Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 15
Peripheral Blood circRNA Microarray Profiling Identities hsa_circ_0001831 and hsa_circ_0000867 as Two Novel circRNA Biomarkers for Early Type 2 Diabetic Nephropathy
Authors Zhang K , Wan X, Khan MA , Sun X, Yi X, Wang Z, Chen K , Peng L
Received 28 July 2022
Accepted for publication 5 September 2022
Published 10 September 2022 Volume 2022:15 Pages 2789—2801
DOI https://doi.org/10.2147/DMSO.S384054
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Juei-Tang Cheng
Keke Zhang,1 Xinxing Wan,1 Md Asaduzzaman Khan,2 Xiaoying Sun,1 Xuan Yi,1 Zhouqi Wang,1 Ke Chen,1 Lin Peng3
1Department of Endocrinology, the Third Xiangya Hospital of Central South University, Changsha, People’s Republic of China; 2The Research Centre for Preclinical Medicine, Southwest Medical University, Luzhou, People’s Republic of China; 3Department of Nephrology, the First Hospital of Changsha, Changsha, People’s Republic of China
Correspondence: Lin Peng, Department of Nephrology, the First Hospital of Changsha, Changsha, People’s Republic of China, Tel +86-731-8466-7510, Email [email protected] Ke Chen, Department of Endocrinology, the Third Xiangya Hospital of Central South University, Changsha, People’s Republic of China, Tel +86-731-8861-8239, Email [email protected]
Purpose: Type 2 diabetes mellitus (T2DM) increases the incidence of diabetic nephropathy (DN) and eventually progresses to end-stage renal disease. Circular RNAs (circRNAs) are a class of non-coding RNAs that are promising as diagnostic biomarkers and therapeutic targets for human diseases. The aim of this study was to analyze the differential expression of circRNAs (DECs) in peripheral blood from patients with early type 2 diabetic nephropathy (ET2DN), T2DM and controls, which will facilitate to discover some new biomarkers for ET2DN.
Patients and Methods: Twenty ET2DN patients, 20 T2DM patients, and 20 normal controls were included in this study. Blood samples from 3 random subjects of age- and sex-matched patients in each group, respectively, were used to detect circRNA expression profiles by circRNA microarray, and the circRNA expression of remaining subjects was validated by real-time quantitative polymerase chain reaction (qRT-PCR). Further functional assessment was performed by bioinformatic tools.
Results: There were 586 DECs in ET2DN vs T2DM group (249 circRNAs were upregulated and 337 circRNAs were downregulated); 176 circRNAs were upregulated and 101 circRNAs were downregulated in T2DM vs control group; 57 circRNAs were upregulated and 5 circRNAs were downregulated in ET2DN vs control group. The functional and pathway enrichment of DECs were analyzed by GO and KEGG. qRT-PCR results revealed that hsa_circ_0001831 and hsa_circ_0000867 were significantly upregulated in ET2DN group compared to both of T2DM and control group. The ROC curve demonstrated that hsa_circ_0001831 and hsa_circ_0000867 have high sensitivity and specificity associated with ET2DN.
Conclusion: Our study showed the expression profiles of circRNAs in ET2DN patients and demonstrated that hsa_circ_0001831 and hsa_circ_0000867 can be used as novel diagnostic biomarkers for ET2DN.
Keywords: circRNAs microarray, diagnostic biomarkers, early type 2 diabetic nephropathy, inflammation
Introduction
Diabetic nephropathy (DN) occurs in approximately 40% of Type 2 diabetes mellitus (T2DM),1 with the progress of T2DM; early type 2 diabetic nephropathy (ET2DN) eventually develops into end-stage renal disease.2,3 Thus, it is particularly crucial to enhance the prevention and treatment of ET2DN and discover valid biological diagnostic biomarkers.
