Back to Journals » Journal of Inflammation Research » Volume 16
Pan-Cancer Analysis of Oncogenic Role of RAD54L and Experimental Validation in Hepatocellular Carcinoma
Authors Zhou Y, Qiu C, Fu Q, Li T, Zhang X, Zhu C, Qin X, Wu B
Received 8 July 2023
Accepted for publication 1 September 2023
Published 11 September 2023 Volume 2023:16 Pages 3997—4017
DOI https://doi.org/10.2147/JIR.S426558
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Adam D Bachstetter
Yongzhen Zhou,1,2,* Chenjie Qiu,3,* Qingsheng Fu,2 Tao Li,1 Xudong Zhang,1 Chunfu Zhu,1 Xihu Qin,1 Baoqiang Wu1,2
1Department of General Surgery, the Affiliated Changzhou No. 2 People’s Hospital of Nanjing Medical University, Changzhou, 213000, People’s Republic of China; 2Graduate School, Bengbu Medical College, Bengbu, 233003, People’s Republic of China; 3Department of General Surgery, Changzhou Hospital of Traditional Chinese Medicine, Changzhou, 213000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Baoqiang Wu, Email [email protected]
Background: RAD54L is a prominent member of the SWI2/SNF2 protein family, primarily involved in the homologous recombination repair (HRR) process, thereby playing a pivotal role in the repair of DNA double-strand breaks (DSBs). RAD54L has been implicated in the development of numerous tumors. Consequently, we aimed to investigate the potential contribution of RAD54L in pan-cancer.
Methods: Various databases and analytical tools were employed for bioinformatics analysis. Moreover, in vitro experiments were conducted to corroborate the findings from the bioinformatics analysis and delve deeper into the role of RAD54L in hepatocellular carcinoma (HCC).
Results: RAD54L expression demonstrated a significant elevation in the majority of tumors, and its overexpression was strongly associated with unfavorable survival outcomes. RAD54L displayed robust correlations with the infiltration levels of various immune cells, including cancer associated fibroblasts (CAFs), endothelial cells, and myeloid-derived suppressor cells (MDSCs). Additionally, associations were observed between RAD54L and key factors such as tumor mutation burden (TMB), microsatellite instability (MSI), multiple immune checkpoints, and immune cell infiltration. Moreover, a close relationship was observed between RAD54L expression levels in HCC and clinicopathological characteristics, as well as immune cell infiltration. Experimental techniques including qRT-PCR, Western blotting, colony-forming, Cell Counting Kit-8 (CCK-8), wound-healing, and transwell assays were employed, which collectively demonstrated that RAD54L promoted the proliferation and migration of HCC cells.
Conclusion: RAD54L exhibits robust expression in both pan-cancer and HCC, exerting a significant influence on the proliferation and migration of HCC cells. These findings highlight its potential as a promising biomarker for pan-cancer and a prospective target for immunotherapy.
Keywords: RAD54L, pan-cancer, tumor microenvironment, malignant behavior
Introduction
RAD54L is a member of the SWI2/SNF2 chromatin remodeling protein family, which plays a crucial role in mobilizing nucleosomes and DNA-associated proteins within mammalian cells.1 Similar to other SWI2/SNF2 remodeling enzymes, RAD54L possesses the ability to translocate along DNA, inducing superhelical torsion and promoting accessibility of nucleosomal DNA.2,3 The binding of RAD54L protein significantly enhances the stability of RAD51 nucleoprotein filaments formed on single-strand DNA (ssDNA) or double-strand DNA (dsDNA), thereby facilitating DNA strand exchange.4 Furthermore, RAD54L actively promotes the formation of specific dsDNA structures that effectively bind to RAD51, facilitating the efficient repair and translocation of damaged DNA. Homologous recombination (HR), which involves the accurate repair of DNA double-strand breaks (DSBs) by utilizing intact sister chromatids as repair templates, relies heavily on the central role played by RAD54L.5–7 Together with RAD51, RAD54L constitutes essential components of the eukaryotic HR machinery. In vitro, RAD51 proteins assemble with ssDNA to form helical nucleoprotein filaments that facilitate the crucial step of DNA strand exchange during HR.4 In addition to its critical involvement in DNA damage repair (DDR), the absence of RAD54L leads to a significant increase in cell death resulting from DNA damage.8 Notably, RAD54L has been implicated in promoting the development and progression of various tumor types, including pancreatic cancer (PAAD),9 bladder cancer (BLCA),10 glioblastoma (GBM),11 and head and neck squamous cell carcinoma (HNSC),12 among others. Consequently, RAD54L emerges as a potentially important protein in tumorigenesis. However, existing studies on the role of RAD54L in tumors have primarily focused on specific cancer types, limiting our comprehensive understanding of its involvement in oncogenesis.
In recent years, numerous studies have been conducted to analyze the development of pan-cancer, aiming to uncover both the commonalities and differences among tumors. Further investigation into the gene profile of pan-cancer is crucial for a comprehensive understanding of its underlying mechanisms. In this study, we extensively searched multiple data sources, including The Cancer Genome Atlas (TCGA), Cancer Cell Line Encyclopedia (CCLE), Genotype-Tissue Expression (GTEx), and Human Protein Atlas (HPA), from which we extracted relevant data. These data were utilized to evaluate the expression pattern of RAD54L and perform correlation analyses concerning its association with prognosis, as well as its relationship with immune cell infiltration, immune checkpoints, microsatellite instability (MSI), and tumor mutational burden (TMB). Our analysis revealed that RAD54L expression exhibited alterations in the majority of tumors. Particularly noteworthy were the findings from our preliminary bioinformatics analysis, demonstrating the pronounced and prognostic significance of RAD54L expression in HCC compared to normal tissues. Building upon these observations, we designed a series of in vitro experiments to investigate the potential role of RAD54L in HCC. Through comprehensive data analysis and essential cellular assays, we evaluated the expression of RAD54L in HCC cell lines and explored its potential biological function in HCC, focusing on aspects such as proliferation and metastatic potential.
