Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 16
Pancreatic Cancer-Derived Exosomal miR-Let-7b-5p Stimulates Insulin Resistance in Skeletal Muscle Cells Through RNF20/STAT3/FOXO1 Axis Regulation
Authors Wang L , Li X, Wu J, Tang Q
Received 4 August 2023
Accepted for publication 28 September 2023
Published 9 October 2023 Volume 2023:16 Pages 3133—3145
DOI https://doi.org/10.2147/DMSO.S430443
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Konstantinos Tziomalos
Lantian Wang,1,2 Xiawei Li,1,2 Jian Wu,1,2 Qiang Tang1,2
1Department of Surgery, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, People’s Republic of China; 2Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education, Cancer Institute, Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, People’s Republic of China
Correspondence: Lantian Wang, Department of Surgery, Second Affiliated Hospital, Zhejiang University School of Medicine, 88 Jiefang Road, Hangzhou, 310009, People’s Republic of China, Email [email protected]
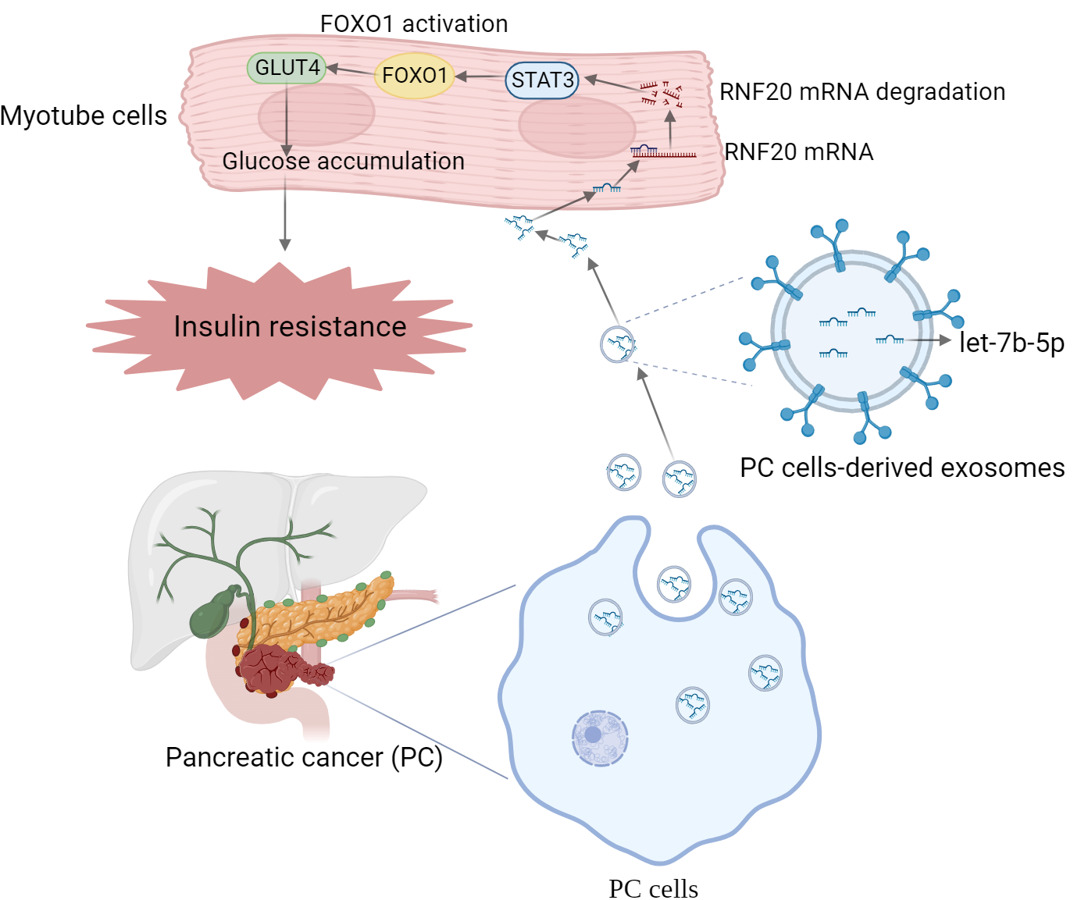
Background: Cancers trigger systemic metabolic disorders usually associated with glucose intolerance, which is an initially apparent phenomenon. One of the features of pancreatic cancer (PC) metabolic reprogramming is the crosstalk between PC and peripheral tissues (skeletal muscle and adipose tissues), emphasized by insulin resistance (IR). Our previous study reported that mice pancreatic cancer-derived exosomes could induce skeletal muscle cells (C2C12) IR, and exosomal microRNAs (miRNAs) may exert an important effect. However, the underlying mechanism remains to be further elucidated.