Circular RNAs (circRNAs) are a group of newly discovered non-coding RNAs.4 In recent years, there has been accumulating evidence that circRNAs can regulate host gene’s splicing and transcription and act as microRNAs and protein sponges, as well as moderate protein translation and protein functions.5 CircRNAs have been identified to play an active role in treating a broad range of diseases.6–8
The pathological manifestations associated with DN include glomerular thylakoid cells hypertrophy and hyperplasia, extracellular matrix (ECM) accumulation, podocytes function impairment and renal fibrosis.9,10 Some studies have been partially demonstrated that circRNAs can act as microRNA sponges to influence the pathogenesis of DN, for instance, An et al have shown that hsa_circ_0003928 can bind to miR-151-3p and moderate Annexin A2 (ANXA2) expression via the microRNAs sponge, thereby attenuating high glucose-induced apoptosis in podocytes and glomerular thylakoid cells.11 circRNA_0080425 is able to target miR-24-3p to regulate fibroblast growth factor 11 (FGF11) expression levels, which led to fibrosis and promoted DN progression.12 miR-24-3p play an important role in regulating cell proliferation, apoptosis and fibrosis.13–15 A study in mice with DN showed that circRNA_15698, which is highly expressed in DN mice, functions as a sponge for miR-185 and upregulates the expression of TGFB1, thereby promoting the production of ECM proteins.9
In addition, some circRNAs have been reported to be particular in DN via inflammatory pathways, for example, circRNA_ITCH improved the renal inflammation in streptozotocin-induced diabetic mice,16 knockdown of circLRP6 decreased high glucose-induced NF-κB signaling pathway in mesangial cells,17 circRNA_WBSCR17 accelerated inflammatory responses in high glucose-induced kidney tubular cells.18
There has been a study on the expression profile of circRNAs using kidney tissue from DN mice, and differential expression circRNAs (DECs) may be candidated as some biomarkers for DN.19 However, there has been no study on circRNAs as diagnostic biomarkers for ET2DN in humans yet. The aim of this study was to find new diagnostic biomarkers for ET2DN by profiling the DECs in peripheral blood from patients with ET2DN.
In this study, we performed circRNA microarray assay on peripheral blood samples from patients with ET2DN and T2DM without DN and age-sex matched normal controls to screen for DECs. Partial circRNAs were validated by quantitative real‐time reverse transcription‐PCR (qRT‐PCR), and further bioinformatic analysis was performed to explore their biological functions. ROC curves were made to evaluate clinical diagnostic value of DECs.
Subjects and Methods
Study Subjects
A total of 60 subjects, including 20 ET2DN patients, 20 age- and sex-matched T2DM without DN patients and 20 age- and sex-matched normal control subjects, were enrolled in this study. All ET2DN and T2DM groups were presented in the Department of Endocrinology of the Third Xiangya Hospital of Central South University from November 2021 to June 2022, and all control subjects volunteered to participate in this study.
The ET2DN inclusion criteria included patients who were firstly tested by urinary albumin excretion rate (UAER) in 20–200 μg/min and were further verified by UAER in 30–300 mg/24h for twice in different days. Patients in ET2DN and T2DM group were confirmed according to World Health Organization criteria.20 All subjects with serious infectious diseases, autoimmune diseases, hematological diseases, malignant diseases, fever were excluded, and control subjects had no history of diabetes mellitus or renal disease. The clinical characteristics of the participants included age, gender, duration of diabetes, systolic blood pressure (SBP), diastolic blood pressure (DBP), height, weight, BMI, waist circumference (WC), hip circumference (HC). In addition, biochemical parameters including fasting blood glucose (FBG), HbA1c, creatinine (Cr), UAER, total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), triglycerides (TG) and haemoglobin (Hb) levels were measured.
This study was approved by the Ethics Committee of The Third Xiangya Hospital of Central South University, Changsha, China, and was performed according to the guidelines outlined in the Declaration of Helsinki. All participants provided written informed consent form.
Peripheral Blood Samples Collection
Two milliliters of peripheral venous blood samples from different groups’ patients were collected. The samples were then randomly selected from 3 ET2DN, 3 age- and sex-matched T2DM and 3 age- and sex-matched control subjects for RNA extraction and circRNA microarray analysis.
CircRNA Microarray Analysis
Total RNA was isolated from peripheral blood samples with the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The NanoDrop ND-1000 (Nanodrop Technologies, Wilmington, DE) was used to quantify the purity and concentration of total RNA samples. Preparation of samples and microarray hybridization was conducted following Arraystar’s standard protocol. Briefly, digestion of total RNA with RNase R (Epicentre, Madison, WI, USA) was performed to remove linear RNA and enriched circRNAs. The abundant circRNA is in turn amplified and transcribed into fluorescent cRNA by the random initiation method (Arraystar Super RNA Label Kit; Arraystar), and tagged cRNAs were hybridized to Arraystar Human CircRNA arrays (8x15K, Arraystar). Once the slides were cleaned, the arrays were scanned using an Agilent G2505C scanner (Agilent Technology, Santa Clara, CA, USA).
