Back to Journals » Pathology and Laboratory Medicine International » Volume 14
Phenotypic Analyses of Blood Culture Contaminants in COVID-19 Intensive Care Unit Using Hierarchical Clustering During the Pandemic First Wave in Surabaya
Authors Edbert D , Mertaniasih NM , Endraswari PD
Received 29 December 2021
Accepted for publication 23 March 2022
Published 12 April 2022 Volume 2022:14 Pages 7—13
DOI https://doi.org/10.2147/PLMI.S356299
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Paul Zhang
Daniel Edbert,1,2 Ni Made Mertaniasih,3– 5 Pepy Dwi Endraswari3– 5
1Clinical Microbiology Specialist Residency Program, Faculty of Medicine, Universitas Airlangga, Surabaya, Indonesia; 2Department of Microbiology, Faculty of Medicine and Health Science, Atma Jaya Catholic University of Indonesia, Jakarta, Indonesia; 3Department of Medical Microbiology, Faculty of Medicine, Universitas Airlangga, Surabaya, Indonesia; 4Department of Clinical Microbiology, Dr. Soetomo Academic Hospital, Surabaya, Indonesia; 5Departement of Clinical Microbiology, Airlangga Hospital, Surabaya, Indonesia
Correspondence: Daniel Edbert; Ni Made Mertaniasih, Email [email protected]; [email protected]
Introduction: Coronavirus disease-2019 (COVID-19) has been reported as an epidemic in December 2019 in Wuhan, Hubei Province, People’s Republic of China. Hospitalized COVID-19 patients, especially those with worse clinical appearance and on a ventilator, need thorough diagnostic tests as COVID-19 patients tend to mimic bacterial infections. Positive blood culture in COVID-19 patients is more likely to be contaminants and hospital-associated infections than primary co-infections. This research aims to phenotypically group data of contaminant coagulase negative staphylococci (CoNS) isolates using hierarchical clustering.
Methods: This is a descriptive study presenting a collection of CoNS culture data performed by a microbiology laboratory of a COVID-19 referral hospital from 26 March 2020 to 31 March 2021. Hierarchical clustering was performed using statistical software.
Results: Hierarchical clustering was performed on Staphylococcus epidermidis (n = 26), S. haemolyticus (n = 19), and S. hominis (n = 16). Two dominant ID clusters and one dominant MIC cluster were found in each species.
Conclusion: Dominant CoNS isolates were found. The isolates may have been transferred into the COVID-19 isolation room by chance. This method can be applied to facilities with limited-resource settings.
Keywords: coagulase-negative staphylococci, blood culture contaminations, COVID-19
Introduction
Since being reported in Wuhan, Hubei Province, People’s Republic of China, Coronavirus disease-2019 (COVID-19) has ruled every aspect of human life. Uncontrolled virus traffic and low mask compliance have prolonged this pandemic. As per 24th September 2021; 231,479,120 confirmed cases have been found and the death toll has reached 4,744,315 (2.04%). In Asia, India reported the most cases (33,594,803 confirmed cases). In the South-East Asia Region, Indonesia has the highest number of confirmed cases (4,204,116 confirmed cases) with a death toll as high as 141,258 (3.4%). The death proportion in Indonesia is higher than the world death proportion.1
Death by COVID-19 is not directly caused by the course of viral infection, but most likely due to multi-organ damages by severe cytokine storm and systemic fibrosis. Multi-organ failure by acute inflammation and activation of fibroblasts is the main pathophysiology that happens in COVID-19 systemic syndromes.2 Hyaline membrane production and fibrosis in the respiratory system plus oxygen receptors decreases brain ability to detect hypoxia and worsens the ability to take oxygen as a source of energy.2,3
Severe and critical COVID-19 patients need thorough diagnostic tests as COVID-19 patients tend to mimic bacterial infections.4 Earlier studies about COVID-19 reported a small proportion of bacterial co-infection during admission. Ventilator-associated pneumonia is the most occurring infection in healthcare-associated infections (HAI). A true positive culture is rare in the early course of COVID-19.4–9 True secondary infection in COVID-19 patients occurs in 50% of severe patients resulting in death.10 The number of true bacteremia in severe and critical patients is 1.6%.9 Most of the studies showed a high rate of positive bacterial culture in COVID-19 patients describes more likely of culture contaminations or invasive device colonization associated with HAI than primary co-infections rather than primary co-infections.8,11
Managements of COVID-19 patients are performed in special isolation wards and the highest level of personal protective equipments are used in patient care thus limiting caregiver movements in delicate procedures.8,12,13 This contributes to high levels of aerobic culture contamination. Positive blood culture can reach 42%-65%, very high compared to the low number of true bacteremia previously stated.9
Species detection and antimicrobial susceptibility profiling in HAI are important in isolated tracing and intrahospital epidemiological study. On the other hand, contaminants are rarely traced. Contaminant tracing has the same purpose and outcome, that is to guide regulation and compliance evaluation. Genotypic tracing has been done in many studies and has developed into predictive microbiology, but high-end instruments and trained professionals are not readily available in most facilities and routine tracing might be costly. There is no standard method for phenotypic tracing other than antibiogram or resistance profiling. Thus a simple and credible phenotypic tracing method is needed for isolated tracing in limited resource facilities. This study aims to describe coagulase negative staphylococci (CoNS) species isolated from blood cultures of COVID-19 intensive care unit (ICU) patients admitted to public hospitals in Surabaya using hierarchical clustering.
