Back to Journals » Infection and Drug Resistance » Volume 16
Pathogenic Profile Characteristics and Clinical Risk Factor Analysis of Patients Who Died from Sepsis Combined with Pulmonary Infection by Metagenomic Next-Generation Sequencing
Authors Chen SX, Lin R, Shi JL, Lin W, Yu XF, Chen JY
Received 19 April 2023
Accepted for publication 13 December 2023
Published 19 December 2023 Volume 2023:16 Pages 7695—7705
DOI https://doi.org/10.2147/IDR.S415503
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Shao-Xiong Chen, Ri Lin, Jiang-Long Shi, Wei Lin, Xing-Feng Yu, Jia-Yi Chen
Department of Critical Care Medicine, Fuqing Hospital Affiliated to Fujian Medical University, Fuqing, Fujian, People’s Republic of China
Correspondence: Jia-Yi Chen; Xing-Feng Yu, Fuqing Hospital Affiliated to Fujian Medical University, Qingrong Road No. 267, Fuqing, Fujian, 350300, People’s Republic of China, Tel +86 139 5912 0913, Email [email protected]; [email protected]
Introduction: Sepsis is one of the major diseases that seriously threatens human health, and its incidence and in-hospital morbidity and mortality rates remain high. Applying metagenomic next-generation sequencing (mNGS) technology to analyze the differences in pathogenic profiles and clinical factors in patients surviving and dying from sepsis combined with pulmonary infections provides diagnostic value and application for clinical purposes.
Methods: Sixty-three BALF samples from patients with sepsis combined with pulmonary infection from Fuqing Hospital Affiliated to Fujian Medical University were collected, and all of them were tested by simultaneous mNGS and conventional microbial combined test (CMT) to compare the pathogenic profiles and clinical indices of patients who survived and died of sepsis combined with pulmonary infection and to further compare the diagnostic differences between mNGS and CMT in patients who survived and died of sepsis combined with pulmonary infection. We analyzed the diagnostic value of mNGS for sepsis combined with pulmonary infection.
Results: A total of 141 strains of pathogens were isolated from 63 samples of patients with sepsis combined with pneumonia at suspected infection sites, Klebsiella pneumoniae, Acinetobacter baumannii, and Stenotrophomonas maltophilia are predominant, and higher ApacheII, LAC, P and PT are all risk factors affecting the death of septic patients.
Conclusion: Applying the mNGS method to patients with sepsis combined with pneumonia can improve the positive detection rate of pathogenic microorganisms and focus on death-related risk factors such as pathogenic bacteria species as well as clinical laboratory indices, which can guide clinicians to take appropriate measures to treat patients with sepsis and reduce the occurrence of death.
Keywords: sepsis, mNGS, clinical diagnosis, death, risk factors
Introduction
Sepsis is one of the major diseases that poses a serious threat to human health, and its incidence and in-hospital mortality rates remain high despite the continuous updating of sepsis guidelines.1 Its main clinical manifestations are elevated body temperature, chills, and rapid heart rate, and it is most common in people with infectious diseases.2,3 If sepsis patients do not receive timely and effective treatment at the early stage of the disease, the disease may worsen to a severe level and induce complications such as shock and multiple organ dysfunction syndrome (MODS), which may lead to death in severe cases.3,4
Clinical studies have found that the earlier antimicrobial drugs are applied after sepsis is confirmed, the greater the benefit to the patient, and intravenous antimicrobial drug therapy should be started as soon as possible within 1 h.5 Before that, microbial cultures should be obtained and microbiologically confirmed, and antimicrobial drugs should be adjusted according to the drug sensitivity results and clinical signs and symptoms.4 Traditional pathogenic microbial culture methods and diagnostic pathogenesis found that the most common clinical pathogenic bacteria are gram-negative bacteria, followed by gram-positive bacteria, fungi, anaerobic bacteria, and parasites. However, the current clinical microbial culture time is long, the positive rate is low, and it cannot detect pathogens that cannot be routinely cultured, such as viruses, intracellular bacteria, and caustic bacteria, thus affecting the subsequent treatment adjustment.6–8
Next-generation sequencing (NGS) technology has evolved from scientific research to clinical applications, initially for individualized tumor treatment and screening of genetic diseases, and in recent years, it has been gradually expanded to the field of infectious diseases for rapid identification of pathogens and tracking the spread and evolution of pathogens.9,10 Among them, mNGS, which does not require culture, is fast and accurate, can identify pathogenic bacteria, and is unique for guiding the diagnosis and anti-infection treatment of sepsis patients in the intensive care unit (ICU). However, there are few related studies.11,12
At present, the clinical treatment of severe sepsis mainly adopts symptomatic treatment, such as infection control and active fluid resuscitation, which can alleviate patients’ clinical symptoms and improve their conditions to a certain extent, but their prognosis is still poor.13 Therefore, clinical studies need to focus on the composition of pathogenic bacteria in patients with severe sepsis and the related risk factors affecting their death and implement corresponding prevention and treatment measures for patients to improve their prognosis. In this study, we aimed to investigate the risk factors affecting their death and to analyze the potential value of mNGS in the prognosis of patients with sepsis combined with pneumonia to provide a scientific basis for the clinical prevention and treatment of severe sepsis and improve the survival rate.