Then, we featured extraction software from Agilent (version 11.0.1.1) for analysis of the acquired array images. The R software limma-package was used for the quantitative normalization and subsequent data processing. CircRNAs with significant differential expression between the three groups were shown by Volcano Plot. The DECs between two samples were identified by fold-change filtering. Hierarchical clustering showed distinguishable expression patterns of circRNAs between samples. The DECs between two groups were defined as | log2 fold change| ≥ 1 and p-value < 0.05.
Gene ontology (GO) analysis of DECs according to their host gene was carried out by GO seq R package, where the ontology covered three areas: biological processes (BP), cellular components (CC) and molecular functions (MF). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of relevant biological pathways was performed by KEGG database (http://www.geneontology.org). The p-value produced by top GO denotes the significance of GO term enrichment in differential circRNAs.
qRT-PCR Validation of Candidate circRNAs
The DECs between the each groups were validated by qRT‐PCR in accordance with the raw data analysis. Total RNA was reversed transcribed into cDNA using Revert Aid TMM-MuLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) with random primers according to the manufacturer’s instructions. RT-PCR was performed at real time on a VIIa 7 real-time polymerase chain reaction system (Applied Biosystems, New York, NY, USA). The thermal cycling conditions for the PCR reaction were 95 °C for 10 min, followed by 45 cycles of 95°C for 10s, 60°C for 60s and 95 °C for 15s. The GAPDH was used as the internal control. The primer list is provided in Supplementary Table 1. The relative expression level was calculated using the 2−∆∆ct method.
The Prediction of circRNA–miRNA–mRNA Networks
To investigate the biological potential function of the target circRNAs, the bioinformation method was used for the prediction of circRNA–microRNA–mRNA interaction network. Firstly, the circRNA–microRNA interaction was predicted with Arraystar’s home-made miRNA target prediction software based on TargetScan (https://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do). DIANA TOOLS (http://diana.imis.athena-innovation.gr/DianaTools/index.php) software was used to predict the microRNA and mRNA interaction; the circRNA–microRNA–mRNA network was displayed using Cytoscape 2.8.2.21
Receiver Operating Characteristic (ROC) Curves
ROC curves were plotted using the ET2DN group as the state variable and the relative expression level of hsa_circ_0001831 and hsa_circ_0000867 as the test variable to assess the diagnostic performance of DECs against ET2DN. The ROC curves and the area under the curve (AUC) were analyzed, and sensitivity as well as specificity was calculated using 95% confidence intervals (Cis), with AUC > 0.5 representing diagnostic value.
Statistics Analysis
Statistical analyses were performed using SPSS 26.0 (SPSS, Inc., IL, USA) and GraphPad Prism 8.0 (GraphPad, Inc., CA, USA). Clinical data and results from experiments were presented as means ± standard deviation or numbers with percentages. Shapiro Wilk test was used to verify whether random sample data conformed to normal distribution. Normally distributed data between groups were calculated by independent sample t-test and one-way analysis of variance (ANOVA), non-normally distributed data were tested using Mann–Whitney U-test. P ≤ 0.05 was considered as statistically significant difference. qRT-PCR results were compared using the unpaired t‐test. Multiple logistic regression was used to analyze the correlation between circRNAs and clinical indicators.
Results
Characteristics of the Study Population
The basic clinical characteristics of the subjects are revealed in Table 1. As shown, the duration of diabetes, SBP, DBP, FBG, HbA1c and HDL-C levels had significant difference in ET2DN vs control group; the ET2DN group had substantially higher UAER than the other two groups (P = 0.014). However, there were no statistical differences in age, height, weight, WC, HC, BMI, Cr, TC, TG, LDL-C and Hb levels across the three groups.
 |
Table 1 Characteristics and Clinical Features of Study Population |
DECs Between Groups
High-throughput human circRNA microarray revealed the circRNA expression profiles in the peripheral blood of three groups’ subjects. By quantile normalization of raw data for further analyses, the Box Plot showed that the distribution of log 2 ratios was similar in all the samples (Figure 1A). Hierarchical clustering analysis showed that the circRNA expression pattern was significantly different between the three groups (Figure 1B). Fold-change filtering of DECs was showed in scatter plot (Figure 1C-E). In addition, Volcano Plots was used to visualize the variation in circRNA expression with statistical significance between the three groups (fold change >1.0 and P < 0.05) (Figure 2A-C).