Materials and Methods
Gene Expression Analysis of RAD54L
The expression data of RAD54L in 31 normal tissues were evaluated using the GTEx database (https://www.gtexportal.org/home/index.html). To obtain information on RAD54L expression in individual tumor cell lines, we accessed the CCLE database (https://portals.broadinstitute.org/ccle) and analyzed the expression levels of RAD54L across 21 tissues based on their tissue origin. To investigate the discrepancies in RAD54L expression levels between tumor and normal tissues, we utilized the GTEx database (https://gtexportal.org/home/) and TCGA database (https://portal.gdc.cancer.gov/). Moreover, the HPA database (https://www.proteinatlas.org/) was utilized to explore the distribution of RAD54L at the subcellular level in tumors and to acquire information on the expression of RAD54L during different cell cycles.
Clinical Correlation Analysis of RAD54L
The diagnostic potential of RAD54L in pan-cancer was assessed using the receiver operating characteristic (ROC) curve analysis. The area under the curve (AUC) was calculated. A higher AUC indicates a better diagnostic performance, with values closer to 1 being more accurate. To examine the relationship between RAD54L expression and patient prognosis, univariate Cox regression analysis was conducted. Prognostic outcomes, including overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI), were evaluated. In HCC, differences in RAD54L expression among clinical subgroups were analyzed based on factors such as grade, stage, T, N, and M stages. We obtained information from the TCGA database (https://portal.gdc.cancer.gov/) for all tumor samples. Of all tumors, 377 samples were used for analysis. All samples were diagnosed as hepatocellular carcinoma. There were 55 patients in G1, 180 in G2, 124 in G3, 13 in G4, 5 in unclear Grade, 175 in Stage I, 87 in Stage II, 86 in Stage III, 5 in Stage IV, and 24 in unclear stage. The prognostic impact of RAD54L in HCC was assessed using the KMplotter, and both univariate and multivariate Cox regression analyses were performed to investigate the independent prognostic value of RAD54L.
Immune-Related and Mutation Sites Characteristics Analysis of RAD54L
The R package “ESTIMATE” was utilized to calculate the immune, stromal, and estimate scores based on the expression matrix. To investigate the relationship between RAD54L and the abundance of various immune-related cells in pan-cancer, we conducted an analysis using the TIMER database (https://cistrome.shinyapps.io/timer/). Furthermore, we extracted the expression data of chemokines, receptors, MHC genes, immunoinhibitors, and immunostimulators in pan-cancer and performed correlation analyses with RAD54L expression. In HCC, we explored the relationship between RAD54L and immune cell markers. Additionally, we downloaded TMB, MSI, homologous recombination deficiency (HRD), and loss of heterozygosity (LOH) data for each tumor from the UCSC database (https://xenabrowser.net/).
Construction of Gene-Gene and Protein-Protein Interaction and Enrichment Analysis
We employed GeneMANIA (https://genemania.org/) to construct a network comprising the 20 genes most strongly associated with RAD54L. Additionally, the RAD54L protein-protein interaction (PPI) network was constructed using the STRING database (https://www.string-db.org/). Functional enrichment analyses, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, were performed using the “Clusterprofiler” package. These analyses provided insights into the biological functions and pathways associated with RAD54L. To explore the impact of RAD54L on tumors, we categorized the samples into high and low expression groups based on RAD54L expression levels. Gene set enrichment analysis (GSEA) was then conducted according to KEGG and HALLMARK pathways, allowing us to identify pathways influenced by RAD54L expression. For HCC, we performed differential analysis using the “limma” package between high and low expression groups, with screening criteria of p < 0.05 and |logFC| > 1. The differentially expressed genes (DEGs) were further subjected to KEGG enrichment analysis to identify the pathways associated with RAD54L in HCC.
Cell Culture
The human HCC cell lines, HCC-LM3 and SMMC-7721, as well as the normal liver cell line LO2, were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RMi-1640 (Gibco, NY) or DMEM (Gibco, NY) medium supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1% penicillin-streptomycin (P/S; Gibco, USA). The cell cultures were maintained in a humidified incubator (Thermo Scientific, Waltham, MA, USA) at 37°C with 5% CO2.
Cell Transfection
Cells were seeded onto 6-well plates at a density of 2×105 cells per well and incubated in complete culture medium for 24 hours. Subsequently, cells were transfected with negative control (NC) or RAD54L-siRNAs using lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Cells were incubated for 48 hours before being used for interference efficiency assays or in vitro experiments. The sequences of the siRNAs targeting RAD54L were as follows: siRNA-RAD54L-001: 5’-GTGCTGGTGTCGAATTACA-3’; siRNA-RAD54L-002: 5’-GCACGATGTCCATTAAGAA-3’; siRNA-RAD54L-003: 5’-GTTGTAGAACGCTTCAATA-3’. The NC sequence was 5’-GGCTCTAGAAAAGCCTATGC-3’.
qRT-PCR Assay
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The extracted RNA was then reverse transcribed into cDNA using HisScript II (Vazyme, Shanghai, China). RAD54L expression was measured using the SYBR Green kit (Vazyme, Shanghai, China). The relative gene expression was calculated using the 2-∆∆Ct method. The primer sequences were as follows: RAD54L-F: 5’-TTACGCCAGAGTCCAGAGTGC-3’; RAD54L-R: 5’-TCCTCCCTCCGAGCCATTT-3’. GAPDH-F: 5’-GAACGGGAAGCTCACTGG-3’; GAPDH-R: 5’-GCCTGCTTCACCACCTTCT-3’.