Materials and Methods
Subjects and Study Design
This prospective study was collected from the Fuqing Hospital Affiliated to Fujian Medical University, Department of Critical Care Medicine, 2021.1–2022.12. The leading investigator determined the sample size based on experience of clinical research and hospital capacity. A total of 63 patients were diagnosed with sepsis combined with pneumonia and were divided into a death group (19 patients) and a survival group (44 patients) according to whether they died within 1 month after the onset of the disease, in which the clinicians determined the criteria for death: 1. deep coma; 2. complete disappearance of brainstem reflexes; and 3. no voluntary breathing. Patients with all of these criteria were judged as dead. Patient information was extracted from their medical records, including age, sex, and underlying disease. Clinical indicators tested within 24 hours after admission were recorded. Patients signed an informed consent form, and the study was approved by the Fuqing Hospital Affiliated to Fujian Medical University ethics committee, and the study was complied with the Declaration of Helsinki.
We performed etiological analysis and analysis of suspected coinfection in 63 patients with mNGS diagnosis of pneumonia infection. Specifically, we performed simultaneous mNGS and conventional combined microbiological test (CMT) tests on 63 bronchoalveolar lavage fluid (BALF) samples from 63 patients. The results of mNGS reports and CMT were also analyzed to determine the positive detection rate (Figure 1).
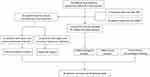 |
Figure 1 Flow chart of this study design. |
Diagnostic Criteria for Severe Pneumonia and Sepsis
Patients met the diagnostic criteria of severe pneumonia in the 2019 Infectious Diseases Society of America/American Thoracic Society (IDSA/ATS) guidelines for the diagnosis and treatment of adult community-acquired pneumonia.14 The main criteria were the need for tracheal intubation and mechanical ventilation, Sequential Organ Failure Assessment (SOFA) score ≥2, and the requirement for vasoactive therapy for septic shock after aggressive fluid resuscitation. Secondary criteria were as follows: respiratory rate ≥30 breaths/min; oxygenation index ≤250; and multiple lobes involved. Disturbance of consciousness; blood urea nitrogen ≥20 mg/dL; WBC count <4×109/L; platelet count <100×109/L; body temperature <36°C; hypotension requiring fluid resuscitation. A diagnosis was made if one major criterion or at least three minor criteria were met. Age ≥18 years old. Sepsis was defined as patients who met the following criteria: (1) with infection or suspected infection; (2) SOFA score ≥2.15 Patients were excluded if any one of the following criteria was met: (1) lacking the results for pathogens from conventional microbiological testing (CMT); (2) the etiological diagnosis was unclear when the patient was discharged from the hospital.