According to the circRNA microarray results, a total of 586 circRNAs were differentially expressed between the ET2DN and T2DM group, of which 249 were upregulated and 337 were downregulated. Top 10 upregulated and downregulated circRNAs were shown in Tables 2 and 3, among them, hsa_circ_0001831 and hsa_circ_0044235 were the most upregulated and downregulated circRNAs between the ET2DN and T2DM group, respectively. In addition, 176 circRNAs were upregulated and 101 circRNAs were downregulated in T2DM vs control group; top 10 upregulated and downregulated circRNAs are shown in Supplementary Table 2 and Supplementary Table 3. Fifty-seven circRNAs were upregulated and 5 circRNAs were downregulated in ET2DN vs control group; top 10 upregulated and 5 downregulated circRNAs are shown in Supplementary Table 4 and Supplementary Table 5, respectively.
 |
Table 2 Upregulated of circRNAs in ET2DN Vs T2DM Group |
 |
Table 3 Downregulated of circRNAs in ET2DN Vs T2DM Group |
We found that majority of the DECs of three groups on human chromosome distribution were mainly located on chromosome 1 (Supplementary Figure 1A, B, C).
GO and KEGG Pathway Analysis for Host Genes of circRNAs
To predict the function enrichment of DECs, GO and KEGG analyses were performed. GO analysis of upregulated circRNAs in ET2DN vs T2DM group showed that the DECs were enriched in small GTPase mediated signal transduction and positive regulation of GTPase activity and so on (Figure 3A). The analysis of downregulated circRNAs in ET2DN vs T2DM group suggested that DECs were more relevant to biological processes such as symbiont process and viral process (Figure 3B). In addition, GO analysis of T2DM vs control group was performed (Supplementary Figure 2A, B); however, GO analysis could not be enriched because too few DECs in ET2DN vs control group. KEGG analysis indicated that the upregulation of circRNAs remarkably enriched pathways including morphine addiction, human cytomegalovirus infection, and downregulation mainly focused on RNA transport, mTOR signaling pathway (Figure 3C and D). Pathway analysis of T2DM vs control group was analyzed (Supplementary Figure 3A, B). As mentioned above, pathway analysis cannot be enriched because there are too few DECs in ET2DN vs control group.
To explore whether special circRNAs were upregulated or downregulated in ET2DN compared to T2DM and control group, the common circRNAs from ET2DN vs T2DM group and ET2DN vs control group were further analyzed. Total 3 circRNAs, named hsa_circ_0000867, hsa_circ_0022430 and hsa_circ_0063812, were upregulated in ET2DN compare to T2DM and control group; however, hsa_circ_0009035 was downregulated (Figure 3E).
Validation of DECs by qRT-PCR
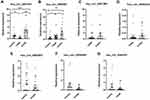
Based on the results of circRNA microarray, six candidate circRNAs were selected for validation by qRT-PCR, including the highest expression (hsa_circ_0001831) and the lowest expression (hsa_circ_0044235 and hsa_circ_0061260) in ET2DN vs T2DM group; the highest expression (hsa_circ_0044235) and the lowest expression (hsa_circ_0001831) in T2DM vs control group; the highest expression (hsa_circ_0000658) and the lowest expression (hsa_circ_0063503) in ET2DN vs control group; the highest expression of circRNAs in ET2DN vs T2DM and control group (hsa_circ_0000867). The Results of RT‑qPCR suggested that hsa_circ_0001831 was significantly upregulated in ET2DN vs T2DM group. In addition, although hsa_circ_0001831 did not be shown in ET2DN vs control group in our circRNA microarray data, our RT-PCR results showed that hsa_circ_0001831 was also upregulated in ET2DN vs control group (Figure 4A). In common expression data, hsa_circ_0000867 was verified as higher expression in ET2DN vs T2DM and control group (Figure 4B). However, hsa_circ_0061260 and hsa_circ_0044235 did not have difference in ET2DN vs T2DM group (Figure 4C and D); hsa_circ_0063503 and hsa_circ_0000658 also had no statistically significant difference between the ET2DN vs control group (Figure 4E and F); hsa_circ_0044235 had no statistically significant difference in T2DM vs control group (Figure 4G). Altogether, consistent with the microarray results, the expression level of hsa_circ_0001831 and hsa_circ_0000867 may be two diagnosis biomarkers in ET2DN in peripheral blood. Therefore, hsa_circ_0001831 and hsa_circ_0000867 were selected for further study.