Western Blotting
Proteins were extracted from HCC cells using RIPA buffer containing protease inhibitors. Then, protein concentrations were determined using a microplate reader (BioTek, Epoch, USA). Proteins were separated by sodium dodecyl sulfate (SDS)-PAGE on a 10% gel and transferred to a polyvinylidene fluoride (PVDF) membrane (Sigma-Aldrich, St. Louis, MO, USA). Incubate overnight at 4°C using anti-RAD54L (1:1000, ImmunoWay, Plano, TX, USA), β-Actin primary antibody (1:5000; 60,008-1-Lg, Proteintech). Rinse three times with TRIS -buffered saline with Tween (TBST) for 10 minutes each time. Subsequently, the secondary antibody was kept at room temperature for 1–2 hours. Finally, protein bands were photographed using a chemiluminescent gel imaging system (FluorChem Q, protein Simple, USA).
Colony-Forming Assay
The cells were seeded into 6-well plates at a density of 500 cells per well. After 14 days of incubation, the number of colony formation were examined. 75% ethanol was added, followed by the addition of crystal violet staining solution to stain the cells. Then, the number of cell colonies was photographed and counted.
Cell Counting Kit-8 (CCK-8) Assay
The cells were seeded in 96-well plates at a density of 2.5×104 cells/mL. Cell viability was assessed using the CCK-8 assay at 24, 48, 72, 96, and 120 hours. The CCK-8 mixture was prepared by combining 10 μL of CCK-8 solution (Beyotime, Shanghai, China) with 90 μL of serum-free medium. The mixture was added to the wells and incubated for 2–3 hours. The absorbance of cells at a wavelength of 450 nm was measured using the microplate reader (BioTek, Epoch, USA).
Transwell Assay
Transwell chambers (Corning Inc., Corning, NY) were employed for conducting the transwell experiments. The cells were seeded into the upper chamber. Migration of cells towards the lower chamber was induced by adding medium containing 10% serum. After 24 hours, the cells that migrated to the lower chamber were collected to evaluate their migratory capacity. To visualize the migrated cells, a 30-minute staining step was performed using a solution of crystalline violet.
Wound-Healing Assay
The cells were seeded in 6-well plates with complete medium. Once the cells reached confluency, the monolayer was carefully disrupted by gently scraping it with a sterile pipette tip. Any resulting cell debris was then removed by rinsing with phosphate-buffered saline (PBS), and serum-free medium was added. Photographic documentation of the cell morphology was performed at both 0 hour and 24 hours after the monolayer disruption.
Statistical Analysis
The statistical analysis was conducted using R software. For the comparison of two groups of continuous variables, the Wilcoxon test was employed. The Kruskal–Wallis test was utilized for comparing continuous variables among multiple groups. Correlation analysis was performed using the Spearman test. Prognostic analyses were conducted using Kaplan-Meier curves and Log rank tests. A significance threshold of p < 0.05 was applied to all statistical analyses.
Results
Gene Expression Analysis of RAD54L in Pan-Cancer
To investigate the mRNA expression of RAD54L in normal tissues, we examined its expression in physiological tissues using the GTEx. Varying levels of RAD54L expression across different types of normal tissues, with the majority exhibiting low expression levels (Figure 1A). Notably, the lowest expression was observed in the liver and pancreas, while higher expression levels were detected in bone marrow and testis. In parallel, we evaluated RAD54L expression in a panel of tumor cell lines and found it to be highly expressed in most of them (Figure 1B). We found that RAD54L was expressed at higher levels in most tumor tissues compared to normal tissues, highlighting its potential oncogenic properties and significant relevance to cancer research. Furthermore, we assessed RAD54L mRNA expression levels in both tumor and normal tissues. Our analysis demonstrated significantly higher RAD54L expression in 22 tumor types, including glioblastoma multiforme (GBM), uterine corpus endometrial carcinoma (UCEC), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), lung adenocarcinoma (LUAD), esophageal carcinoma (ESCA), Stomach and Esophageal carcinoma (STES), kidney renal papillary cell carcinoma (KIRP), Pan-kidney cohort (KICH+KIRC+KIRP)(KIPAN), colon adenocarcinoma (COAD), Colon adenocarcinoma/Rectum adenocarcinoma Esophageal carcinoma (COADREAD), prostate adenocarcinoma (PRAD), stomach adenocarcinoma (STAD), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), lung squamous cell carcinoma (LUSC), liver hepatocellular carcinoma (LIHC), bladder urothelial carcinoma (BLCA), thyroid carcinoma (THCA), rectum adenocarcinoma (READ), pheochromocytoma and paraganglioma (PCPG), and cholangiocarcinoma (CHOL) (Figure 1C). Subsequently, we conducted a pan-cancer analysis using the combined TCGA and GTEx databases. Results indicated significant variations in RAD54L expression across 30 different tumor types. Notably, RAD54L was significantly upregulated in 28 tumor types, while downregulation was observed in brain lower grade glioma (LGG) and THCA (Figure 1D). On the whole, we observed that RAD54L was upregulated in nearly all cancer types, with only a small number of tumors showing downregulation.
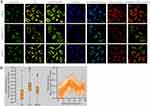
Intracellular Localization of RAD54L
We examined the subcellular localization of RAD54L using the HPA database. Specifically, we investigated its localization in A-431 (human epidermoid carcinoma cells), U-251MG (human astrocytoma cells), and U2 osteosarcoma (OS) cells. To determine the intracellular location of RAD54L, we employed immunofluorescence techniques. RAD54L co-localized with DAPI staining in A-431, U-251MG, and OS cells, indicating its presence in the nuclei (Figure 2A). Moreover, we conducted an analysis to assess the expression of RAD54L throughout the cell cycle. We observed a correlation between RAD54L expression and the cell cycle (Figure 2B). During G1, S, and G2 of the cell cycle, which were critical periods of active material metabolism, DNA replication, and synthesis of RNA and proteins, we found that RAD54L was expressed at a higher level during all of these periods. This supported that RAD54L played an important function during DNA replication. Similarly, there were regular changes in RAD54L expression inside the cell cycle time.