Methods: qPCR was used to determine the expression of let-7b-5p in normal pancreatic islet cells and PC cells. Exosomes were purified from PC cell culture medium by ultracentrifugation. The role let-7b-5p on IR-mediated by PC cells-derived exosomes was asses by Oil Red O staining using miRNA inhibitor. Western blot assay was performed to examine the expression of IR-related genes and the activation of signaling pathways. A Luciferase experiment was applied to confirm how let-7b-5p regulated the expression of RNF20. IP/WB analysis further determined whether RNF20 promoted STAT3 ubiquitination. Rescue experiment using RNF20 overexpression plasmid was performed to confirm the role of RNF20 on IR-mediated using PC cell-derived exosomes in C2C12 myotube cells.
Results: miRNA-let-7b-5p was identified as the key exosomal miRNA, which could promote the IR in C2C12 myotube cells supported the lipid accumulation, the activation of STAT3/FOXO1 axis, and the decreased expression of IRS-1 and GLUT4. RNF20, an E3 ubiquitin ligase, was confirmed as the target gene of let-7b-5p and was found to improve IR by downregulating STAT3 protein expression via ubiquitination-mediated protein degradation. The ectopic expression of RNF20 could effectively attenuate the IR mediated by the pancreatic cancer-derived exosomes in C2C12 myotube cells.
Conclusion: Our data suggest that exosomal miRNA-let-7b-5p may promote IR in C2C12 myotube cells by targeting RNF20 to activate the STAT3/FOXO1 axis.
Keywords: insulin resistance, exosome, let-7b-5p, RNF20, STAT3, FOXO1
Graphical Abstract:
Introduction
Pancreatic cancer (PC) is a morbid disease with intense invasiveness, it can be commonly triggered by multiple risk factors including smoking, diabetes, obesity, and alcohol use.1 Limited by the medical technology and conditions, early diagnosis of PC is difficult to address, which leads to a poor 5-year survival rate for PC patients that is under 9%.2 More worryingly, tumors always grow around critical arteries and metastasize to the para-aorta, as such, surgical excision is extremely challenging.3,4 Therefore, the main goals of PC research lie in looking for markers of the early stage, understanding the biological behaviors of the tumor, and developing novel therapeutic methods and targets.
Multiple clinical studies have shown that pancreatic cancer-associated new-onset diabetes mellitus (PC-DM) may be one of the early indicators of PC.5–8 The fundamental cause is “metabolic reprogramming” and “metabolic crosstalk”,9–11 characterized by PC. In other words, PC affects peripheral tissue (skeletal muscle and adipose tissues) through certain mechanisms, and results in insulin resistance (IR) in these tissues, which is the main pathological component of PC-DM and type II diabetes mellitus (T2DM).12
How PC cells exert their influence on peripheral tissues is still unknown. Our previous study reported that mice pancreatic cancer-derived exosomes could induce (C2C12) IR via the FOXO1 pathway, and exosomal microRNAs (miRNAs) may exert an important effect.13 The effects of exosomal miRNAs exhibited on tumorigenesis and development have been extensively reported. For example, it has been pointed out that the M2 polarization of macrophages can be induced by exosomal miRNA-301a derived from PC cells with hypoxia treatment, owing to the activating of PTEN/PI3Kγ pathway.14 Moreover, exosomal miRNAs also contribute to insulin sensitivity. Yu et al demonstrated that the exosomal miR-27a derived from adipocytes results in insulin resistance of skeletal muscle cells by suppressing PPARγ.15
In this study, we identify that let-7b-5p is the key exosomal miRNA from pancreatic cancer cells, and RNF20, an E3 ubiquitin ligase, is the downstream target gene. Let-7b-5p downregulates RNF20 expression, then decreases the ubiquitination of STAT3, the elevated STAT3 activates FOXO1 and finally suppresses the expression of GLUT4, which contributes to the development of IR in skeletal muscle cells.
Materials and Methods
Cell Culture and Transfection
The insulin-producing β-cell line TC-tet, the murine pancreatic cancer cell line, KPC and Pan02, and C2C12 myotube cells were purchased from American Type Culture Collection (ATCC), The cell lines above were all cultured in a 5% (v/v) CO2-included humidified incubator at 37°C, in the DMEM (Tecono, MI, USA, NO. L111-500) supplemented with 10% FBS.