Prediction of the circRNA–microRNA–mRNA Interaction Network
To further clarify the function of hsa_circ_0001831 and hsa_circ_0000867, TargetScan and miRanda were used to predict circRNA–microRNA interaction network, and DIANA TOOLS software was used to predict the microRNA–mRNA interaction. This indicated that hsa-miR-3151-5p, hsa-miR-6791-5p, hsa-miR-939-5p, hsa-miR-637 and hsa-miR-7974 were predicted as target microRNAs of hsa_circ_0001831, and has-miR-146a-3p and has-miR-877-3p were predicted as target microRNAs of hsa_circ_0000867. The interaction network of hsa_circ_0001831 and hsa_circ_0000867 with microRNA–mRNA is shown in Figure 5A and B.
The Value of hsa_circ_0001831 and hsa_circ_0000867 in the Diagnosis of ET2DN
To determine the diagnostic value of hsa_circ_0001831 and hsa_circ_0000867 in ET2DN, ROC curves were drawn and analyzed. The results showed that hsa_circ_0001831 had a sensitivity of 0.85, a specificity of 0.85, an AUC value of 0.95 and a maximum Jordan index of 0.75 for the diagnosis of ET2DN (Figure 6A). In addition, hsa_circ_0000867 had a sensitivity of 0.6, a specificity of 0.95, an AUC value of 0.95 and a maximum Jordan index of 0.55 for the diagnosis of ET2DN (Figure 6B). Based on the Jordan index, the optimal exploration cut-off point for hsa_circ_0001831 was 1.55 and hsa_circ_0000867 was 1.14, respectively. Together, hsa_circ_0001831 and hsa_circ_0000867 showed high diagnostic value for ET2DN.
 |
Figure 6 ROC curve analysis of diagnostic value in hsa_circ_0001831 (A) and hsa_circ_0000867 (B) in ET2DN patients. |
Discussion
DN is one of the chronic complications of diabetes, which causes end-stage renal disease.22,23 Some studies have revealed that circRNAs can be used for diagnostic biomarkers for some diseases.24–26 CircRNAs may be involved in the development of DN and could be a new therapeutic target for DN, for instance, circ_0000285 caused podocyte injury by sponging miR-654-3p and activating MAPK6 in DN.27 However, there was no study on the expression profile of circRNAs in ET2DN. Therefore, our aim was to explore the expression profile of circRNAs in patients’ peripheral blood with ET2DN, and we tried to find out some new diagnostic biomarkers for ET2DN.
In the present study, circRNA microarray was used for detecting the DECs in peripheral blood of patients with ET2DN. Our results indicate that there were 586 DECs in ET2DN vs T2DM group and 62 DECs in ET2DN vs control group. These results implied that the DECs in ET2DN patients were significantly different from T2DM and control group, and these circRNAs might be strongly associated with ET2DN.
Next, we performed GO and KEGG analysis on the DECs to understand the possible functions of circRNAs in ET2DN. The results suggested that the DECs in ET2DN were closely associated with multiple biological processes such as GTPases and protein modifications, and the pathways were enriched in some biological pathways like RNA transport, mTOR signaling pathway. Several pathways have been reported to be related to DN, including MAPK and mTOR signaling pathway.28 We speculated that some circRNAs might be involved in the modification of ET2DN through these pathways.
