Back to Journals » Infection and Drug Resistance » Volume 14
Overexpression of microRNA-340-5p Ameliorates Inflammatory Response and Intracellular Survival of Mycobacterium Tuberculosis in Alveolar Type II Cells
Authors Zhang B, Li H, Zhang J, Hang Y, Xu Y
Received 12 November 2020
Accepted for publication 13 March 2021
Published 21 April 2021 Volume 2021:14 Pages 1573—1584
DOI https://doi.org/10.2147/IDR.S291867
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Bailing Zhang,1 Honglang Li,2 Jieling Zhang,3 Yaping Hang,1 Yi Xu4
1Department of Laboratory Medicine, The Second Affiliated Hospital of Nanchang University, Nanchang, 330006, Jiangxi, People’s Republic of China; 2Department of Gastrointestinal Surgery, The Second Affiliated Hospital of Nanchang University, Nanchang, 330006, Jiangxi, People’s Republic of China; 3Department of Oncology, The Second Affiliated Hospital of Nanchang University, Nanchang, 330006, Jiangxi, People’s Republic of China; 4Department of Geriatric Medicine, People’s Hospital of Jiangxi Province, Nanchang, 330006, Jiangxi, People’s Republic of China
Correspondence: Yi Xu
Department of Geriatric Medicine, People’s Hospital of Jiangxi Province, No. 92, Aiguo Road, Donghu District, Nanchang, 330006, Jiangxi, People’s Republic of China
Tel/Fax +86-13970805275
Email [email protected]
Background: The importance of microRNAs (miRs) has been documented in infections. This study estimated the role of miR-340-5p in Mycobacterium tuberculosis (Mtb)-infected alveolar type II cells.
Methods: The microarray of GEO database was analyzed to find the differentially expressed miRs caused by Mtb infection, and miR-340-5p was selected as the research object. The effects of Mtb infection on A549 cells were studied by MTT, CFU, EdU, flow cytometry and ELISA assays. miR-340-5p expression was altered in Mtb-infected A549 cells. The downstream target of miR-340-5p was found by bioinformatics analysis and verified by the rescue experiment. The pathways regulated by miR-340-5p and its target gene were further studied.
Results: Mtb infection suppressed the activity of A549 cells and promoted the release of inflammatory factors. Mtb infection inhibited miR-340-5p expression. Overexpression of miR-340-5p enhanced the resistance of A549 cells to Mtb infection. Moreover, miR-340-5p targeted TMED7. Overexpression of TMED7 reversed the protective effect of miR-340-5p on Mtb-infected A549 cells. miR-340-5p inhibited the activation of NF-κB by targeting TMED7.
Conclusion: miR-340-5p inhibits the activation of NF-κB by targeting TMED7, thus alleviating the injury of A549 cells caused by Mtb infection. This study may offer a novel approach to Mtb infection.
Keywords: Mycobacterium tuberculosis, alveolar type II cells, microRNA-340-5p, TMED7, NF-κB
Introduction
Mycobacterium tuberculosis (Mtb) is a specific intracellular pathogen with human beings as the major hosts, which grows most rapidly in organs with high oxygen level.1 The lung represents the most common organ involved in tuberculosis infection among immune active hosts, and the lung involvement rate of active tuberculosis patients is estimated to be 79% to 87%.2 Pulmonary tuberculosis generally progresses slowly without an exact date of onset, and has an extensive spectrum of manifestations from skin positivity with negative X rays to advanced tuberculosis.3 The typical clinical characteristics of pulmonary tuberculosis involve chronic cough, expectoration, anorexia, weight loss, fever, night sweats and hemoptysis.4 Treatment for pulmonary tuberculosis is a costing and prolonged progress, usually accompanied by high incidence of drug-related toxic effects.1 Hence, developing novel and potent therapeutic targets for the treatment and intervention of pulmonary tuberculosis remains an urgent issue to be solved.