Exosome Isolation
The exosomes used in this study were isolated from the KPC cell lines (KPC exosomes) and Pan02 cell lines (Pan02 exosomes) by ultracentrifugation. Briefly, When the above pancreatic cancer cells were confluent to 80%, DMEM was replaced with an exosome-free 10% FBS medium, and the supernatant was collected after 48 h for isolation. The details were referred to the previous description.16 Finally, the exosome pellet was resuspended with 1×PBS after centrifugation for use.
Oil Red O Staining
C2C12 myotube cells were cultured with KPC exosomes or Pan02 exosomes, and then fresh Oil red O was used to examine lipidosis for 10 mins after different treatments for 24h incubation. Finally, the staining results were observed under a light microscope.
Western Blotting
Lysing C2C12 myotube cells with RIPA lysis buffer with 1% proteinase inhibitor (Bimake, TX, USA, NO. B14002) included. And then BCA assay was performed to determine the concentrations of protein, separated by electrophoresis on SDS–PAGE gels (8% or 10%), shifted to PVDF membranes, proteins were incubated for 2 h with 5% skimmed milk at room temperature. After the primary antibodies were co-incubated overnight at a 4°C temperature, the corresponding HPR-conjugated secondary antibody was incubated for another 2 h. The signals were obtained and subsequently reacted with HRP substrate. The information for all antibodies is shown in Table 1
 |
Table 1 Information for Antibodies Used in This Study |
Construction and Transfection of RNF20 Overexpression Plasmid
Synthetic the cDNA coding sequence of RNF20. Digested with HindIII and EcoRI, the cDNA was subcloned into the pcDNA 3.1 vector to yield the overexpression plasmid, and the empty pcDNA 3.1 vector was used as a control. DNA sequencing was used to verify the integrity of the plasmid constructs. The plasmid and LipofectamineTM 2000 (Invitrogen, CA, USA, NO. 11668019) were mixed, left to stand for 30 mins, and then transfection was performed both at 37°C.
Construction of FOXO1-Silenced C2C12 Cell Lines
The siRNAs targeting FOXO1 (siR-FOXO1) and corresponding control siRNAs (siR-NC) were purchased from Tsingke Biotech (Beijing, China). The C2C12 cells (80% confluence) were transfected with siR-FOXO1 and siR-NC using LipofectamineTM 2000 at 37°C, respectively.
RNA Extract and qRT-PCR
TRIzol (Invitrogen, CA, USA, NO.15596026) was used to extract total RNA from the cell samples and the exosome samples. Then, cDNA synthesis was performed by using a cDNA Synthesis Kit (Vazyme Biotech, Nanjing, China). qRT-PCR was implemented using SYBR Real-time PCR I kit (Takara, Japan, NO. RR420L) on the machine of ABI-7500 (Applied Biosystems, Foster City, USA). The primer sequences were as follows:
let-7b-5p forward:5’-TATCCAGTGCAGGGTCCGAG-3’,
let-7b-5p reverse:5’-CATGCTGAGGTAGTAGGTTGT-3’;
STAT3 forward: 5’-ACCTTTGAGACCGAGGTGTA-3’,
STAT3 reverse: 5’-CACCAGGTCCCAAGAGTTTC-3’;
RNF20 forward:5’- CCAGGTTCGCAAGGAGTATGAG-3’,
RNF20 reverse:5’- GGCTGCTAATGAGATGCCGCAT-3’;
Finally, the relative gene expression was analyzed using the 2−ΔΔCT method.
IP/WB Assay for STAT3 Ubiquitination
An immunoprecipitation (IP) experiment was performed using Protein A/G Agarose beads (Pierce Biotechnology) according to the manufacturer’s instructions. In brief, lysis buffer (Pierce Biotechnology) supplemented with 1% protease inhibitor cocktail (Sigma) was used to lyse cells on ice for 15 min to obtain protein. After the protein concentration was determined, 1 μg STAT3 antibody was added to 800 μg protein per IP reaction, and incubated in a rotator at 4°C overnight. Then, 10 μL Protein A/G Agarose beads were added into each reaction, and incubated in rotator at 4°C for another 4 h. After incubation, the beads were washed three times in cold lysis buffer and centrifuged 1000 rpm at 4°C for 5 min. Before the WB assay for detecting STAT3 ubiquitination, 1× sodium dodecyl sulfate (SDS)-containing buffer was added into beads and boiled for 10 min to denature the protein.