Sequencing and Data Processing
Patients were operated on by fibrinoscopists with more than 5 years of experience in performing fibrinoscopic alveolar lavage. Clinical samples were taken in accordance with aseptic processing standards: (1) From each patient, 10 mL or 5 mL of the BALF sample. TIANamp Micro DNA Kit was used for nucleic acid extraction (DP316, Tiangen Biotech Co., Beijing, China). (2) The VAHTS Universal Plus DNA Library Prep Kit for Illumina was used to create DNA libraries (ND617-C2, Vazyme Biotech Co., Nanjing, China). The DNA libraries underwent quality control using an Agilent 2100. The Illumina 550 platform sequenced the qualified libraries. (3) Human host sequences matching to the human reference genome (hg19) were computationally subtracted from the high-quality sequencing data using Kraken 2.1.2 and Burrows-Wheeler Alignment (BWA). Fastp 0.20.1 was used to filter out low-quality and short readings (less than 50 bp in length). The remaining information, which included information on bacteria, viruses, fungi, and parasites, was matched to the Pathogenic Microbial Genome Databases. The mapped data was processed for advanced analysis after the classification reference databases were acquired from the National Center of Biotechnology Information (NCBI) (ftp:/ftp.ncbi.nlm.nih.gov/genomes/).
mNGS Bioinformatic Analysis
Raw reads were filtered by fastp (v0.19.5) and Komplexity v0.3.6 to remove adaptor contamination, low-quality, and low-complexity reads. With Bowtie2 v2.3.4.3, reads that were mapped to the GRCh38 human reference assembly were eliminated. Subsequently, using SNAP v1.0 beta.18, the reads were aligned to the microorganism database, which contains over 12,000 genomes. For each species, either the NCBI GenBank genome database or the NCBI RefSeq genome database was used to categorize the mapped reads. We used Perl programs to count the species or genus abundance after removing false positive organisms.
Observation Indices
1. Distribution of pathogenic bacteria. Counting the pathogenic strains contained in all patient samples, including mNGS and CMT. 2. Single-factor analysis and multifactor analysis affecting the death of patients with severe sepsis. The clinical data of the chosen patients included: surgical operation or not; gender, age, and past medical history (hematological system, rheumatoid immune system, and neoplastic disorders were classified as immune-related diseases in this study); APACHE II (acute physiology and chronic health evaluation) score; The clinical laboratory data of both groups of patients were counted, mainly including albumin (ALB), alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatine kinase (CK), creatinine (Cr), C reactive protein (CRP), cardiac troponin (cTnI), Lactic acid (LAC), lactate dehydrogenase (LDH), LYMPH, LYMPH-2, myoglobin (MYO), NEUT, NEUT-2, NT-proBNP, Serum phosphorus (P), oxygenation index (PaO2/FiO2), Procalcitonin (PCT), Prothrombin time (PT), red blood cell count (RBC), and white blood cell count (WBC). Multifactorial logistic regression analysis affecting death in patients with severe sepsis. Multifactor unconditional logistic regression analysis of death in patients with severe sepsis affecting death in patients with severe sepsis was performed by using whether the patients died as the dependent variable and the indicators with statistically significant differences in the univariate analysis as the independent variables.
mNGS Clinical Performance
The definition and calculation rules of mNGS clinical performance, including sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and diagnostic agreement rate, are shown in Table S1.
Statistical Analysis
Software named SPSS 22.0 was implemented to analyze the data. T-tests were employed to assess group differences in the patient characteristic data, which has been normally distributed and represented as the mean standard deviation (SD). A rank-sum test was used to compare the data since nonnormally distributed data are best characterized by the median as well as the interquartile range (IQR). Wilcoxon test was used, if applicable, for comparative analysis. P values lower than 0.05 were deemed significant. The R package ggpubr was used to build the Random Forests regression model. R programming language or Microsoft Excel software was used to complete all statistical analyses and graphics.
Results
Patient Characteristics
A total of 63 patients with sepsis combined with pneumonia were studied, of whom 44 patients survived (17 women and 27 men) and 19 patients died (4 women and 17 men). In the surviving group, the age ranged from 9 to 92 years old, with an Apache II score of 10–40. Forty-three (68.25%) of them had underlying disease, including 6 (9.52%) with autoimmune disease without diabetes, 13 (20.63%) with diabetes, and 14 (22.22%) with neoplasm. There were 6 (9.52%) patients with previous immunosuppressive use and 26 (41.27%) patients with current immunosuppressive use. Patients in the death group ranged in age from 42 to 93 years old with an Apache II score of 17–50, of whom 17 (26.98%) had underlying disease and 2 (3.17%) were clearly without underlying disease. No autoimmune disease other than diabetes in death group. Diabetic mellitus in 6 (9.52%), and tumors in 5 (7.94%). There were 3 (4.76%) patients with nonsevere pneumonia with previous immunosuppression and 10 (15.87%) patients with current immunosuppression (Table 1).