microRNAs (miRs) are accepted as regulators of gene expression at posttranscriptional level and affect considerable biological processes, including mammalian immunity.5 miRs bear potential applications as biomarkers of non-communicable and communicable diseases.6 A recent study has revealed that host immune response to Mtb is modulated by a variety of miRs, and differential expression of miRs contributes to distinguishing active tuberculosis patients from healthy individuals or latent tuberculosis patients.7 For example, miR-155 and miR-132 are implicated in the progression of pulmonary tuberculosis, thus serving as potential biomarkers for diagnosis of pulmonary tuberculosis.8 Inhibition of miR-27a or injection of miR-27a antagomir possesses resistant effects on Mtb infection in mice.9 Moreover, miR-124 played a negative regulatory role in fine-tuning inflammatory response in macrophages upon mycobacterial infection.10 miR-223 was critical for the control of tuberculosis by regulating leukocyte chemotaxis.11 miR-340-5p is located in 5q35 and embedded in the second intron of host gene RNF130, which has been demonstrated to function as a tumor suppressor of human tumors, including myeloma and diffuse large B-cell lymphoma.12,13 Intriguingly, a previous literature has pointed out that knockdown of endogenous miR-340 can prevent cells from being infected with influenza A virus, and enforced miR-340 expression promotes the replication of influenza A virus.14 However, whether miR-340-5p exerts an effect on Mtb infection remains unknown yet. We herein investigated the role of miR-340-5p in the resistance of alveolar type II cells to Mtb infection, along with its downstream mechanism, which shall provide impetus for the determination of new therapeutic targets for patients with pulmonary tuberculosis.
Materials and Methods
Cell Culture and Transfection
Human alveolar type II cell A549 (CCL-185) from American type culture collection (ATCC, Manassas, VA, USA) were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum and 100 U/mL penicillin (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) and 100 mg/mL streptomycin (Sigma-Aldrich) at 37°C and 5% CO2.
The overexpression (oe)-TMED7 plasmids, miR-340-5p mimic and their respective controls were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). All transfections were carried out with Lipofectamine 3000 (Thermo Fisher Scientific Inc., Waltham, MA, USA).
Mtb Infection
Mtb H37Rv (27294) purchased from ATCC was cultured in Middlebrook 7H9 liquid medium (BD Biosciences, Franklin Lakes, NJ, USA) containing oleic acid albumin dextrose catalase enrichment (Sigma-Aldrich) at 37°C. Mtb infection was conducted according to a previous report.15 Mtb culture (1 mL) was precipitated for 2 minutes and then resuspended in RPMI-1640 medium. Next, the suspension was whirled for 2 minutes and then ultrasonically treated in a bath ultrasound (Thomas scientific, Swedesboro, USA) for 5 minutes. After that, the top layer of suspension was allowed to stand for 5 minutes, and then the suspension was used to infect A549 cells. For infection, H37Rv and A549 cells seeded in 24-well plates were infected with Mtb at a multiplicity of infection (MOI) of 10 and further cultured to the indicated time (0, 12, 24 and 48 hours) to examine the effect of infection duration on miR-340-5p expression in cells. Cells were infected with Mtb at different MOI values (2, 4, 6, 8, 10) to examine the effect of different MOI values on miR-340-5p expression in cells.
Colony Forming Unit (CFU)
CFU analysis was adopted to evaluate the survival rate of Mtb-infected A549 cells. In short, the transfected A549 cells (1 × 104) were infected with Mtb at 10 MOI for a indicated time, and lysed with sterile distilled water. Ten-fold serial diluents were used for quantitative culture, and each diluent was seeded on the Middlebrook 7h10 agar plate containing 10% oleate albumin glucose catalase enrichment (Sigma-Aldrich). After 3 weeks of culture, Mycobacterium colony was observed, and CFU was calculated.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) Assay
MTT kit (Beyotime Biotechnology Co., Ltd., Shanghai, China) was adopted as per the protocol provided by the manufacturer for A549 cell viability assessment. A549 cells grew overnight in 96-well plates at 3×103 cells/well to an 80% confluence. After transfection and/or Mtb infection, 10 μL MTT (10 mg/mL) was added to 200 μL cell suspension for a 4-hour incubation at 37°C. The medium was aspirated, and the purple formazan crystal was dissolved by adding 100 μL dimethyl sulfoxide. After incubation in the dark at 37°C for 30 minutes, the optical density (OD) value was measured at 570 nm using a spectrophotometer (Thermo Fisher).
5-ethynyl-2ʹ-deoxyuridine (EdU) Labeling Assay
The BeyoClick EdU kit (Beyotime) was adopted as per the protocol provided by the manufacturer to detect DNA synthesis in cells. In short, 1×105 cells after transfection and/or Mtb infection were incubated with EdU (10 μM) for 3 hours, and then fixed for 15 minutes with 4% paraformaldehyde. The cells were washed with phosphate-buffered saline (PBS) for 5 minutes and treated for 15 minutes with 0.3% Triton X-100. Then cells were incubated for 30 minutes with Click Additive solution in the dark. The nuclei were stained with 4ʹ,6-diamidino-2-phenylindole. The cells were observed under a fluorescence microscope. The proportion of EdU-positive cells was counted by ImageJ.