Dual-Luciferase Reporter
To detect the relationship of targeting relationship between let-7b-5p and RNF20, a dual-luciferase reporter assay was applied. Mutant sequences were constructed on the predicted target binding site of RNF20 to the SRC gene. Briefly, the cDNA fragments of RNF20 containing the let-7b-5p targeted site were inserted into pGLO vectors (Promega, Madison, WI, USA), treated with NC mimics, and let-7b-5p, the C2C12 myotube cells were co-incubated with pGLO vectors, pGLO vectors+ RNF20 WT or pGLO vectors+RNF20 MUT for 24 h. After being collected and lysed, the relative luciferase activity was expressed as the ratio of Firefly luciferase activity to Renilla luciferase activity.
Statistical Analysis
Results are expressed as the means ± standard error. Analyses between different groups were performed using the two-tailed Student’s t-test or one-way analysis of variance. Statistical significance was set as follows: a P value of 0.05 was significant, and a value of 0.01 was highly significant.
Results
PC Cells-Derived Exosomal Let-7b-5p Induced IR in Myotube Cells
We previously reported that miRNAs of mice PC cell-derived exosomes induce IR of skeletal muscle cells but the key miRNA is unknown. In this study, we found that the expression levels of let-7b-5p in PC cell lines (KPC and Pan02) were significantly higher than that in insulin-producing β-cell line TC-tet (Figure 1A). Besides, we confirmed that let-7b-5p was expressed in PC cell-derived exosomes (Figure 1B and C). This showed us that let-7b-5p may be the key exosomal miRNA that contributes to insulin resistance in skeletal muscle cells. To further verify whether let-7b-5p was involved in PC cells-derived exosome-induced IR in skeletal muscle cells, a miRNA inhibitor was included. The result revealed that the relative expression of let-7b-5p was significantly reduced in C2C12 myotube cells after 48h transfection with let-7b-5p inhibitor (Figure 1D). In addition, insulin resistance occurs with the hydrolysis of triglycerides, which causes an increase in plasma-free fatty acid levels.17 Oil Red O staining was used to detect the level of lipidation in C2C12 myotube cells, and the results were as we hypothesized, a large number of lipid particles were observed in cells treated with exosomes from both KPC and Pan02 cells, while let-7b-5p inhibitor significantly reversed the above phenomenon (Figure 1E). These data suggested that PC cell-derived exosomes, precisely, the exosomal let-7b-5p, are closely associated with insulin resistance in skeletal muscle cells.
Exosomal Let-7b-5p Exerted Its Role on IR by Activating the STAT3/FOXO1 Pathway
The expression of some proteins, including IRS-1, GLUT4, STAT3, p-STAT3, and FOXO1, which play a key role in glucolipid metabolism and insulin activation, were detected by Western blotting. As illustrated in Figure 2A and B, as a receptor substrate for insulin, there was a significant decrease in IRS-1 expression in C2C12 myotube cells after exosome treatment, and let-7b-5p inhibitor invalidated the inhibitory effect of exosomes on IRS-1. The expression of GLUT4, a protein related to sugar translocation, varied similarly to that of IRS-1, which was significantly reduced after exosome treatment, but let-7b-5p inhibitor again significantly upregulated its protein expression. Western blotting results showed a striking elevation of FOXO1 expression in the nucleus upon exosome treatment of C2C12 myotube cells, and not surprisingly, let-7b-5p inhibitor still reversed the exosome-induced increase in FOXO1 expression. For STAT3 and p-STAT3, both KPC exosomes and Pan02 exosomes treatment caused an increase in their expression. Further, we examined the expression levels of STAT3 mRNA after KPC exosomes and Pan02 exosomes treatment by qPCR, and interestingly, exosome treatment had no significant effect on STAT3 mRNA expression (Figure 2C and D). Our data indicated that PC cell-derived exosome might induce IR in C2C12 myotube cells by activating STAT3/FOXO3 axis via let-7b-5p, which upregulated STAT3 expression at the post-transcriptional level.