Identification of the Diagnostic and Prognostic Value of RAD54L in Pan-Cancer
We evaluated the diagnostic value of RAD54L through ROC curves, which provide sensitivity and specificity for various tumors. Our results demonstrated that RAD54L exhibited high accuracy in predicting the diagnosis of 24 tumor types, as indicated by the AUC values. Specifically, RAD54L showed high accuracy (AUC > 0.8) in predicting the following tumor types: Acute Lymphoblastic Leukemia (ALL) (AUC = 0.91), BLCA (AUC = 0.94), BRCA (AUC = 0.96), CESC (AUC = 1.00), CHOL (AUC = 1.00), COAD (AUC = 0.97), COADREAD (AUC = 0.96), ESCA (AUC = 0.89), GBM (AUC = 0.81), HNSC (AUC = 0.96), acute myeloid leukemia (LAML) (AUC = 0.99), LIHC (AUC = 0.91), LUAD (AUC = 0.86), LUSC (AUC = 0.98), Ovarian serous cystadenocarcinoma (OV) (AUC = 0.98), pancreatic adenocarcinoma (PAAD) (AUC = 0.97), pheochromocytoma and paraganglioma (PCPG) (AUC = 0.95), READ (AUC = 0.91), skin cutaneous melanoma (SKCM) (AUC = 0.95), STAD (AUC = 0.93), STES (AUC = 0.86), UCEC (AUC = 0.96), uterine carcinosarcoma (UCS) (AUC = 1.00), and Wilms Tumor (WT) (AUC = 1.00). On the whole, RAD54L demonstrated high accuracy (AUC > 0.8) in diagnosing 24 cancer types, and even higher accuracy (AUC > 0.9) in 20 tumor types (Figure 3).
 |
Figure 3 Receiver operating characteristic (ROC) analysis of RAD54L to distinguish tumors in pan-cancer. |
Furthermore, to assess the prognostic value of RAD54L in tumors, we conducted univariate Cox analyses for OS, DSS, DFI, and PFI. High RAD54L expression significantly predicted poor prognosis for patients with lower grade glioma and glioblastoma (GBMLGG), KIPAN, LGG, KIRC, adrenocortical carcinoma (ACC), mesothelioma (MESO), LIHC, KIRP, kidney chromophobe (KICH), PAAD, sarcoma (SARC), LUAD, and PRAD. Conversely, high RAD54L expression was associated with a protective effect on OS in patients with thymoma (THYM), STAD, and lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) (Figure 4A). Similarly, the DSS analysis revealed that RAD54L had a significant impact on the prognosis of GBMLGG, KIPAN, LGG, KIRP, KIRC, ACC, MESO, KICH, LIHC, PRAD, PAAD, SARC, and LUAD patients, while conferring a protective effect in DLBC patients (Figure 4B). The DFI analysis confirmed that high RAD54L expression was a risk factor for KIRP, LIHC, KIPAN, THCA, SARC, and BRCA (Figure 4C). Additionally, RAD54L was identified as a risk factor for GBMLGG, KIPAN, LGG, PRAD, KIRP, ACC, KICH, LIHC, KIRC, MESO, PAAD, SARC, BRCA, and uveal melanoma (UVM) patients in the PFI analysis, while being protective in STAD patients (Figure 4D).
 |
Figure 4 Univariate Cox regression analysis of RAD54L. Forest plot showing univariate Cox regression results for OS (A), DSS (B), DFI (C), and PFI (D) of RAD54L in pan-cancer. |
Immune-Related Characteristics of RAD54L in Pan-Cancer
To further explore the relationship between RAD54L expression and tumor microenvironment (TME), we utilized the ESTIMATE algorithm to evaluate the association of RAD54L expression with immune, stromal, and estimate scores. In terms of immune scores, positive correlations were observed between RAD54L expression and immune scores in KIRC and LGG, while negative correlations were found in ACC, CESC, ESCA, GBM, KIRP, LAML, LUAD, LUSC, READ, SARC, SKCM, STAD, testicular germ cell tumors (TGCT), and UCEC (Figure 5). In LGG, RAD54L expression exhibited a positive correlation with stromal score, while negative correlations were observed in ACC, BRCA, CESC, COAD, GBM, HNSC, LIHC, LUAD, LUSC, READ, SARC, SKCM, STAD, TGCT, THYM, and UCEC (Figure S1). Furthermore, RAD54L expression displayed a negative correlation with the estimate score in most tumor types (Figure S2).
 |
Figure 5 Correlation of RAD54L expression levels with immune score in pan-cancer. |
To investigate the association between RAD54L expression and immune cell infiltration in different tumors, we utilized the TIMER database. Among the various immune cell types, we focused on cancer associated fibroblasts (CAFs), endothelial cells, and myeloid-derived suppressor cells (MDSCs). The heatmap revealed that RAD54L expression was predominantly positively correlated with immune infiltration by MDSCs in most tumors, while negative correlations were observed in CAFs and endothelial cell (Figure 6A). Moreover, we examined the potential correlations between RAD54L expression and various immunomodulators genes, including chemokines, receptors, MHC molecules, immunoinhibitors, and immunostimulators, across different tumor types. In most tumors, significant correlations were observed between RAD54L expression and these genes (Figure 6B). Notably, positive correlations were detected between RAD54L expression and most checkpoint genes in LGG, UVM, and LIHC, while a negative correlation was observed in THYM. Additionally, we performed correlation analyses between RAD54L expression and TMB and MSI across various tumors. The results revealed positive correlations between RAD54L expression and TMB or MSI in several tumors such as ACC, STAD, LUAD, LUSC, SARC, HNSC, and STES, suggesting a potential association between RAD54L and immunotherapy efficacy (Figures 6C and D). However, THYM exhibited a negative correlation with TMB, and GBMLGG displayed a negative correlation with MSI and a positive correlation with TMB. Furthermore, most tumors showed a positive correlation between RAD54L expression and HRD as well as LOH (Figures 6E and F), with notable correlations observed in BRCA, LUAD, and LIHC.