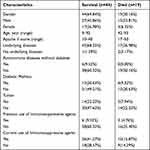 |
Table 1 Comparison of Underlying Clinical Data in Patients Surviving and Dying from Sepsis Combined with Pulmonary Infection |
Distribution of Pathogens in the Survival and Died Disease Groups Detected by mNGS and CMT
The spectrum of mNGS pathogens was very broad. We tested samples from 63 patients and found more bacterial species than viruses and fungi (Figure 2). A total of 141 strains of bacteria and viruses were detected in the 63 patients enrolled, including 49 pathogens, namely, bacteria (32 species), fungi (10 species), and viruses (7 species). mNGS and CMT together detected 49 pathogens, and mNGS alone detected 48 pathogens (Figure 2). mNGS identified 48 dominant species, including 31 of the 5 samples of bacteria, 10 fungi, and 7 viruses, and CMT identified a total of 11 dominant species, including 10 bacteria, 1 fungi, and 0 viruses in 63 samples (Figure 2). CMT was much less able to detect pathogens than mNGS (11 vs 48).
 |
Figure 2 The pathogens by mNGS and CMT based on the final diagnosis between survival and died severe pneumonia patients with sepsis. |
Among the microorganisms isolated by mNGS and CMT methods, Klebsiella pneumoniae, Acinetobacter baumannii, Stenotrophomonas maltophilia, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus pneumoniae and Enterococcus faecium were the dominant bacteria detected by both methods. Candida albicans and Candida tropicalis were the dominant fungi detected by both methods. No viral pathogens were detected by CMT, and the seven viruses detected were all detected by mNGS (Figure 2).
Among the results of mNGS detection in the surviving patient group, Klebsiella pneumoniae, Acinetobacter baumannii, Stenotrophomonas maltophilia, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus pneumoniae, Enterococcus faecium, Escherichia coli, Legionella pneumophila, Serratia marcescens, and Haemophilus influenzae were the most commonly emerged bacteria, with 31.25% (15/48), 14.58% (7/48), 16.67% (8/48), 12.5% (6/48), 10.42% (5/48), 12.5% (6/48), 10.42% (5/48), 6.25% (3/48), 4.16% (2/48), 8.331% (4/48), and 6.25% (3/48), respectively. Similarly, Candida albicans was the most frequently detected fungus in this group of patients, at 8.331% (4/48). Thirteen patients tested positive for viruses, with human betaherpesvirus 1 being the most frequently detected virus at 8.331% (4/48) and human alpha herpesvirus 5 and human betaherpesvirus 7 both at 6.25% (3/48). In the deceased patient group mNGS test results, Klebsiella pneumoniae, Acinetobacter baumannii, Stenotrophomonas maltophilia, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus pneumoniae, and Enterococcus faecium were the most commonly detected bacteria, with 63.64% (7/11), 63.64% (7/11), 63.64% (7/11), 27.27% (3/11), 27.27% (3/11), 18.18% (2/11), and 27.27% (3/11), respectively. Candida albicans was the most frequently detected fungus at 36.36% (4/11). Twelve patients tested positive for viruses, and the most frequently detected viruses were human betaherpesvirus 5 (18.75%, 4/11), human alpha herpesvirus 1, and human betaherpesvirus 7 with (6.25%, 2/11) and (6.25%, 3/11) (Figure 2).
Comparison of the Diagnostic Efficacy of mNGS and CMT
A total of 58 positive results were obtained from 63 included patient samples tested by the mNGS method, and 35 positive results were obtained from samples tested by CMT; the percentage of mNGS-positive samples was significantly higher than that of CMT-positive samples, and the sensitivity and specificity of mNGS for diagnosis were 0.906 (0.801–0.961, 58/64) and 0.069 (0.021–0.168, 4/61), respectively, while the positive predictive value (PPV) and negative predictive value (NPV) were 50.4% (58/115) and 40% (4/10), respectively (Table S2). The sensitivity and specificity of CMT for diagnosis were 0.389 (0.29–0.498, 35/90) and 0.25 (0.045–0.644, 2/8), while the positive predictive value (PPV) and negative predictive value (NPV) were 85.37% (35/41) and 3.75% (2/57), respectively (Table S2). According to the mNGS test results, a total of 41 patients (65.1%) were adjusted for anti-infective treatment (Table S3).