Flow Cytometry
The cells (1 × 104) were fixed on ice with 70% ethanol and stained with 50 μg/mL propidium iodide (Thermo Fisher) at 4°C for 30 minutes. Flow cytometer (BD FACScalibur ™) and ModFit LT 3.1 software were used for cell cycle analysis.
Enzyme-Linked Immunosorbent Assay (ELISA)
The supernatant of 1×105 A549 cells after transfection and/or Mtb infection was collected for ELISA analysis of inflammatory cytokines. The concentrations of interleukin-6 (IL-6) (ab178013, Abcam, Cambridge, MA, USA) and tumor necrosis factor-α (TNF-α) (ab181421) in cell supernatant were analyzed by ELISA kits (Abcam).
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
TRIzol kit was adopted for total RNA extraction. The cDNA was synthesized using TaqMan MicroRNA reverse transcription kit (Thermo Fisher) or RevertAid RT reverse transcription kit (Thermo Fisher). RT-qPCR was performed on LightCycler 480 Real-Time PCR system (Roden Applied Sciences, Mannheim, Baden Wuerttemberg, Germany). U6 or GAPDH served as the internal reference. Quantitative expression was calculated by 2−ΔΔCT. The primers are shown in Table 1.
 |
Table 1 Primer Sequences for RT-qPCR |
Dual-Luciferase Reporter Gene Assay
The potential binding sites of miR-340-5p and TMED7 were obtained by Starbase. The wild type of TMED7 gene containing miR-340-5p binding sequence (WT-TMED7) and the mutant of the binding sites of TMED7 and miR-340-5p (MT-TMED7) were amplified by PCR and inserted into pGL3 luciferase vector (Promega, Madison, WI, USA). Then, A549 cells were co-transfected with WT-TMED7 or MT-TMED7 luciferase plasmids with NC mimic or miR-340-5p mimic by Lipofectamine 3000. The cells were harvested at 48 hours post-transfection, and the luciferase activity was measured by a dual-luciferase reporter kit (Promega), and the luciferase activity was standardized.
RNA Immunoprecipitation (RIP)
The binding relationship between miR-340-5p and TMED7 was detected by RIP kit (Merck Millipore, Billerica, MA, USA). After the cells (1 × 104) were lysed with RIP lysis buffer (Beyotime), the supernatant was incubated with magnetic beads containing anti-Ago2 (ab32381, 1:50) or IgG (ab109489, 1:100) after centrifugation. Then, protease K buffer was added to the sample. Finally, the target RNA was extracted and purified by RT-qPCR.
Western Blotting (WB)
Total protein was extracted from whole cells using RIP lysis buffer. NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher) were used to obtain soluble protein fractions in the nucleus and cytoplasm. Protein concentration was tested using the bicinchoninic acid assay kit (Thermo Fisher). Then, the protein was subjected to electrophoresis separation and transferred onto nitrocellulose membranes. The membranes were blocked with 5% bovine serum albumin for 1 hour and probed with primary antibodies to TMED7 (1:1000, ab126094, Abcam), GAPDH (1:1000, ab181603, Abcam), H3 (1:1000, ab1791, Abcam), NFκB p65 (1:1000, #8242, Cell Signaling Technology, CST), and p-NFκB p65 (Ser536) (1:1000, #3033, CST) at 4°C overnight. Afterwards, the membranes were cultured for 2 hours with the secondary horseradish peroxidase-conjugated goat anti-rabbit antibody to IgG (1:5000, ab205718, Abcam). Next, the membranes were visualized using the enhanced chemiluminescence reagent (Millipore). The image of protein blotting was analyzed by ImageJ.
Statistical Analysis
All data were processed using SPSS 22.0 (IBM Corp., Armonk, NY, USA). Data of three independent experiments were expressed in the form of mean ± standard deviation. Unpaired t test was adopted to compare the two groups. One-way or two-way analysis of variance (ANOVA) was applied for comparison among multiple groups, followed by Tukey’s multiple comparisons test. The p < 0.05 indicated the statistically significant difference.