Construction of PPI and Genes Interaction Network, and Enrichment Analysis
We constructed a gene-gene interaction network comprising 21 genes and analyzed their functions using the GeneMANIA database (Figure 7A). Among these genes, the five most associated genes with RAD54L were EME1, FEN1, RAD51, MAPRE1, and DMC1. Furthermore, we investigated RAD54L-interacting proteins, and conducted pathway enrichment analyses to gain insights into the molecular mechanisms underlying tumor progression. We established a PPI network consisting of 50 RAD54L-interacting proteins (Figure 7B). Subsequently, we performed functional enrichment analysis (Figures 7C-F). The results highlighted several significantly enriched pathways, including the Fanconi anemia pathway, homologous recombination, mismatch repair, and cell cycle, based on the KEGG. Analysis of biological processes (BP) indicated that RAD54L-coexpressed genes were mainly involved in DNA metabolic processes, DNA repair, and double-strand break repair. The cellular component (CC) analysis revealed enrichment of RAD54L in the nucleoplasm, nuclear lumen, and nuclear part. The molecular function (MF) analysis demonstrated that RAD54L was primarily associated with nucleic acid binding, DNA binding, and DNA-dependent ATPase activity. Subsequently, we performed GSEA to investigate potential signaling pathways related to tumors (Figures 7G-J). Results showed that high RAD54L expression was associated with pathways such as cell cycle, oocyte meiosis, and one-carbon pool by folate. High RAD54L expression was also mainly associated with E2F targets, G2M checkpoint, and mTORC1 signaling based on HALLMARK.
Clinical Characteristics of RAD54L Expression in HCC
We performed an analysis to examine the correlation between RAD54L and clinicopathological features in HCC. Our results revealed that RAD54L was significantly upregulated in HCC compared to normal tissues (Figure 8A). Furthermore, RAD54L exhibited a significant correlation with various clinical features of HCC patients (Figures 8B-F). As the grade increased, RAD54L also showed an elevation. Moreover, RAD54L expression was significantly higher in advanced stage tumors compared to early-stage tumors. Additionally, RAD54L expression demonstrated a positive correlation with T stage, although the correlation with N and M stages was not statistically significant. The Sankey diagram illustrated the distribution trend between RAD54L expression, clinicopathological features, and survival status in HCC patients (Figure 8G). KM curves indicated that high expression of RAD54L was significantly associated with a poorer prognosis in HCC, regardless of OS, DSS, Progression-free survival (PFS), and Relapse-free survival (RFS) (Figures 8H-K). Univariate and multivariate Cox regression analyses confirmed that RAD54L was an independent risk factor for HCC patients (Figures 8L and M).
To gain insights into the relationship between RAD54L expression and immune infiltration, we focused on investigating the association between RAD54L and immune cell markers. After adjusting for purity, we observed that RAD54L expression was positively correlated with most immune cell markers, including B cells, monocytes, M1 macrophages, M2 macrophages, natural killer cells, Th1 cells, Th2 cells, Treg cells, and exhausted T cells. For instance, in the M2 subtype of tumor-associated macrophages (M2-TAM) in HCC, RAD54L expression exhibited a significant correlation with immune markers such as CD163, VSIG4, and MS4A4A. Additionally, the expression of CD86 and CSF1R on monocytes, CD19 and CD79A on B cells, CD3D, CD3E, and CD2 on T cells, Treg markers CCR8 and TGFB1, and exhaustion markers PDCD1, CTLA4, and LAG3 on T cells, all displayed close correlations with RAD54L expression (Table 1).
 |
Table 1 Correlation Analysis Between Rad54L and Relate Genes and Markers of Immune Cells in TIMER |
To uncover the biological significance of RAD54L in HCC progression, we performed differential analysis between the RAD54L-low and RAD54L-high groups, and a total of 3288 upregulated genes and 235 downregulated genes were identified (Figure 8N). The heatmap displayed the top 50 upregulated and downregulated genes (Figure 8O). Furthermore, KEGG enrichment analysis revealed their involvement in pathways such as cell cycle, DNA replication, and HR, among others (Figure 8P).
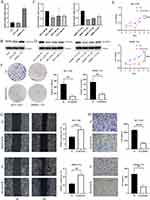
RAD54L Promoted the Proliferation and Migration of HCC Cells
We initially found a significant upregulation of RAD54L in the HCC cell lines compared to LO2 (Figure 9A). To further explore the functional implications of RAD54L in HCC, we employed siRNA-mediated knockdown of RAD54L in HCC-LM3 and SMMC-7721 cells. RAD54L-Si1 exhibited the most efficient knockdown effect and was chosen for subsequent assays (Figure 9C). Western blotting analysis of human normal hepatocytes (LO2) and HCC cells (HCC-LM3 and SMMC-7721 cells) confirmed that RAD54L was highly expressed in HCC cell lines (Figure 9B). We performed Western blotting using siRNA-mediated knockdown of RAD54L in HCC-LM3 and SMMC-7721 cells (Figure 9D). CCK-8 assays revealed a substantial decrease in the proliferative capacity of HCC cells upon RAD54L knockdown (Figure 9E). Moreover, we performed a colony-forming assay, which indicated a significant reduction in the clonogenic potential of HCC-LM3 and SMMC-7721 cells upon disturbance of RAD54L expression (Figure 9F). We also investigated the effect of RAD54L on the migratory ability of HCC cells. A significant decrease was observed in cell migration following RAD54L knockdown in HCC-LM3 and SMMC-7721 cells (Figure 9G). Similarly, we could see a notable reduction in cell migration ability after RAD54L interference (Figure 9H). Taken together, these findings underscore the critical role of RAD54L in HCC.