RNF20 was Identified as the Target Gene of Let-7b-5p in C1C12 Myotube Cells and Contributed to the STAT3 Protein Degradation
We have already observed abnormal expression of STAT3 protein in C2C12 myotube cells after exosome treatment. Next, we tried to further illustrate the underlying mechanisms by which let-7b-5p influenced the STAT3 protein stability. By using the database of TargetScan, we found an E3 ubiquitin ligase RNF20 is a potential target gene of let-7b-5p, and the most important is RNF20 has been reported that played critical roles in the control of white and brown adipose tissue development18 and physiological function of islet β-cell.19 In the present study, we first examined the role of exosome on RFN20 expression, and whether let-7b-5p could attenuate this function. As shown in Figure 3A–D, both KPC exosomes and Pan02 exosomes treatment down-regulated the protein expression level of RNF20 in C2C12 myotube cells, and however, let-7b-5p inhibitor significantly recovered the inhibited RNF20 expression. Similar results were observed in the data of qPCR, in which RNF20 mRNA expression was inhibited by PC cell-derived exosomes and recovered by the let-7b-5p inhibitor (Figure 3E and F).
Subsequently, we transfected C2C12 myotube cells with let-7b-5p mimics and examined the expression of RNF20. As shown in Figure 4A–C, overexpression of let-7b-5p effectively inhibited the mRNA and protein expression of RNF20. These findings further supported that RNF20 was a target gene of let-7b-5p. Next, we constructed the luciferase reporter plasmids containing wildtype or mutant binding site of let-7b-5p on RNF20 3’UTR (Figure 4D) to confirm the potential binding site. As shown in Figure 4E, let-7b-5p significantly suppressed the luciferase activity of the wild-type plasmid, while failing in the mutant one. Finally, an RNF20 overexpression plasmid was constructed to upregulate its expression in C2C12 myotube cells (Figure 4F and G), and more importantly, the ectopic expression of RNF20 promoted the ubiquitination of STAT3 protein and inhibited its expression, while let-7b-5p mimics exerted an opposite function (Figure 4H and I). Taken together, our data illustrated that PC cell-derived exosomes activated the STAT3 pathway upon let-7b-5p mediated STAT3 stabilization by targeting RNF20 to inhibit protein ubiquitination.
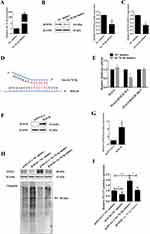 |
Figure 4 Validation of the targeting relationship between let-7b-5p and RNF20. (A) qRT-PCR assay was performed to validate the transfection efficiency of let-7b-5p mimics in C2C12 myotube cells; (B) Western blotting results of RNF20 expression in C2C12 myotube cells transfected with let-7b-5p mimics or NC for 24h, and quantification of the protein bands, and β-actin was used as a control; (C) qRT-PCR assay data RNF20 mRNA expression in C2C12 myotube cells treated as in A; (D) TargetScan Human 8.0 (https://www.targetscan.org) was used for predicting the targeting relationship and binding sites between let-7b-5p and RNF20; (E) Dual-luciferase activity reporter assay results of in C2C12 myotube cells co-transfected with let-7b-5p mimics and luciferase reporter plasmids containing wildtype or mutant RNF20 3’UTR; (F–G) Western blotting results of RFN20 overexpression (F), and quantification of protein band, β-actin was used as a control (G); (H and I) IP/WB experiment was used to detect the ubiquitination and expression of STAT3 in C2C12 myotube cells co-transfected with let-7b-5p mimics and/or RFN20 overexpression plasmid (H), and quantification of protein band, β-actin was used as a control (I). *P<0.05, **P<0.01. |
Overexpression of RNF20 Reverses PC Cells-Derived Exosome-Induced Insulin Resistance
To examine the role of RNF20 on IR, we performed oil red O staining again on C2C12 myotube cells transfected with RNF20 overexpression plasmid or co-treated with KPC exosomes or Pan02 exosomes. The results indicated that overexpression of RNF20 did reduce the lipid accumulation level of C2C12 myotube cells upregulated by exosomes (Figure 5A). Moreover, Western blotting results revealed that after exosome treatment, both KPC exosomes and Pan02 exosomes, those proteins with elevated expression, including STAT3, p-STAT3, total protein FOXO1 and nuclear FOXO1, had their expression significantly decreased after RNF20 overexpression; While IRS-1 and GLUT4, proteins with decreased expression after exosome treatment, overexpression of RNF20 also caused reversion (Figure 5B and C). Subsequently, we silenced FOXO1 for further validation. As shown in Figure 6A, the effect of FOXO1 knockdown on the lipid accumulation level of C2C12 cells was similar to that of RNF20 overexpression, and the WB data also showed us that exosome treatment-induced down-regulation of IRS-1 and GLUT4 were reversed by knocking FOXO1 down (Figure 6B). In summary, our results indicated that PC cells-derived exosomes can reduce the expression of RNF20 through let-7b-5p, thereby increasing the expression of STAT3, and activating FOXO1, which in turn inhibits GLUT4, a glucose transporter protein, leading to abnormal glucose accumulation and ultimately causing insulin resistance.