Methods
This is a descriptive study presenting a collection of coagulase negative staphylococci culture data performed by a microbiology laboratory of a COVID-19 referral hospital from 26th March 2020–31st March 2021. Data was collected using non-consecutive sampling methods from patients admitted to COVID-19 ICU. Blood culture contamination is defined as positive growth of normal skin flora (CoNS, Bacillus spp., Viridans Streptococci, Corynebacterium spp., Micrococcus spp., etc) in one of two sets of blood culture bottles14–16 and is not related to any new episode of fever and device-related infections.17 Contaminant CoNS are defined as CoNS that grew from one of two sets of blood culture bottles. Three Staphylococcus species were found to be dominant in number ie Staphylococcus epidermidis, Staphylococcus haemolyticus, and Staphylococcus hominis. Hierarchical cluster analyses were performed to analyze the similarity of isolates using biochemistry profile and Minimum Inhibitory Concentration (MIC) from an automated identification system (BD Phoenix, Becton-Dickinson).18
Ethical Statement
This research has been reviewed by dr. Soetomo Public Hospital Ethical Committee. Letter of exemption no. 0457/LOE/301.4.2/V/2021. All the secondary data and patient information are kept in a private and secure database.
Data Collection Method and Analysis
Biochemistry and MIC data were gathered using the BD Epicenter (Becton-Dickinson) program and clinical information was collected using the hospital information system. Hierarchical clustering was performed using statistical software, with Euclidean distance analysis, between-group linkage. A binary data setting was used to analyze biochemistry profiles.
Results
The number of contamination is as high as 79% (Note that this data is obtained from the first wave of COVID-19 before interventions were applied). The number of positive blood cultures is 38%.14 The top three CoNS that were diagnosed as contaminants are Staphylococcus epidermidis (n=26), S. haemolyticus (n= 19), and S. hominis (n=16) (Table 1). Hierarchical cluster analyses were performed on each species.
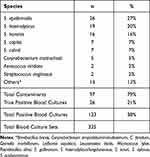 |
Table 1 Distribution of Blood Culture Contaminants in COVID-19 ICU |
The cluster clades are divided based on ID clusters (Biochemistry profile) and MIC clusters (MIC profile) which are shown in Figure 1. ID clusters are coded in numbers and MIC clusters are coded in capital letters. There are a total of 5 cluster combinations for S. epidermidis, 4 cluster combinations for S. haemolyticus, and 4 cluster combinations for S. hominis. S. epidermidis is dominated by cluster 1A (38.5%) and 2A (38.5%) which are Nitrofurantoin sensitive. The differences between S. epidermidis cluster ID 1 and 2 isolates are phenylalanine and proline utilizing and glucoside producing ability of cluster ID 1. S. haemolyticus is dominated by cluster 1A (57.8%) and 2A (13.6%) which are Teicoplanin intermediate. S. haemolyticus cluster ID 1 is glucoside-producing isolates. S. hominis is dominated by cluster 1A (37.5%) and 2A (50%) which are Vancomycin-resistant. S. hominis cluster ID 2 isolates are trehalose fermenters. Interpretation of MIC cut-off was performed using CLSI M-100 document.18 The distribution of each cluster is described in Table 2. Almost all isolates within the species fall into the same MIC cluster (Cluster A). It is important to note that this grouping nomenclature only applies to this data. Different data sets can result in different groupings.
 |
Table 2 Crosstabulation of Clusters According to ID Clusters and MIC Clusters |
The significant clusters were identified more in November to December, when the highest number of COVID-19 incidence happen in Surabaya, and it is dominated by MIC cluster A for the three species (Figure 2). There were two periods of COVID-19 incidence increase in Surabaya, that is June-July and November-December. The variations of cluster within species are high in the period of increased number of cases (1–2 cluster variations per species in April, May, October, September, January, February, and March; compared to 3 cluster variations in June, July, August, November, and December).