Discussion
RAD54L belongs to the SWI2/SNF2 protein family, which comprises DNA-dependent ATPases.13 It has been demonstrated to interact with RAD51 recombinase and participate in the repair of DSBs through the process of HR.14 HR is a critical pathway for DNA double-strand break (DSB) repair, and proteins involved in HR play a crucial role in maintaining genomic stability, suppressing tumor development, and repairing DSBs and damaged cross-replication.15 Both endogenous (spontaneous) and exogenous (environmental) DNA damage significantly contribute to tumorigenesis and progression, as they can promote genetic instability.16,17 DSBs pose a particularly serious threat to genomic stability, as the lost sequence information cannot be recovered from the same DNA molecule. To prevent errors in genetic information and disruptions in essential processes like replication and transcription, cells have robust mechanisms to repair DSBs.18 When recombination is required, a series of ordered reactions initiates, and DSBs are an inevitable consequence of DNA replication. Homologous recombination repair (HRR) is essential for the repair of these single-end breaks.19 Thus, HR is a vital process, with RAD54L considered a key gene within the HRR pathway. Our GO and KEGG enrichment analyses strongly support the involvement of RAD54L in HRR and DSBs. This finding aligns with the aforementioned understanding of RAD54L’s function. RAD54L not only plays a critical role in DDR but has also been implicated in various tumors. For example, it has been identified as a potential mechanism influencing T1 stage progression in LUAD patients20 and as a therapeutic target in multiple myeloma through its involvement in HRR regulation.21 Moreover, RAD54L has been suggested as a significant factor in HCC prognosis.22 Despite these observations, a comprehensive pan-cancer analysis of RAD54L and its potential diagnostic, prognostic, and molecular mechanisms within the TME remains unexplored. Therefore, it is of paramount importance to conduct a comprehensive pan-cancer analysis to investigate the potential biological functions of RAD54L in diverse tumor types.
In this study, we conducted a comprehensive and systematic analysis of RAD54L in human tumors to elucidate its expression and prognostic significance in pan-cancer. To evaluate the diagnostic and prognostic value of RAD54L, we performed ROC curve analysis across various cancer types. These robust AUC values suggest that RAD54L exhibits high sensitivity and specificity in distinguishing tumor patients. Furthermore, we found that higher RAD54L expression was associated with shorter survival time in most tumor types. The varying prognostic implications of RAD54L across different tumor types highlight the significant heterogeneity in its function and underscore its potential as a biomarker for predicting tumor patient prognosis.23 In summary, our findings warrant further investigation into the molecular mechanisms and therapeutic potential of RAD54L in diverse cancer contexts.
Tumor development is a multifaceted and dynamic process intricately linked to the TME.24,25 Increasing evidence has highlighted the crucial role of the TME in promoting tumor drug resistance, supporting tumor progression, invasiveness, metastasis, and even the maintenance of a cancer stem-like phenotype.26 Our study investigated the involvement of RAD54L in modulating immune cell infiltration and its potential therapeutic implications in tumor treatment. One of the major mechanisms employed by tumor cells to evade immune destruction is the suppression of antitumor immune responses. The TME harbors a population of immunosuppressive cells known as MDSCs that contribute to the maintenance of tumor progression.27 Additionally, CAFs interact with tumor-infiltrating immune cells and other immune components within the TME by releasing various cytokines, thereby fostering an immunosuppressive microenvironment that facilitates immune evasion by tumor cells.28 Furthermore, endothelial cells are involved in the detachment and metabolism of tumor cells, promoting tumor formation and metastasis.29 We established a robust correlation between RAD54L and the infiltration levels of diverse immune cell types, including CAFs, endothelial cells, and MDSCs. Interestingly, we observed a negative correlation between RAD54L expression and the infiltration of CAFs and endothelial cells in certain tumor types. Conversely, MDSCs infiltration exhibited a positive correlation across almost all tumor types studied, contributing to the establishment of an immunosuppressive TME. These findings collectively suggest that RAD54L may play a direct or indirect role in tumor initiation, metastasis, and subsequent induction of immunosuppressive responses. In summary, our study highlights the intricate interplay between RAD54L and immune cell infiltration. The identified correlations between RAD54L expression and the presence of specific immune cell subsets shed light on its potential involvement in tumor generation, metastasis, and the establishment of an immunosuppressive microenvironment. Further investigations are warranted to elucidate the precise mechanisms by which RAD54L influences immune cell infiltration and to explore its therapeutic implications in tumor treatment.
We observed a significant positive correlation between RAD54L expression and the infiltration levels of various immune cell types. These findings suggest that RAD54L may play a role in tumor immunity by influencing the presence of these immune cell subsets. Furthermore, we conducted correlation analysis between RAD54L expression and immune checkpoint genes. Our results revealed a positive correlation between RAD54L and the expression of several immune checkpoint genes, such as LGG, KIRC, LIHC, UVM, MESO, and others. This suggests that RAD54L may be involved in immune escape mechanisms in human tumor immunotherapy. These findings provide predictive evidence for the potential use of RAD54L as a robust biomarker for immune checkpoint blockade therapy in the future. In recent years, studies have increasingly demonstrated that TMB and MSI are predictive biomarkers for immunotherapy and can predict the response to immune checkpoint inhibitors.30–32 Our results showed a positive correlation between RAD54L expression and TMB or MSI in various tumor types. This indicates that RAD54L expression levels may impact TMB and MSI in tumors, consequently influencing the response of patients to immune checkpoint inhibition therapy. Based on these compelling findings, we propose that RAD54L may exert a direct or indirect effect on tumor immunity, potentially leading to the subsequent induction of an immunosuppressive response.
Through our comprehensive bioinformatics analysis, we discovered that RAD54L exhibited higher expression levels and close association with clinicopathological features in HCC. Overexpression of RAD54L in HCC tissues was closely correlated with clinicopathological features. Analysis of clinical data showed that RAD54L expression increased with tumor stage and was associated with poor OS. Therefore, our study aimed to elucidate the role of RAD54L in the development of HCC. Previous studies have reported increased RAD54L expression in HCC tissues using Western blotting and quantitative immunohistochemistry (IHC) analyses, further supporting its oncogenic role.33 However, the functional implications of RAD54L in HCC cells have not yet been validated through in vitro experiments. Therefore, our subsequent focus was to conduct in vitro experiments. Subsequently, we verified the elevated expression of RAD54L in HCC cell lines using Western blotting, functional effects in HCC cells. As anticipated, our experimental findings corroborated the bioinformatics analysis and confirmed that RAD54L may promote HCC cell proliferation and metastasis. In summary, we found that RAD54L, a representative gene of HRR in HCC, was upregulated in tumors. RAD54L was also associated with poor prognosis and can be used as a marker to predict HCC progression.