Univariate Analysis of Factors Affecting Death in Patients with Sepsis Combined with Pneumonia
In patients who died from sepsis combined with pneumonia, we analyzed the association between patient information, medical history, disease and severity assessment scores and biochemical indicators and patient death using univariate analysis. The results showed no statistically significant association between patient age and gender, as well as the presence of underlying diseases, tumors and diabetes, and the risk of death. Patients’ ApacheII, had a statistically significant P value of 0.005. Among the biochemical indicators, there were no statistically significant differences in ALB, ALT, AST, Cr, CRP, cTnI, LDH, LY, LY%, MYO, NEUT, NEUT%, NT-proBNP, PaO2/FiO2, PCT, RBC, and WBC. The differences in LAC, P, and PT of patients were statistically significant (all P<0.05) (Table 2).
 |
Table 2 Comparative Analysis of Clinical Factors in Patients Who Died of Sepsis Combined with Pulmonary Infection (Univariate) |
Multifactor Logistic Regression Analysis Affecting Mortality in Patients with Sepsis Combined with Pneumonia
Whether the patients with sepsis combined with pneumonia died was used as the dependent variable, and the indicators significantly associated in the univariate analysis were included as independent variables in a multifactorial unconditional logistic regression model analysis. The results showed that LAC, P, and PT were not significantly different in patients who died of sepsis combined with pneumonia, and ApacheII was statistically significant (P<0. 05) (Table 3).
 |
Table 3 Comparative Analysis of Clinical Factors in Patients Dying from Sepsis Combined with Pulmonary Infection (Multifactorial) |
Further, the results of ROC curve analysis of the predictive value of ApacheII score on the risk of death in patients with sepsis showed that the AUC value of APACHEII score for predicting the risk of death in patients with sepsis was 0.740 (Figure 3).
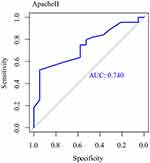 |
Figure 3 ROC curve analysis of the predictive value of ApacheII score on the risk of death in patients with sepsis. |
Discussion
Sepsis is a common complication of severe infection, trauma, and postsurgical major surgery that can progress to severe disease and cause multiple organ dysfunction or even failure, resulting in a poor prognosis and a high morbidity and mortality rate for patients.16 Timely diagnosis and identification of the causative agent of infection in patients and clarification of its pathogenesis are the keys to controlling the disease. Blood culture is the “gold standard” for the clinical diagnosis of bloodstream infection, but the test is time-consuming and easily contaminated during the culture process, which affects the accuracy of the results and causes patients to miss the best treatment time, thus affecting the diagnosis and control of patients’ conditions.8,17,18
The results of this study showed that 141 strains of pathogenic bacteria were isolated from 63 samples of suspected infection sites of patients with sepsis combined with pneumonia, among which Klebsiella pneumoniae, Acinetobacter baumannii, Stenotrophomonas maltophilia, Pseudomonas aeruginosa, Staphylococcus aureus, Streptococcus pneumoniae, Enterococcus faecium, Escherichia coli, Legionella pneumophila, Serratia marcescens, and Haemophilus influenzae were predominant, which may be related to personal hygiene problems, bacterial multidrug resistance, extensive use of antibiotics, low immune function and impaired defense of the body.19–21 Moreover, our results showed that mNGS has good clinical performance than CMT. This result was consistent with several studies in mNGS for sepsis patients, Hao Wang et al reported that mNGS test performed better than traditional BC in detecting causative microorganisms in sepsis.22 In this study, 41 patients (65.1%) had their anti-infective treatment adjusted according to mNGS results. This outcome underscores the significant role played by mNGS in clinical management, enabling physicians to more accurately identify