Results
MTB Infection Inhibits miR-340-5p Expression in A549 Cells
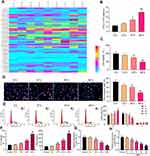
Peripheral blood mononuclear cells from six patients with active tuberculosis (GSM722294-GSM722299) and three normal volunteers (GSM722306-GSM722308) were selected from GEO database. The differentially expressed miRs were analyzed. The 50 most representative miRs are shown in the heatmap (Figure 1A). The relative expression of these genes is shown in the Supplementary Document. The miR-340-5p expression in tuberculotic patients was lower than that in normal volunteers, and miR-340-5p in pulmonary tuberculosis has not been studied.
First, A549 cells were infected with Mtb (MOI = 10). With the prolongation of infection time, the number of Mtb in the cells was increased (Figure 1B), and cell viability was decreased gradually (Figure 1C). EdU staining elicited that Mtb infection significantly prevented the DNA synthesis of A549 cells (Figure 1D). Mtb infection resulted in G1 phase arrest of A549 cells and a significant decrease in S phase (Figure 1E). In addition, the effect of Mtb infection on the concentrations of pro-inflammatory factors IL-6 and TNF-α in the supernatant of A549 cells was detected by ELISA, and cells without Mtb infection were used as controls (Figure 1F). We found that the concentrations of pro-inflammatory factors in the control group maintained a relatively stable level, while Mtb infection resulted in the increases of IL-6 and TNF-α levels. The aforementioned results exhibited that we successfully infected A549 cells with Mtb, which decreased cell viability and induced the levels of pro-inflammatory factors.
The miR-340-5p expression in A549 cells with or without Mtb infection was detected by RT-qPCR. Mtb notably inhibited miR-340-5p expression in a time-dependent manner (Figure 1G). The infection time of Mtb was selected as 48 hours for the later experiment. We then infected cells with Mtb at different MOI values (2, 4, 6, 8, 10) for 48 hours. Cells without Mtb infection were used as controls. RT-qPCR results revealed that Mtb infection inhibited the expression of miR-340-5p in cells in an MOI value-dependent manner (Figure 1H).
Overexpression of miR-340-5p Attenuates the Damage of Mtb Infection to A549 Cells
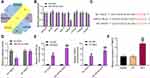
To study the effect of miR-340-5p on Mtb infection, we transfected miR-340-5p mimic and its control into A549 cells, and the effective transfection was detected after 24 hours (Figure 2A). The transfected A549 cells were infected with Mtb for 48 hours. The viability of A549 cells after miR-340-5p mimic transfection was clearly increased (Figure 2B), which occurred concomitant with repressed survival of Mtb (Figure 2C) and enhanced DNA synthesis (Figure 2D) and cell cycle entry (Figure 2E). Additionally, the increases of IL-6 and TNF-α induced by Mtb infection were significantly reversed by overexpression of miR-340-5p (Figure 2F).
miR-340-5p Targets TMED7
To probe the potential downstream of miR-340-5p in A549 cells, we predicted the target genes of miR-340-5p in the bioinformatics websites Starbase, Targetscan, RNA22 and miRWalk. There were 8 overlapping target genes (NFE2L2, SOX4, NPAT, RNF11, TMED7, PTP4A1, FRMD6, and RUNX1) (Figure 3A). The expression of these genes was detected in A549 cells transfected with NC mimic or miR-340-5p mimic. Only TMED7 was found to have significant difference (Figure 3B).
The binding sites of miR-340-5p and TMED7 were obtained and mutated to construct WT-TMED7 and MT-TMED7 plasmids (Figure 3C). These plasmids were co-transfected with NC mimic or miR-340-5p mimic into A549 cells. miR-340-5p mimic clearly inhibited the luciferase activity of WT-TMED7, but had no effect on MT-TMED7 (Figure 3D). RIP experiment further validated the binding relation between miR-340-5p and TMED7 (Figure 3E). Specifically, compared with anti-IgG, anti-Ago2 significantly enriched the expression of miR-340-5p and TMED7, and after miR-340-5p mimic transfection, the enrichment ability of both was enhanced. Finally, we detected TMED7 expression in Mtb-infected A549 cells. We found that the expression of TMED7 was significantly higher in cells after 48 h of Mtb infection than that in cells without Mtb infection (Figure 3F).
