol Biosyst. 2007;3(7):495–506. doi:10.1039/b705113a
22. Evans MJ, Saghatelian A, Sorensen EJ, Cravatt BF. Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat Biotechnol. 2005;23(10):1303–1307. doi:10.1038/nbt1149
23. Zhang D, Jin N, Sun W, et al. Phosphoglycerate mutase 1 promotes cancer cell migration independent of its metabolic activity. Oncogene. 2017;36(20):2900–2909. doi:10.1038/onc.2016.446
24. Durany N, Joseph J, Campo E, Molina R, Carreras J. Phosphoglycerate mutase, 2,3-bisphosphoglycerate phosphatase and enolase activity and isoenzymes in lung, colon and liver carcinomas. Br J Cancer. 1997;75(7):969–977. doi:10.1038/bjc.1997.168
25. Durany N, Joseph J, Jimenez OM, et al. Phosphoglycerate mutase, 2,3-bisphosphoglycerate phosphatase, creatine kinase and enolase activity and isoenzymes in breast carcinoma. Br J Cancer. 2000;82(1):20–27. doi:10.1054/bjoc.1999.0871
26. Buhrens RI, Amelung JT, Reymond MA, Beshay M. Protein expression in human non-small cell lung cancer: a systematic database. Pathobiology. 2009;76(6):277–285. doi:10.1159/000245893
27. Tang CE, Li C, Xiao ZQ, et al. Comparative proteome analysis of human lung squamous cell carcinoma. Chin J Cancer. 2006;28(4):274–279.
28. Hitosugi T, Zhou L, Elf S, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22(5):585–600. doi:10.1016/j.ccr.2012.09.020
29. Rose ZB, Dube S. Phosphoglycerate mutase. Kinetics and effects of salts on the mutase and bisphosphoglycerate phosphatase activities of the enzyme from chicken breast muscle. J Biol Chem. 1978;253(23):8583–8592.
30. Grisolia S, Cleland WW. Influence of salt, substrate, and cofactor concentrations on the kinetic and mechanistic behavior of phosphoglycerate mutase. Biochemistry. 1968;7(3):1115–1121. doi:10.1021/bi00843a032
31. Sasaki R, Utsumi S, Sugimoto E, Chiba H. Subunit structure and multifunctional properties of yeast phosphoglyceromutase. Eur J Biochem. 1976;66(3):523–533. doi:10.1111/ejb.1976.66.issue-3
32. Rose ZB, Dube S. Rates of phosphorylation and dephosphorylation of phosphoglycerate mutase and bisphosphoglycerate synthase. J Biol Chem. 1976;251(16):4817–4822.
33. Sasaki R, Hirose M, Sugimoto E, Chiba H. Studies on a role of the 2,3-diphosphoglycerate phosphatase activity in the yeast phosphoglycerate mutase reaction. Biochim Biophys Acta. 1971;227(3):595–607. doi:10.1016/0005-2744(71)90010-6
34. Omenn GS, Cheung SC. Phosphoglycerate mutase isozyme marker for tissue differentiation in man. Am J Hum Genet. 1974;26(3):393–399.
35. Shanske S, Sakoda S, Hermodson MA, DiMauro S, Schon EA. Isolation of a cDNA encoding the muscle-specific subunit of human phosphoglycerate mutase. J Biol Chem. 1987;262(30):14612–14617.