Discussion
PC is one of the most invasive carcinomas worldwide, which causes distinct symptoms of malnutrition and altered glucose homeostasis. The only curative treatments rely on surgical resection, yet the efficiency of this approach is limited by the highly aggressive behaviors and lack of inceptive diagnosis. About 90% of PC patients who lost the chance of surgery eventually show cachexia, which is a severe complication involving pathological weight loss due to the wasting of skeletal muscle and adipose tissue.20,21 The clinical syndrome also indicates a high metastasis rate, dangerous condition, poor prognosis, and low survival rate.22 During the disease progression, the severity of IR is closely related to the development of cancer cachexia.23,24 In the state of IR, the normal signal pathway is inhibited, and the proteolysis system is activated, leading to the degradation of muscle protein,10 which is the primary characteristic of cancer cachexia. However, how PC exerts its influence on peripheral tissues IR is still poorly understood.
The tumor microenvironment (TME), which consists of immune cells, stromal cells, blood vessels, and extracellular matrix, is an important target for anti-cancer therapy.25 Current research has revealed that cancer cells themselves are the most important regulators of the TME and that they act on various other components of the TME through intrinsic genetic alterations, metabolic reprogramming, and aberrant regulation of signaling, which are key determinants for tumors to shape their microenvironment.26 Exosomes are also key components of the TME and are important mediators in transmitting information.27 They also play an equally important role in the malignant progression of pancreatic cancer, for example, hypoxia regulates the miR-628-3p/BPTF axis through circPDK1 delivered by exosome in PC cells to activate c-myc, and ultimately, promotes glycolysis in PC.28
Our previous publication reported that exosomal microRNAs (miRNAs) derived from PC cells may exert an important effect on skeletal muscle cells (C2C12) IR.13 In the present study, we found that miRNA-let-7b-5p was more highly expressed in the PC cells than that in insulin-producing β-cell line TC-tet, and more importantly, the inhibitor of let-7b-5p effectively attenuated the lipid accumulation in C2C12 myoblast cells induced by the exosome treatment, which is an important biomarker of IR.29 Furthermore, the data from the Western blot experiment showed that exosomal let-7b-5p effectively suppressed the expression of IRS-1 and GLUT4, the downregulation of which are the critical indicators for IR in skeletal muscle cells.30 However, the inhibitor of let-7b-5p obviously restored the expression of these two proteins. These findings further supported let-7b-5p played a critical role in the development of IR in C2C12 myoblast cells.
Our previous data proved that FOXO1 was involved in the IR process mediated by the PC cells-derived exosome in C2C12 myoblast cells.13 Multiple signaling pathways are reported to take part in the activation of FOXO1, such as STAT3 signaling pathway.31 On the other hand, the STAT3 signaling pathway is also reported to play important roles in muscle wasting in cancer cachexia32 and in the process of IR.33 Although little is known about whether STAT3 promoted IR by activating FOXO1, our data both p-STAT3 and total FOXO1 levels, as well as the nuclear FOXO1 were up-regulated by PC cells-derived exosome in C2C12 myoblast cells, and the inhibitor of let-7b-5p can attenuate these functions. Furthermore, we found that the basal expression of STAT3 protein was also upregulated, and inhibited by the inhibitor of let-7b-5p, but its mRNA level remained generally stable These findings suggested exosomal let-7b-5p might regulate the stability of STAT3 protein. Multiple studies have reported that STAT3 can be ubiquitinated to degradation, and several E3 ubiquitin ligases were involved.34,35 Interestingly, the dataset of Targetscan indicates that RNF20, an E3 ubiquitin ligase, was a target of let-7b-5p, and our data revealed that exosomal let-7b-5p significantly decreased the expressions of mRNA and protein of RNF20, which were also reversed by its inhibitor. The data from the luciferase assay further supported RNF20 a target gene of let-7b-5p. Next, we also tried to determine whether exosomal let-7b-5p could influence the ubiquitination of STAT3. The results presented that the overexpression of RNF20 obviously increased the ubiquitination of STAT3 and decreased its protein level, while let-7b-5p mimics almost blocked this function. All these findings implied that exosomal let-7b-5p promoted STAT3 activation by inhibiting the ubiquitination and degradation of STAT3 by targeting RFN20. Finally, rescue experiments were performed by overexpressing RNF20 in C2C12 myoblast cells. The data indicated that the ectopic expression of RNF20 effectively improved IR mediated by PC cells-derived exosomal let-7b-5p in C2C12 myoblast cells. More importantly, knocking FOXO1 down by siRNAs also exerted similar reverse effects on IR induced by PC cell-derived exosomes, further demonstrating that the exosomal let-7b-5p induced IR by finally targeting to up-regulating FOXO1, at least partially.