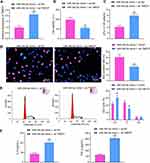
Overexpression of TMED7 Attenuates the Protective Effect of miR-340-5p Mimic on Mtb-Infected A549 Cells
To prove that TMED7 is the downstream gene of miR-340-5p in Mtb-infected A549 cells, we transfected oe-TMED7 plasmids into A549 cells overexpressing miR-340-5p, and the transfection was verified by RT-qPCR (Figure 4A). The transfected A549 cells were infected with Mtb for 48 hours. The viability of A549 cells after miR-340-5p mimic + oe-TMED7 treatment was clearly decreased (Figure 4B), along with increased survival of Mtb (Figure 4C) and weakened DNA synthesis (Figure 4D) and cell cycle entry (Figure 4E). Additionally, the IL-6 and TNF-α levels reduced by miR-340-5p overexpression were significantly reversed by oe-TMED7 (Figure 4F).
miR-340-5p Targets TMED7 to Mediate the NF-κB Pathway
miR-340-5p can inhibit inflammation in pulmonary hypertension, neuroinflammation and heart injury by inhibiting the activation of NF–κB,16–18 while TMED7 is a cis-related gene of NF-κB.19 Therefore, we speculated that miR-340-5p can reduce the Mtb infection-induced inflammatory responses by inhibiting TMED7 and NF-κB pathway.
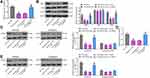
A549 cells transfected with miR-340-5p mimic or miR-340-5p mimic + oe-TMED7 were collected for Mtb infection. By detecting changes in TMED7 mRNA expression using RT-qPCR, we found that miR-340-5p mimic significantly repressed the expression of TMED7 mRNA, while the expression of TMED7 mRNA was significantly restored after transfection with oe-TMED7 (Figure 5A). WB test was conducted to examine the protein level of TMED7 and the activation of NF-κB in A549 cells transfected with miR-340-5p mimic + oe-TMED7. Mtb infection enhanced TMED7 expression and NF-κB phosphorylation. By contrast, overexpression of miR-340-5p significantly annulled the increases of TMED7 and NF-κB protein levels induced by Mtb infection, which was reversed by overexpression of TMED7 (Figure 5B) (all p < 0.05). We also performed subcellular fractionation and subjected the nuclear and cytoplasmic components to WB experiments to detect NFκB nuclear translocation (phosphorylated NFκB p65). We observed that the expression of p-NFκB in the nucleus of cells transfected with miR-340-5p mimic was significantly reduced compared to cells transfected with NC mimic, while overexpression of TMED7 significantly promoted the expression of p-NFκB in the nucleus (Figure 5C).
To demonstrate that the miR-340-5p/TMED7/NFκB axis that regulates Mtb infection still holds under the condition of a low MOI value, we infected A549 cells transfected with miR-340-5p mimic, miR-340-5p mimic + oe-TMED7 and their respective controls with Mtb at a low MOI value (4). The expression of TMED7 in cells was detected by RT-qPCR (Figure 5D). miR-340-5p mimic suppressed TMED7 expression even in cells with low MOI levels. Transfection of oe-TMED7 significantly reversed the inhibitory effect of miR-340-5p mimic on TMED7 expression. By WB detection of nuclear p-NFκB expression, we noted that overexpression of miR-340-5p still suppressed nuclear p-NFκB expression at a low MOI, while p-NFκB nuclear translocation was significantly increased after overexpression of TMED7 (Figure 5E).
Discussion
Tuberculosis remains a conspicuous public health problem worldwide and ranks the second leading cause of infectious diseases-related death in the world.20 In view of the critical function of miRs in anti-Mtb infection, differentially expressed miRs can affect the results of tuberculosis infection and become biomarkers for diagnosis and treatment of tuberculosis.7 To our knowledge, we were the first to clarify that miR-340-5p played a regulatory role in Mtb infection via the TMED7/NF-κB axis.