Discussion
Contaminants are attributed to the transfer of microorganisms from the immediate environment or caregivers to the patients and vice versa. The diversity of the contaminants reflects the source of the bacteria that can be assumed to reside in stratum corneum, clothing, and, more recently, high level of personal protective equipment (Hazmat, gown, face shield, apron, etc) that is not changed between patients. Hand hygiene and antiseptic use are effective in reducing the amount of resident flora above stratum corneum, but not below, as more than 20% of resident flora might be beyond disinfectant reach.15 However, the use of antiseptic in hand gloves and other protective equipment that cannot be changed regularly is not yet studied. Inappropriate disinfection of culture site – that is, human error - might lead to blood culture specimen contamination as shown in Figure 2. The number of S. epidermidis cluster 1A first showed in July and rise in November, in the time frame of which the number of COVID-19 cases also risen. The variation of clusters within species also expanded during the increase of blood culture contamination. This increase of cluster variation in the timeline might indicate that the contamination is brought from various sources within the hospital possibly carried or transmitted by new recruits with less experiences. The findings also correlate with the workload thus supporting that the isolates found are from human factors.
A study in New York city hospitals claimed that the rate of bacteremia was significantly lower among COVID-19 patients (3.8%) than non-COVID-19 patients (8%). COVID-19 patients had a higher proportion of organisms reflective of skin commensals, indicating blood culture contamination. Most of the growth was detected within 4 days of incubation.9 This also correlates with this study, where CoNS were found to be phenotypically related. This false-positive blood culture might lead to unnecessary antibiotic courses for more than a week. Fever, blood culture positive growth <48 hours, high leukocyte count, C-reactive Protein, procalcitonin, high Sequential Organ Failure Assessment (SOFA) score, or Simplified Acute Physiology Score II (SAPS II) score are commonly used in the diagnosis of sepsis.19–22 However, these biomarkers or scores might also be high in COVID-19 patients.2–5 Therefore, further typing of the isolated bacteria is often needed, such as molecular typing. This study method provides an alternative to isolate typing that is cheap and vastly available. As this is the first study to elaborate clinical features to define contamination and holistic bacteriological features.
CoNS can also be significant as a pathogen in the installment of central vein catheter, intra-arterial blood pressure monitor, or other invasive devices, but a true “gold standard” for differentiating contaminant CoNS from pathogens does not exist.19,22 This duality of CoNS complicates the diagnostic and interpretation of positive blood culture specimens. Some studies compare “antibiotype” or susceptibility patterns to identify the similarity of CoNS isolates,23 but the metabolic aspects and biological fitness costs are often abandoned. By combining the use of clinical features and isolate grouping, a more accurate diagnosis might be made according to the bacteriological features of the isolate and its cluster.
High internal homogeneity and high external heterogeneity are the basis of grouping in hierarchical clustering. In clinical settings, if these isolates are truly from the same clone, this phenotype had been circulating inside the hospital that transferred into the COVID-19 isolation ward. On the other hand, if the isolates are not from the same clone, knowing that phenotype might be triggered or suppressed due to external stressor such as antimicrobial agents, these isolates have undergone similar stressor that elicits the same phenotype, especially when the MIC is grouped into the same cluster (Cluster A). Molecular tracing of resistance genes might not be available vastly, especially in limited-resource settings. In this method, using MIC instead of susceptibility test interpretation reduces bias because of the changes in susceptibility cut-off. Also, the analyses are not affected by levels of genetic expression or biological fitness cost that can bias molecular study. Similar clustering results occur when using Ward’s method and neighbor clustering; this explains the consistency of the result. Continuous data gathering and analysis are needed to make this simple analysis applicable for local epidemiological study. This simple method can be applied to group hospital-acquired infection isolates and multi-drug resistance isolates outbreak management.
Conclusion
S. epidermidis, S. haemolyticus, and S. hominis that were clustered in this research are divided into significant MIC groups that might indicate similarity in previous antibiotic exposure thus indicating a common source for each contaminant and may have been transferred into the isolation room by chance. The spread of the isolates and its cluster variations is also coherent with the increase of COVID-19 incidence in November to December 2021. This method can be used to trace common characteristics of isolates in bacterial outbreaks or be used as proof for regulations by phenotypic tracing, especially in limited-resource settings.