36. Liu X, Tan X, Liu P, Wu Y, Qian S, Zhang X. Phosphoglycerate mutase 1 (PGAM1) Promotes Pancreatic Ductal Adenocarcinoma (PDAC) Metastasis by acting as a novel downstream target of the PI3K/Akt/mTOR pathway. Oncol Res. 2018;26(7):1123–1131. doi:10.3727/096504018X15166223632406
37. Schrade A, Kyronlahti A, Akinrinade O, et al. GATA4 regulates blood-testis barrier function and lactate metabolism in mouse sertoli cells. Endocrinology. 2016;157(6):2416–2431. doi:10.1210/en.2015-1927
38. Burk D, Schade AL. On respiratory impairment in cancer cells. Science. 1956;124(3215):270–272.
39. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi:10.1126/science.1160809
40. Vander Heiden MG, Locasale JW, Swanson KD, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329(5998):1492–1499. doi:10.1126/science.1188015
41. Xu Q, Tu J, Dou C, et al. HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma. Mol Cancer. 2017;16(1):178. doi:10.1186/s12943-017-0748-y
42. Hamanaka RB, Chandel NS. Targeting glucose metabolism for cancer therapy. J Exp Med. 2012;209(2):211–215. doi:10.1084/jem.20120162
43. Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi:10.1146/annurev-cellbio-092910-154237
44. Hitosugi T, Zhou L, Fan J, et al. Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation. Nat Commun. 2013;4:1790. doi:10.1038/ncomms2759
45. Engel M, Mazurek S, Eigenbrodt E, Welter C. Phosphoglycerate mutase-derived polypeptide inhibits glycolytic flux and induces cell growth arrest in tumor cell lines. J Biol Chem. 2004;279(34):35803–35812. doi:10.1074/jbc.M402768200
46. Jacobowitz DM, Jozwik C, Fukuda T, Pollard HB. Immunohistochemical localization of Phosphoglycerate mutase in capillary endothelium of the brain and periphery. Microvasc Res. 2008;76(2):89–93. doi:10.1016/j.mvr.2008.04.001
47. Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9(8):563–575. doi:10.1038/nrc2676
48. Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136(3):521–534. doi:10.1016/j.cell.2008.11.044
49. Li C, Shu F, Lei B, Lv D, Zhang S, Mao X. Expression of PGAM1 in renal clear cell carcinoma and its clinical significance. Int J Clin Exp Pathol. 2015;8(8):9410–9415.
50. Rigden DJ, Walter RA, Phillips SE, Fothergill-Gilmore LA. Polyanionic inhibitors of phosphoglycerate mutase: combined structural and biochemical analysis. J Mol Biol. 1999;289(4):691–699. doi:10.1006/jmbi.1999.2848
51. Huang K, Jiang L, Li H, Ye D, Zhou L. Development of anthraquinone analogues as phosphoglycerate mutase 1 inhibitors. Molecules. 2019;24(5):845.
52. Li X, Tang S, Wang QQ, et al. Identification of epigallocatechin-3- gallate as an inhibitor of phosphoglycerate mutase 1. Front Pharmacol. 2017;8:325. doi:10.3389/fphar.2017.00325
53. Wang P, Jiang L, Cao Y, et al. Xanthone derivatives as phosphoglycerate mutase 1 inhibitors: design, synthesis, and biological evaluation. Bioorg Med Chem. 2018;26(8):1961–1970. doi:10.1016/j.bmc.2018.02.044
54. Ma YC, Li C, Gao F, et al. Epigallocatechin gallate inhibits the growth of human lung cancer by directly targeting the EGFR signaling pathway. Oncol Rep. 2014;31(3):1343–1349. doi:10.3892/or.2013.2933
55. Wen CL, Huang K, Jiang LL, et al. An allosteric PGAM1 inhibitor effectively suppresses pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2019;116(46):23264–23273. doi:10.1073/pnas.1914557116
56. Huang K, Liang Q, Zhou Y, et al. A novel allosteric inhibitor of phosphoglycerate mutase 1 suppresses growth and metastasis of non-small-cell lung cancer. Cell Metab. 2019;30(6):1107–1119 e1108. doi:10.1016/j.cmet.2019.09.014
57. Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1), W296–W303.
58. Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis. 2009;30, S162–S173.
59. Berman HM, Westbrook J, Fengu Z, et al. The Protein Data Bank. Nucleic Acids Research. 2000;28:235–242.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.