To further validate the DECs, six candidate DECs were detected by qRT-PCR. The expression of hsa_circ_0001831 and hsa_circ_0000867 were significantly upregulated in ET2DN compared to both of T2DM and control group. Therefore, hsa_circ_0001831 and hsa_circ_0000867 may be two diagnostic biomarkers in ET2DN. hsa_circ_0001831 was produced by its host gene, scribble planar cell polarity protein (SCRIB), which is a tumor suppressor, hsa_circ_0001831 could decrease SCRIB mRNA expression but did not affect mRNA splicing, in addition, hsa_circ_0001831 was upregulated in breast cancer and promoted breast cancer cell proliferation, migration and invasion.29 Moreover, SCRIB, as an essential gene, was upregulated in podocytes according to the Human Protein Atlas (HPA) database (https://www.proteinatlas.org).30
However, there were no study about hsa_circ_0000867, but its host gene, ornithine decarboxylase antizyme 1 (OAZ1) was significantly associated with T2DM duration and correlated with HbA1c.31,32
CircRNAs indirectly promoted target mRNA expression through binding to microRNAs and repress microRNAs expression.33 Therefore, we predicted the hsa_circ_0001831 and hsa_circ_0000867-microRNA–mRNA interaction network. In predicted target microRNAs of hsa_circ_0000867, hsa-miR-877-3p were upregulated in urinary exosomal of T2DN.34 In addition, hsa-miR-146a-3p was strongly upregulated in DN patients compared to control group;35 however, another study showed that inhibition of hsa-miR-146a increases podocyte injury in diabetic glomerulopathy36 and inhibited inflammatory and reactive oxygen species in mice DN.37,38 Moreover, inflammatory pathways play a critical role in T2DN, except for hsa-miR-146a, some predicted microRNAs were related to inflammatory process in other diseases, for instance, hsa-miR-939 negatively regulated pro-inflammatory genes in human monocytic leukaemia THP-1 cells.39 Furthermore, miR-939-5p was able to attenuate PM2.5-induced endothelial injury via NF-κB p65 signaling pathway in human aortic endothelial cells,40 and hsa-miR-637 suppressed vascular inflammation in hypertension through inflammatory mediators.41 Besides, miR-877-3p negatively regulated inflammatory factor including IL-1β in mesangial cells.42 Together, hsa_circ_0001831 and hsa_circ_0000867 might induce ET2DN through these predicted microRNAs. However, more research is needed on the mechanisms by which these DECs affect T2DN.
An assessment of the potential diagnostic value of hsa_circ_0001831 and hsa_circ_0000867 for ET2DN was made. Analysis using ROC curves based on disease diagnosis showed that hsa_circ_0001831 and hsa_had a larger AUC value, better sensitivity and strength, as well as higher predictive accuracy. Therefore, hsa_circ_0001831 and hsa_circ_0000867 could be the two potential new biomarkers for ET2DN screening and diagnosis.
There are some limitations in our study. Firstly, the sample size of this study was relatively small. Secondly, only a few circRNAs from the DECs were validated. Thirdly, the specific mechanism of these DECs in ET2DN remains to be further investigated.
Conclusion
In summary, our study revealed the circRNA expression profiles of ET2DN patients utilizing circRNA microarrays indicating that hsa_circ_0001831 and hsa_circ_0000867 may be used as two biomarkers of ET2DN, which provides a new idea for the diagnosis of ET2DN.
Funding
This study was supported in part by grants from the Hunan Province Natural Science Foundation of China (2022JJ70045 and 2022JJ30881).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Shi S, Ni L, Gao L, et al. Comparison of Nonalbuminuric and Albuminuric Diabetic Kidney Disease Among Patients With Type 2 Diabetes: a Systematic Review and Meta-Analysis. Front Endocrinol. 2022;13:871272. doi:10.3389/fendo.2022.871272