Data Sharing Statement
All data underlying the results are available as part of the article and no additional source data are required.
Acknowledgments
We thank Dr. Soetomo Academic Hospital for providing all necessary support in this research.
Author Contributions
All authors made a significant contribution to the work reported, in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no financial or personal conflicts of interests in this work.
References
1. World Health Organization. Situation Report COVID-19. World Health Organization; 2021.
2. Tobin MJ, Laghi F, Jubran A. Why COVID-19 silent hypoxemia is baffling to physicians. Am J Respir Crit Care Med. 2020;202(3):356–360. doi:10.1164/rccm.202006-2157CP
3. Wilkerson RG, Adler JD, Shah NG, Brown R. Silent hypoxia: a harbinger of clinical deterioration in patients with COVID-19. Am J Emerg Med. 2020;38(10):
4. Rawson TM, Moore LSP, Zhu N, et al. Bacterial and fungal coinfection in individuals with Coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020;71(9):2459–2468. doi:10.1093/cid/ciaa530
5. Dudoignon E, Caméléna F, Deniau B, et al. Bacterial Pneumonia in COVID-19 critically ill patients: a case series. Clin Infect Dis. 2020. doi:10.1093/cid/ciaa762
6. Adler H, Ball R, Fisher M, Mortimer K, Vardhan MS. Low rate of bacterial co-infection in patients with COVID-19. Lancet Microbe. 2020;1(2):e62. doi:10.1016/S2666-5247(20)30036-7
7. Contou D, Claudinon A, Pajot O, et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann Intensive Care. 2020;10(119). doi:10.1186/s13613-020-00736-x
8. Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect. 2020;26(10):1395–1399. doi:10.1016/j.cmi.2020.06.025
9. Sepulveda J, Westblade LF, Whittier S, et al. Bacteremia and blood culture utilization during COVID-19 surge in New York City. J Clin Microbiol. 2020;58(8):e00875–20. doi:10.1128/JCM.00875-20
10. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi:10.1016/S0140-6736(20)30566-3
11. Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81(2):266–275. doi:10.1016/j.jinf.2020.05.046
12. Siegel JD, Rhinehart E, Jackson M, Chiarello L, and the Healthcare Infection Control Practices Advisory Committee. Guideline for isolation precautions: preventing transmission of infectious agents in healthcare setting; 2007. Available from: https://www.cdc.gov/infectioncontrol/guidelines/isolation/index.ht.
13. World Health Organization. Interim Guidance: Clinical Management of COVID-19. World Health Organization; 2020.
14. Leber AL. Clinical Microbiology Procedures Handbook.
15. Dargère S, Cormier H, Verdon R. Contaminants in blood cultures: importance, implications, interpretation and prevention. Clin Microbiol Infect. 2018;24(9):964–969. doi:10.1016/j.cmi.2018.03.030
16. Krisanapan P, Chaiwarith R. Time to blood cultures positivity of microorganisms using a continuous-monitoring automated blood cultures system. Asian Biomed. 2019;13(2):61–69. doi:10.1515/abm-2019-0041
17. National Healthcare Safety Network. 2021 NHSN Patient Safety Component Manual. Centers for Disease Control and Prevention; 2021.
18. Clinical and Laboratory Standards Institute. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2021.
19. Elzi L, Babouee B, Vögeli N, et al. How to discriminate contamination from bloodstream infection due to coagulase-negative staphylococci: a prospective study with 654 patients. Clin Microbiol Infect. 2012;18(9):E355–61. doi:10.1111/j.1469-0691.2012.03964.x
20. Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22(7):707–710. doi:10.1007/BF01709751
21. Beck DH, Smith GB, Pappachan JV, Millar B. External validation of the SAPS II, APACHE II and APACHE III prognostic models in South England: a multicentre study. Intensive Care Med. 2003;29(2):249–256. doi:10.1007/s00134-002-1607-9
22. Previsdomini M, Gini M, Cerutti B, Dolina M, Perren A. Predictors of positive blood cultures in critically ill patients: a retrospective evaluation. Croat Med J. 2012;53(1):30–39. doi:10.3325/cmj.2012.53.30
23. Sidhu SK, Malhotra S, Devi P, Tuli AK. Significance of coagulase negative Staphylococcus from blood cultures: persisting problems and partial progress in resource constrained settings. Iran J Microbiol. 2016;8(6):366–371.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