pathogenic microorganisms and make targeted adjustments to antibiotic therapy. Such personalized treatment adjustments can enhance the chances of patient recovery while reducing unnecessary antibiotic use, thereby helping to prevent the further development of antibiotic resistance. Furthermore, this finding highlights the potential benefits of mNGS in disease management, especially in the diagnosis and treatment of infectious diseases. The results of this study showed that higher ApacheII, LAC, P and PT were all risk factors affecting the mortality of patients with severe sepsis. The reason for this analysis may be that septic shock, MODS, and acute renal failure are all common complications of severe sepsis, which can accelerate the progression of the patient’s disease and lead to his or her death.23 Therefore, the occurrence of common complications can be reduced clinically by adopting targeted treatment. Chronic underlying diseases can easily lead to disorders of the body’s internal environment and damage renal function, which can induce renal failure in patients with severe sepsis24 and speed up the process of death.25 Therefore, clinics can take targeted treatment and care measures according to patients’ chronic underlying diseases to reduce the symptoms of chronic underlying diseases, thereby improving the prognosis and avoiding patient death.26,27
Klebsiella pneumoniae, Acinetobacter baumannii, and Stenotrophomonas maltophilia are predominant, and higher ApacheII, LAC, P and PT are all risk factors affecting the death of septic patients. Therefore, clinical measures can be taken accordingly. Clinical measures can be taken to prevent and treat patients with severe sepsis to improve prognosis and reduce the occurrence of death.
Nevertheless, there were some deficiencies in our study. Firstly, the limited sample size may affect the accuracy of the experimental study. Secondly, both CMT and mNGS lack unified standards to identify whether detected pathogenic microorganisms are derived from infection, colonization, or contamination. For practical applications of this technique, the subjective judgment of clinicians is still needed, which is highly dependent on clinical experience. Although consensus was reached in this study by three experienced senior physicians and was based on the clinical manifestations of patients combined with other laboratory results, subjective bias is still possible. In addition, the sample size of this study was small, and there were some limitations, which may also affect the accuracy of the results. In the future, we will consider expanding the sample size and conducting multicenter studies to obtain more accurate results.
Conclusion
In conclusion, the application of the mNGS method for testing patients with sepsis combined with pneumonia can improve the positive detection rate of pathogenic microorganisms and focus on death-related risk factors such as pathogenic species and ApacheII, LAC, P and PT.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Faix JD. Biomarkers of sepsis. Crit Rev Clin Lab Sci. 2013;50(1):23–36. doi:10.3109/10408363.2013.764490
2. Evans T. Diagnosis and management of sepsis. Clin Med. 2018;18(2):146–149. doi:10.7861/clinmedicine.18-2-146
3. Suetrong B, Walley KR. Lactic acidosis in sepsis: it’s not all anaerobic: implications for diagnosis and management. Chest. 2016;149(1):252–261. doi:10.1378/chest.15-1703
4. Moriyama K, Nishida O. Targeting cytokines, pathogen-associated molecular patterns, and damage-associated molecular patterns in sepsis via blood purification. Int J Mol Sci. 2021;22(16):8882. doi:10.3390/ijms22168882
5. Lo AH, Kee AC, Li A, Rubulotta F. Controversies in sepsis management-what is the way forward? Ann Acad Med Singap. 2020;49(9):661–668. doi:10.47102/annals-acadmedsg.202090
6. Ventetuolo CE, Levy MM. Sepsis: a clinical update. Clin J Am Soc Nephrol. 2008;3(2):571–577. doi:10.2215/CJN.01370307
7. Hiong A, Thursky KA, Teh BW, Haeusler GM, Slavin MA, Worth LJ. Sepsis following cancer surgery: the need for early recognition and standardised clinical care. Expert Rev Anti Infect Ther. 2016;14(4):425–433. doi:10.1586/14787210.2016.1154787