Conclusion
This work suggested that PC cells-derived exosomal miRNA-let-7b-5p may promote IR in C2C12 myotube cells by targeting RNF20, which promoted the activation of the STAT3/FOXO1 axis. Our data paves the way for discovering potential targets for correcting metabolic disorders and improving the diagnosis/treatment of PC.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province (no. LY19H160049).
Disclosure
All authors declare that no conflict of interest could be perceived as prejudicing the impartiality of this work.
References
1. Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol. 2021;18(7):493–502. doi:10.1038/s41575-021-00457-x
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
3. Paiella S, Sandini M, Gianotti L, Butturini G, Salvia R, Bassi C. The prognostic impact of para-aortic lymph node metastasis in pancreatic cancer: a systematic review and meta-analysis. Eur J Surg Oncol. 2016;42(5):616–624. doi:10.1016/j.ejso.2016.02.003
4. Li Z, Shang H, Zhang X, Zhang H, Bao J, Hao C. Surgical treatment for locally advanced pancreatic cancer localized in the pancreatic body and tail (report of 11 cases). Int J Clin Exp Med. 2015;8(3):4676–4681.
5. Chari ST, Leibson CL, Rabe KG, et al. Pancreatic cancer-associated diabetes mellitus: prevalence and temporal association with diagnosis of cancer. Gastroenterology. 2008;134(1):95–101. doi:10.1053/j.gastro.2007.10.040
6. Pannala R, Leirness JB, Bamlet WR, Basu A, Petersen GM, Chari ST. Prevalence and clinical profile of pancreatic cancer-associated diabetes mellitus. Gastroenterology. 2008;134(4):981–987. doi:10.1053/j.gastro.2008.01.039
7. Pannala R, Basu A, Petersen GM, Chari ST. New-onset diabetes: a potential clue to the early diagnosis of pancreatic cancer. Lancet Oncol. 2009;10(1):88–95. doi:10.1016/S1470-2045(08)70337-1
8. Singh S, Singh PP, Singh AG, Murad MH, McWilliams RR, Chari ST. Anti-diabetic medications and risk of pancreatic cancer in patients with diabetes mellitus: a systematic review and meta-analysis. Am J Gastroenterol. 2013;108(4):510–519. doi:10.1038/ajg.2013.7
9. Halbrook CJ, Lyssiotis CA. Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell. 2017;31(1):5–19. doi:10.1016/j.ccell.2016.12.006
10. Porporato PE. Understanding cachexia as a cancer metabolism syndrome. Oncogenesis. 2016;5(2):e200. doi:10.1038/oncsis.2016.3
11. Wagner EF, Petruzzelli M. Cancer metabolism: a waste of insulin interference. Nature. 2015;521(7553):430–431. doi:10.1038/521430a
12. Sah RP, Nagpal SJ, Mukhopadhyay D, Chari ST. New insights into pancreatic cancer-induced paraneoplastic diabetes. Nat Rev Gastroenterol Hepatol. 2013;10(7):423–433. doi:10.1038/nrgastro.2013.49
13. Wang L, Zhang B, Zheng W, et al. Exosomes derived from pancreatic cancer cells induce insulin resistance in C2C12 myotube cells through the PI3K/Akt/FoxO1 pathway. Sci Rep. 2017;7(1):5384. doi:10.1038/s41598-017-05541-4
14. Wang X, Luo G, Zhang K, et al. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kgamma to promote pancreatic cancer metastasis. Cancer Res. 2018;78(16):4586–4598. doi:10.1158/0008-5472.CAN-17-3841
15. Yu Y, Du H, Wei S, et al. Adipocyte-derived exosomal MiR-27a induces insulin resistance in skeletal muscle through repression of PPARgamma. Theranostics. 2018;8(8):2171–2188. doi:10.7150/thno.22565