In summary, our initial pan-cancer analysis of RAD54L revealed its differential expression in tumor and normal tissues, with high RAD54L expression correlating with poor prognosis in patients. Additionally, our investigation into the clinical features of RAD54L in HCC, along with validation from in vitro assays, confirmed its upregulation in HCC cell lines. Our results suggest that RAD54L can serve as an independent prognostic factor in various tumors, with the level of its expression contributing to distinct prognostic outcomes in different tumor types. Further exploration of the specific role of RAD54L in each tumor is warranted. However, it is important to acknowledge that these results are based on data analyses from various databases, introducing potential systematic bias. Therefore, our future plans involve collecting tissue samples for validation and conducting prospective experimental studies to delve deeper into the mechanistic aspects and to continue unraveling the specific role of RAD54L in HCC.
Conclusion
The high expression of RAD54L was found in most tumors, and was closely associated with poor prognosis. It could provide an important reference value for tumor diagnosis and prognosis.
Abbreviation
ACC, Adrenocortical carcinoma; ALL, Acute Lymphoblastic Leukemia; AUC, Area under the curve; BLCA, Bladder cancer; BLCA, Bladder urothelial carcinoma; BP, Biological processes; BRCA, Breast invasive carcinoma; CAFs, Cancer associated fibroblasts; CC, Cellular component; CCK-8, Cell Counting Kit-8; CCLE, Cancer Cell Line Encyclopedia; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangiocarcinoma; COAD, Colon adenocarcinoma; COADREAD, Colon adenocarcinoma/Rectum adenocarcinoma Esophageal carcinoma; DDR, DNA damage repair; DEGs, Differentially expressed genes; DFI, Disease-free interval; DLBC, Lymphoid neoplasm diffuse large B-cell lymphoma; DSB, DNA double-strand break; DSBs, DNA double-strand breaks; dsDNA, Double-strand DNA; DSS, Disease-specific survival; ER, Endoplasmic reticulum; ESCA, Esophageal carcinoma; FBS, Fetal bovine serum; GBM, Glioblastoma multiforme; GBMLGG, Lower grade glioma and glioblastoma; GO, Gene Ontology; GSEA, Gene set enrichment analysis; GSEA, Gene set enrichment analysis; GTEx, Genotype-Tissue Expression; HCC, Hepatocellular carcinoma; HNSC, Head and neck squamous cell carcinoma; HPA, Human Protein Atlas; HR, Homologous recombination; HRD, Homologous recombination deficiency; HRR, Homologous recombination repair; IHC, Immunohistochemistry; KEGG, Kyoto Encyclopedia of Genes and Genomes; KICH, Kidney chromophobe; KIPAN, Pan-kidney cohort; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LAML, Acute myeloid leukemia; LGG, Brain lower grade glioma; LIHC, Liver hepatocellular carcinoma; LOH, Loss of heterozygosity; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; M2-TAM, M2 subtype of tumor-associated macrophages; MDSCs, Myeloid-derived suppressor cells; MESO, Mesothelioma; MF, Molecular function; MSI, Microsatellite instability; NC, Negative control; OS, Overall survival; OV, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic cancer; PAAD, Pancreatic adenocarcinoma; PBS, Phosphate-buffered saline; PCPG, Pheochromocytoma and paraganglioma; PCPG, Pheochromocytoma and paraganglioma; PFI, Progression-free interval; PFS, Progression-free survival; PPI, Protein-protein interaction; PRAD, Prostate adenocarcinoma; READ, Rectum adenocarcinoma; RFS, Relapse-free survival; ROC, Receiver operating characteristic; SARC, Sarcoma; SKCM, Skin cutaneous melanoma; ssDNA, Single-strand DNA; STAD, Stomach adenocarcinoma; STES, Stomach and Esophageal carcinoma; TCGA, The Cancer Genome Atlas; TGCT, Testicular germ cell tumors; THCA, Thyroid carcinoma; THYM, Thymoma; TMB, Tumor mutation burden; TME, Tumor microenvironment; UCEC, Uterine corpus endometrial carcinoma; UCS, Uterine carcinosarcoma; UVM, Uveal melanoma; WT, Wilms Tumor.
Data Sharing Statement
The original data can be provided by the corresponding author upon reasonable request.
Ethical Approval
The data of patients are from public database. Ethical approval is not applicable for this article. This article does not contain any studies with human or animal subjects.
Acknowledgments
We thank the relevant websites mentioned in the article for providing the platform and meaningful datasets. Sincere thanks to The Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University central laboratory and all participants involved in this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
Open access funding supported and organized by the Wu Jieping Medical Foundation (No. 320.6750.2021-16-23).
Disclosure
The authors have no relevant financial or non-financial interests to disclose for this work.