8. Peri AM, Harris PNA, Paterson DL. Culture-independent detection systems for bloodstream infection. Clin Microbiol Infect. 2022;28(2):195–201. doi:10.1016/j.cmi.2021.09.039
9. Long Y, Zhang Y, Gong Y, et al. Diagnosis of sepsis with cell-free DNA by next-generation sequencing technology in ICU patients. Arch Med Res. 2016;47(5):365–371. doi:10.1016/j.arcmed.2016.08.004
10. Cao XG, Zhou SS, Wang CY, Jin K, Meng HD. The diagnostic value of next-generation sequencing technology in sepsis. Front Cell Infect Microbiol. 2022;12:899508. doi:10.3389/fcimb.2022.899508
11. Sun L, Zhang S, Yang Z, et al. Clinical application and influencing factor analysis of Metagenomic Next-Generation Sequencing (mNGS) in ICU patients with sepsis. Front Cell Infect Microbiol. 2022;12:905132. doi:10.3389/fcimb.2022.905132
12. Jing C, Chen H, Liang Y, et al. Clinical evaluation of an improved metagenomic next-generation sequencing test for the diagnosis of bloodstream infections. Clin Chem. 2021;67(8):1133–1143. doi:10.1093/clinchem/hvab061
13. Chien JY, Yu CJ, Hsueh PR, Bekal S. Utility of metagenomic next-generation sequencing for etiological diagnosis of patients with sepsis in intensive care units. Microbiol Spectr. 2022;10(4):e0074622. doi:10.1128/spectrum.00746-22
14. Metlay JP, Waterer GW, Long AC, et al. Diagnosis and treatment of adults with community-acquired pneumonia. an official clinical practice guideline of the American Thoracic Society and infectious diseases Society of America. Am J Respir Crit Care Med. 2019;200(7):e45–e67. doi:10.1164/rccm.201908-1581ST
15. Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. on behalf of the working group on sepsis-related problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996;22(7):707–710. doi:10.1007/BF01709751
16. Jacobi J. Sepsis: a frequent, life-threatening syndrome. Pharmacotherapy. 2002;22(12 Pt 2):169s–181s. doi:10.1592/phco.22.18.169S.33705
17. Lee IK, Chang JP, Huang WC, Tai CH, Wu HT, Chi CH. Comparative of clinical performance between next-generation sequencing and standard blood culture diagnostic method in patients suffering from sepsis. J Microbiol Immunol Infect. 2022;55(5):845–852. doi:10.1016/j.jmii.2022.07.011
18. Dubourg G, Raoult D. Emerging methodologies for pathogen identification in positive blood culture testing. Expert Rev Mol Diagn. 2016;16(1):97–111. doi:10.1586/14737159.2016.1112274
19. Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9). doi:10.1128/JCM.00776-18
20. Yang Y, Yang Y, Chen G, et al. Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg Microbes Infect. 2021;10(1):700–709. doi:10.1080/22221751.2021.1906163
21. Panpetch W, Phuengmaung P, Hiengrach P, et al. Candida worsens Klebsiella pneumoniae induced-sepsis in a mouse model with low dose dextran sulfate solution through gut dysbiosis and enhanced inflammation. Int J Mol Sci. 2022;23(13). doi:10.3390/ijms23137050
22. Yin M, Zheng Y, Zhang L, et al. The real-life performance of metagenomic next-generation sequencing in sepsis. J Infect. 2022;84(3):418–467. doi:10.1016/j.jinf.2021.11.018
23. Rhee C, Dantes R, Epstein L, et al. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009–2014. JAMA. 2017;318(13):1241–1249. doi:10.1001/jama.2017.13836
24. Shapiro NI, Douglas IS, Brower RG, et al. Early restrictive or liberal fluid management for sepsis-induced hypotension. N Engl J Med. 2023;388(6):499–510.
25. Mainali R, Zabalawi M, Long D, et al. Dichloroacetate reverses sepsis-induced hepatic metabolic dysfunction. Elife. 2021;1:10.
26. Rhee C, Jones TM, Hamad Y, et al. Prevalence, underlying causes, and preventability of sepsis-associated mortality in US acute care hospitals. JAMA Network Open. 2019;2(2):e187571. doi:10.1001/jamanetworkopen.2018.7571
27. Alrawashdeh M, Klompas M, Simpson SQ, et al. Prevalence and outcomes of previously healthy adults among patients hospitalized with community-onset sepsis. Chest. 2022;162(1):101–110. doi:10.1016/j.chest.2022.01.016
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