2. Zhang W, Liu X, Dong Z, et al. New Diagnostic Model for the Differentiation of Diabetic Nephropathy From Non-Diabetic Nephropathy in Chinese Patients. Front Endocrinol. 2022;13:913021. doi:10.3389/fendo.2022.913021
3. Yuan CM, Nee R, Ceckowski KA, et al. Diabetic nephropathy as the cause of end-stage kidney disease reported on the medical evidence form CMS2728 at a single center. Clin Kidney J. 2017;10(2):257–262. doi:10.1093/ckj/sfw112
4. Hsiao KY, Sun HS, Tsai SJ. Circular RNA - New member of noncoding RNA with novel functions. Exp Biol Med. 2017;242(11):1136–1141. doi:10.1177/1535370217708978
5. van Zonneveld AJ, Kölling M, Bijkerk R, et al. Circular RNAs in kidney disease and cancer. Nat Rev Nephrol. 2021;17(12):814–826. doi:10.1038/s41581-021-00465-9
6. Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res Commun. 2017;487(4):769–775. doi:10.1016/j.bbrc.2017.04.044
7. Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi:10.1038/srep35576
8. Wu H, Wu S, Zhu Y, et al. Hsa_circRNA_0054633 is highly expressed in gestational diabetes mellitus and closely related to glycosylation index. Clin Epigenetics. 2019;11(1):22. doi:10.1186/s13148-019-0610-8
9. Hu W, Han Q, Zhao L, et al. Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-beta1. J Cell Physiol. 2019;234(2):1469–1476. doi:10.1002/jcp.26959
10. He X, Kuang G, Zuo Y, et al. The Role of Non-coding RNAs in Diabetic Nephropathy-Related Oxidative Stress. Front Med. 2021;8:626423. doi:10.3389/fmed.2021.626423
11. An L, Ji D, Hu W, et al. Interference of Hsa_circ_0003928 Alleviates High Glucose-Induced Cell Apoptosis and Inflammation in HK-2 Cells via miR-151-3p/Anxa2. Diabetes Metab Syndr Obes. 2020;13:3157–3168. doi:10.2147/DMSO.S265543
12. Liu H, Wang X, Wang ZY, et al. Circ_0080425 inhibits cell proliferation and fibrosis in diabetic nephropathy via sponging miR-24-3p and targeting fibroblast growth factor 11. J Cell Physiol. 2020;235(5):4520–4529. doi:10.1002/jcp.29329
13. Wang J, Huang W, Xu R, et al. MicroRNA-24 regulates cardiac fibrosis after myocardial infarction. J Cell Mol Med. 2012;16(9):2150–2160. doi:10.1111/j.1582-4934.2012.01523.x
14. Yan L, Ma J, Zhu Y, et al. miR-24-3p promotes cell migration and proliferation in lung cancer by targeting SOX7. J Cell Biochem. 2018;119(5):3989–3998. doi:10.1002/jcb.26553
15. Zhou N, Yan HL. MiR-24 promotes the proliferation and apoptosis of lung carcinoma via targeting MAPK7. Eur Rev Med Pharmacol Sci. 2018;22(20):6845–6852. doi:10.26355/eurrev_201810_16153
16. Liu J, Duan P, Xu C, et al. CircRNA circ-ITCH improves renal inflammation and fibrosis in streptozotocin-induced diabetic mice by regulating the miR-33a-5p/SIRT6 axis. Inflamm Res. 2021;70(7):835–846. doi:10.1007/s00011-021-01485-8
17. Chen B, Li Y, Liu Y, et al. circLRP6 regulates high glucose-induced proliferation, oxidative stress, ECM accumulation, and inflammation in mesangial cells. J Cell Physiol. 2019;234(11):21249–21259. doi:10.1002/jcp.28730
18. Li G, Qin Y, Qin S, et al. Circ_WBSCR17 aggravates inflammatory responses and fibrosis by targeting miR-185-5p/SOX6 regulatory axis in high glucose-induced human kidney tubular cells. Life Sci. 2020;259:118269. doi:10.1016/j.lfs.2020.118269
19. Xiong X, Liu C, Shen M, et al. Circular RNA expression profile in transgenic diabetic mouse kidneys. Cell Mol Biol Lett. 2021;26(1):25. doi:10.1186/s11658-021-00270-z
20. Lazo-Porras M, Bernabe-Ortiz A, Ruiz-Alejos A, et al. Regression from prediabetes to normal glucose levels is more frequent than progression towards diabetes: the CRONICAS Cohort Study. Diabetes Res Clin Pract. 2020;163:107829. doi:10.1016/j.diabres.2019.107829
21. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
22. Badal SS, Danesh FR. New insights into molecular mechanisms of diabetic kidney disease. Am J Kidney Dis. 2014;63(2 Suppl 2):S63–S83. doi:10.1053/j.ajkd.2013.10.047