References
1. Thomä NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol. 2005;12(4):350–356. doi:10.1038/nsmb919
2. Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat Struct Biol. 2003;10(3):182–186. doi:10.1038/nsb901
3. Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol Cell. 2000;6(3):563–572. doi:10.1016/s1097-2765(00)00055-1
4. Mazin AV, Alexeev AA, Kowalczykowski SC. A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J Biol Chem. 2003;278(16):14029–14036. doi:10.1074/jbc.M212779200
5. Wright WD, Heyer WD. Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D loop formation. Mol Cell. 2014;53(3):420–432. doi:10.1016/j.molcel.2013.12.027
6. Tong Y, Merino D, Nimmervoll B, et al. Cross-Species Genomics Identifies TAF12, NFYC, and RAD54L as Choroid Plexus Carcinoma Oncogenes. Cancer Cell. 2015;27(5):712–727. doi:10.1016/j.ccell.2015.04.005
7. Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi:10.1146/annurev.genet.40.110405.090451
8. Wang Y, Zhou T, Chen H, Wen S, Dao P, Chen M. 54L promotes bladder cancer progression by regulating cell cycle and cell senescence. Med Oncol. 2022;39(12):185. doi:10.1007/s12032-022-01751-7
9. Li D, Frazier M, Evans DB, et al. Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J Clin Oncol. 2006;24(11):1720–1728. doi:10.1200/JCO.2005.04.4206
10. Mun JY, Baek SW, Park WY, et al. E2F1 Promotes Progression of Bladder Cancer by Modulating RAD54L Involved in Homologous Recombination Repair. Int J Mol Sci. 2020;21(23):9025. doi:10.3390/ijms21239025
11. Li Q, Xie W, Wang N, Li C, Wang M. CDC7-dependent transcriptional regulation of RAD54L is essential for tumorigenicity and radio-resistance of glioblastoma. Transl Oncol. 2018;11(2):300–306. doi:10.1016/j.tranon.2018.01.003
12. Nathansen J, Lukiyanchuk V, Hein L, et al. Oct4 confers stemness and radioresistance to head and neck squamous cell carcinoma by regulating the homologous recombination factors PSMC3IP and RAD54L. Oncogene. 2021;40(24):4214–4228. doi:10.1038/s41388-021-01842-1
13. Flaus A, Martin DMA, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34(10):2887–2905. doi:10.1093/nar/gkl295
14. Selemenakis P, Sharma N, Uhrig ME, et al. RAD51AP1 and RAD54L Can Underpin Two Distinct RAD51-Dependent Routes of DNA Damage Repair via Homologous Recombination. Front Cell Dev Biol. 2022;10:866601. doi:10.3389/fcell.2022.866601
15. Talens F, Jalving M, Gietema JA, Van Vugt MA. Therapeutic targeting and patient selection for cancers with homologous recombination defects. Expert Opin Drug Discov. 2017;12(6):565–581. doi:10.1080/17460441.2017.1322061
16. Friedberg EC, McDaniel LD, Schultz RA. The role of endogenous and exogenous DNA damage and mutagenesis. Curr Opin Genet Dev. 2004;14(1):5–10. doi:10.1016/j.gde.2003.11.001
17. Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2016;19(1):1–9. doi:10.1038/ncb3452
18. Wright WD, Shah SS, Heyer WD. Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem. 2018;293(27):10524–10535. doi:10.1074/jbc.TM118.000372
19. Provasek VE, Mitra J, Malojirao VH, Hegde ML, Double-Strand DNA. Breaks as Pathogenic Lesions in Neurological Disorders. Int J Mol Sci. 2022;23(9):4653. doi:10.3390/ijms23094653
20. Zheng S, Yao L, Li F, et al. Homologous recombination repair rathway and RAD54L in early-stage lung adenocarcinoma. PeerJ. 2021;9:e10680. doi:10.7717/peerj.10680
21. Bong IPN, Ng CC, Othman N, Esa E. Gene expression profiling and in vitro functional studies reveal RAD54L as a potential therapeutic target in multiple myeloma. Genes Genomics. 2022;44(8):957–966. doi:10.1007/s13258-022-01272-7
22. Li C, Ding J, Mei J. Comprehensive Analysis of Epigenetic Associated Genes on Differential Gene Expression and Prognosis in Hepatocellular Carcinoma. J Environ Pathol Toxicol Oncol. 2022;41(1):27–43. doi:10.1615/JEnvironPatholToxicolOncol.2021039641
23. Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17(3):139–152. doi:10.1038/s41575-019-0229-4
24. Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting Tumor Microenvironment for Cancer Therapy. Int J Mol Sci. 2019;20(4):840. doi:10.3390/ijms20040840
25. Belli C, Trapani D, Viale G, et al. Targeting the microenvironment in solid tumors. Cancer Treat Rev. 2018;65:22–32. doi:10.1016/j.ctrv.2018.02.004
26. Hernández-Camarero P, López-Ruiz E, Marchal JA, Perán M. Cancer: a mirrored room between tumor bulk and tumor microenvironment. J Exp Clin Cancer Res. 2021;40(1):217. doi:10.1186/s13046-021-02022-5
27. De Cicco P, Ercolano G, Ianaro A. The New Era of Cancer Immunotherapy: targeting Myeloid-Derived Suppressor Cells to Overcome Immune Evasion. Front Immunol. 2020;11:1680. doi:10.3389/fimmu.2020.01680
28. Mao X, Xu J, Wang W, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. 2021;20(1):131. doi:10.1186/s12943-021-01428-1
29. Jiang Z, Zhou J, Li L, et al. Pericytes in the tumor microenvironment. Cancer Lett. 2023;556:216074. doi:10.1016/j.canlet.2023.216074
30. Palmeri M, Mehnert J, Silk AW, et al. Real-world application of tumor mutational burden-high (TMB-high) and microsatellite instability (MSI) confirms their utility as immunotherapy biomarkers. ESMO Open. 2022;7(1):100336. doi:10.1016/j.esmoop.2021.100336
31. Luchini C, Bibeau F, Ligtenberg MJL, et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol. 2019;30(8):1232–1243. doi:10.1093/annonc/mdz116
32. Rizzo A, Ricci AD, Brandi GPD, Li TMB. MSI, and Other Predictors of Response to Immune Checkpoint Inhibitors in Biliary Tract Cancer. Cancers. 2021;13(3):558. doi:10.3390/cancers13030558
33. Li H, Zhuang H, Gu T, et al. RAD54L promotes progression of hepatocellular carcinoma via the homologous recombination repair pathway. Funct Integr Genomics. 2023;23(2):128. doi:10.1007/s10142-023-01060-w
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.