16. Crescitelli R, Lasser C, Szabo TG, et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013;2:1.
17. Yazici D, Sezer H. Insulin resistance, obesity and lipotoxicity. Adv Exp Med Biol. 2017;960:277–304.
18. Liang X, Tao C, Pan J, et al. Rnf20 deficiency in adipocyte impairs adipose tissue development and thermogenesis. Protein Cell. 2021;12(6):475–492. doi:10.1007/s13238-020-00770-2
19. Wade AK, Liu Y, Bethea MM, Toren E, Tse HM, Hunter CS. LIM-domain transcription complexes interact with ring-finger ubiquitin ligases and thereby impact islet beta-cell function. J Biol Chem. 2019;294(31):11728–11740. doi:10.1074/jbc.RA118.006985
20. Mueller TC, Burmeister MA, Bachmann J, Martignoni ME. Cachexia and pancreatic cancer: are there treatment options? World J Gastroenterol. 2014;20(28):9361–9373. doi:10.3748/wjg.v20.i28.9361
21. Tisdale MJ. Mechanisms of cancer cachexia. Physiol Rev. 2009;89(2):381–410. doi:10.1152/physrev.00016.2008
22. Fearon KC, Glass DJ, Guttridge DC. Cancer cachexia: mediators, signaling, and metabolic pathways. Cell Metab. 2012;16(2):153–166. doi:10.1016/j.cmet.2012.06.011
23. Honors MA, Kinzig KP. The role of insulin resistance in the development of muscle wasting during cancer cachexia. J Cachexia Sarcopenia Muscle. 2012;3(1):5–11. doi:10.1007/s13539-011-0051-5
24. Dugnani E, Balzano G, Pasquale V, et al. Insulin resistance is associated with the aggressiveness of pancreatic ductal carcinoma. Acta Diabetol. 2016;53(6):945–956. doi:10.1007/s00592-016-0893-6
25. Anderson NM, Simon MC. The tumor microenvironment. Curr Biol. 2020;30(16):R921–R925. doi:10.1016/j.cub.2020.06.081
26. Huang J, Zhang L, Wan D, et al. Extracellular matrix and its therapeutic potential for cancer treatment. Signal Transduct Target Ther. 2021;6(1):153. doi:10.1038/s41392-021-00544-0
27. Maacha S, Bhat AA, Jimenez L, et al. Extracellular vesicles-mediated intercellular communication: roles in the tumor microenvironment and anti-cancer drug resistance. Mol Cancer. 2019;18(1):55. doi:10.1186/s12943-019-0965-7
28. Lin J, Wang X, Zhai S, et al. Hypoxia-induced exosomal circPDK1 promotes pancreatic cancer glycolysis via c-myc activation by modulating miR-628-3p/BPTF axis and degrading BIN1. J Hematol Oncol. 2022;15(1):128. doi:10.1186/s13045-022-01348-7
29. Sun N, Shen C, Zhang L, et al. Hepatic Kruppel-like factor 16 (KLF16) targets PPARalpha to improve steatohepatitis and insulin resistance. Gut. 2021;70(11):2183–2195. doi:10.1136/gutjnl-2020-321774
30. Li B, Ye J, Liu R, et al. Programmed cell death 5 improves skeletal muscle insulin resistance by inhibiting IRS-1 ubiquitination through stabilization of MDM2. Life Sci. 2021;285:119918. doi:10.1016/j.lfs.2021.119918
31. Yao X, Takayama H, Kamoshita K, et al. Cyclosporine A downregulates selenoprotein P expression via a STAT3-FoxO1 pathway in hepatocytes in vitro. J Pharmacol Exp Ther. 2022;382:199–207. doi:10.1124/jpet.121.001175
32. Niu M, Song S, Su Z, et al. Inhibition of heat shock protein (HSP) 90 reverses signal transducer and activator of transcription (STAT) 3-mediated muscle wasting in cancer cachexia mice. Br J Pharmacol. 2021;178(22):4485–4500. doi:10.1111/bph.15625
33. Zhang L, Chen Z, Wang Y, Tweardy DJ, Mitch WE. Stat3 activation induces insulin resistance via a muscle-specific E3 ubiquitin ligase Fbxo40. Am J Physiol Endocrinol Metab. 2020;318(5):E625–E635. doi:10.1152/ajpendo.00480.2019
34. Chen W, Patel D, Jia Y, et al. MARCH8 suppresses tumor metastasis and mediates degradation of STAT3 and CD44 in breast cancer cells. Cancers. 2021;13(11):1.
35. Bhattacharjee D, Kaveti S, Jain N. APC/C CDH1 ubiquitinates STAT3 in mitosis. Int J Biochem Cell Biol. 2023;154:106333. doi:10.1016/j.biocel.2022.106333
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