23. Natesan V, Kim SJ. Diabetic Nephropathy - a Review of Risk Factors, Progression, Mechanism, and Dietary Management. Biomol Ther. 2021;29(4):365–372. doi:10.4062/biomolther.2020.204
24. Noureddine L, Hajarnis S, Patel V. MicroRNAs and Polycystic Kidney Disease. Drug Discov Today Dis Models. 2013;10(3):e137–e1743. doi:10.1016/j.ddmod.2013.10.001
25. Zhou Q, Chung AC, Huang XR, et al. Identification of novel long noncoding RNAs associated with TGF-β/Smad3-mediated renal inflammation and fibrosis by RNA sequencing. Am J Pathol. 2014;184(2):409–417. doi:10.1016/j.ajpath.2013.10.007
26. Kato M, Wang M, Chen Z, et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun. 2016;7:12864. doi:10.1038/ncomms12864
27. Yao T, Zha D, Hu C, et al. Circ_0000285 promotes podocyte injury through sponging miR-654-3p and activating MAPK6 in diabetic nephropathy. Gene. 2020;747:144661. doi:10.1016/j.gene.2020.144661
28. Bhattacharjee N, Barma S, Konwar N, et al. Mechanistic insight of diabetic nephropathy and its pharmacotherapeutic targets: an update. Eur J Pharmacol. 2016;791:8–24. doi:10.1016/j.ejphar.2016.08.022
29. Ma J, Du WW, Zeng K, et al. An antisense circular RNA circSCRIB enhances cancer progression by suppressing parental gene splicing and translation. Mol Ther. 2021;29(9):2754–2768. doi:10.1016/j.ymthe.2021.08.002
30. Song H, Zhuang L, Xu X, et al. MCC Regulator of WNT Signaling Pathway (MCC) Is a Podocyte Essential Gene. Front Med. 2021;8:777563. doi:10.3389/fmed.2021.777563
31. Yamamoto CM, Murakami T, Oakes ML, et al. Uromodulin MRNA from Urinary Extracellular Vesicles Correlate to Kidney Function Decline in Type 2 Diabetes Mellitus. Am J Nephrol. 2018;47(5):283–291. doi:10.1159/000489129
32. Wilson KH, Eckenrode SE, Li QZ, et al. Microarray analysis of gene expression in the kidneys of new- and post-onset diabetic NOD mice. Diabetes. 2003;52(8):2151–2159. doi:10.2337/diabetes.52.8.2151
33. Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
34. Xie Y, Jia Y, Cuihua X, et al. Urinary Exosomal MicroRNA Profiling in Incipient Type 2 Diabetic Kidney Disease. J Diabetes Res. 2017;2017:6978984. doi:10.1155/2017/6978984
35. Huang Y, Liu Y, Li L, et al. Involvement of inflammation-related miR-155 and miR-146a in diabetic nephropathy: implications for glomerular endothelial injury. BMC Nephrol. 2014;2(15):142. doi:10.1186/1471-2369-15-142
36. Lee HW, Khan SQ, Khaliqdina S, et al. Absence of miR-146a in Podocytes Increases Risk of Diabetic Glomerulopathy via Up-regulation of ErbB4 and Notch-1. J Biol Chem. 2017;292(2):732–747. doi:10.1074/jbc.M116.753822
37. Bhatt K, Lanting LL, Jia Y, et al. Anti-Inflammatory Role of MicroRNA-146a in the Pathogenesis of Diabetic Nephropathy. J Am Soc Nephrol. 2016;27(8):2277–2288. doi:10.1681/ASN.2015010111
38. Wan RJ, Li YH. MicroRNA-146a/NAPDH oxidase4 decreases reactive oxygen species generation and inflammation in a diabetic nephropathy model. Mol Med Rep. 2018;17(3):4759–4766. doi:10.3892/mmr.2018.8407
39. Ramanathan S, Shenoda BB, Lin Z, et al. Inflammation potentiates miR-939 expression and packaging into small extracellular vesicles. J Extracell Vesicles. 2019;8(1):1650595. doi:10.1080/20013078.2019.1650595
40. Liang S, Ning R, Zhang J, et al. MiR-939-5p suppresses PM2.5-induced endothelial injury via targeting HIF-1a in HAECs. Nanotoxicology. 2021;15(5):706–720. doi:10.1080/17435390.2021.1917716
41. He X, Bao X, Tao Z, et al. The microarray identification circular RNA hsa_circ_0105015 up-regulated involving inflammation pathway in essential hypertension. J Clin Lab Anal. 2021;35(2):e23603. doi:10.1002/jcla.23603
42. Liang Y, Zhao G, Tang L, et al. MiR-100-3p and miR-877-3p regulate overproduction of IL-8 and IL-1β in mesangial cells activated by secretory IgA from IgA nephropathy patients. Exp Cell Res. 2016;347(2):312–321.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.