Mtb possesses a recognized ability to evade the host’s immune system, which makes it survive in cells for a long time.7 The multifactorial characteristics of miRs facilitate the study of their roles in complicated host reactions, including immunity against chronic bacterial infections.21 GEO database showed that miR-340-5p expression in peripheral blood mononuclear cells from tuberculosis patients was notably lower than that in healthy volunteers. Much information about the benefit of miR-340–5p in human malignancy can be obtained from previous literatures.22–24 For instance, miR-340-5p can act as a predictor for the prognosis of lung adenocarcinoma.25 miR-340-5p expression was downregulated during Hepatitis B virus infection, thus accelerating the hepatocellular carcinoma cell migration.26 Nevertheless, the role of miR-340-5p in Mtb infection has not been clarified before. To determine the function of miR-340-5p in Mtb infection, we transfected miR-340-5p mimic into A549 cells and found that miR-340-5p mimic clearly suppressed the survival of Mtb in A549 cells. Consistently, upregulated miR-340 can prevent Hepatitis B virus-induced cell growth, making it a promising therapeutic strategy for hepatocellular carcinoma.27 Susceptibility to Mtb may be due to poor regulation of the immune response, including systemic inflammation and excessive inflammation under certain conditions.28 Disease-induced systemic inflammation is characterized by upregulated levels of cytokines.29 Tuberculosis patients show elevated levels of systemic inflammation-associated markers such as IL-6 and TNF-α.30 Our ELISA results exhibited that miR-340-5p mimic could notably reverse the upregulated levels of IL-6 and TNF-α upon Mtb infection. Overexpressing miR-340-5p also reduced the levels of TNF-α and IL-6 in rats with chronic constriction injury.31 Enhanced miR-340-5p expression can inhibit the inflammatory response of pulmonary artery smooth muscle cells by declining the levels of IL-1β and IL-6.18 It was suggested that overexpression of miR-340-5p attenuated the damage of Mtb to A549 cells by mitigating inflammatory responses and reducing Mtb survival.
Subsequently, the target genes of miR-340-5p were predicted by bioinformatics website. We detected the expressions of these genes in A549 cells transfected with NC mimic or miR-340-5p mimic and found that only TMED7 showed significant difference. Dual-luciferase assay and RIP assay confirmed the binding relation between miR-340-5p and TMED7. TMED proteins are a crucial family of transmembrane adaptor proteins, and deletions of some TMEDs may result in defects in embryonic development, which can even be fatal.32 Whether TMED7 has influences on Mtb infection remains unknown yet. Our results revealed that TMED7 expression was elevated after Mtb infection. Overexpression of TMED7 attenuated the protective effects of miR-340-5p mimic on A549 cells infected with Mtb.
We then shifted to investigating the signaling pathway regulated by miR-340-5p/TMED7. Upregulated miR-340 can alleviate neuroinflammation in lipopolysaccharides-induced microglia cells via the NF-κB pathway.16 NF-κB pathway is implicated in the inhibitory effects of miR-340-5p in hypoxia/reoxygenation-induced myocyte apoptosis and oxidative stress.17 Importantly, TMED7 is reported to be the cis-related gene of NF-κB.19 We speculated that miR-340-5p reduced the Mtb infection-induced inflammatory response via TMED7/NF-κB. NF-κB is indispensable for innate and humoral immunity, but the activated NF-κB pathway in organs may trigger various diseases.33 Notably, NF-κB pathway is demonstrated to participate in the innate immune responses against Mtb infection.19 We exhibited that Mtb infection promoted NF-κB phosphorylation. Moreover, overexpression of miR-340-5p inhibited the phosphorylation and nuclear translocation of NF-κB induced by Mtb infection, which could be reversed by overexpression of TMED7. Briefly, miR-340-5p impaired the NF-κB axis by targeting TMED7. However, the detailed mechanism of TMED7 involvement in the NF-κB axis regulation awaits our further investigation.
Conclusion
To sum up, overexpression of miR-340-5p relieved A549 cell injury induced by Mtb infection via the TMED7/NF-κB axis (Figure 6). This study might hint the possibility of miR-340-5p as a potential target for patients with pulmonary tuberculosis. In the future, we shall conduct more researches on the feasibility and safety of miR-340-5p in the management of pulmonary tuberculosis, with the aim to transform miR-340-5p from a gene tool into a clinical means. Even though human alveolar type II cell A549 is a cell type with stem cell potential for epithelial injury repair and regeneration and more available than the primary cells, the altered phenotypes make it does not fully resemble primary cells. Therefore, more suitable cells should be applied in our further works.
 |
Figure 6 Mtb infection resulted in a significant decrease in miR-340-5p expression in A549 cells to promote TMED7/NFκB axis expression, causing cell cycle arrest and inflammation. |
Abbreviations
ANOVA, analysis of variance; CFU, colony forming unit; EDU, 5-ethynyl-2ʹ-deoxyuridine; ELISA, enzyme-linked immunosorbent assay; IL-6, interleukin-6; miRs, microRNAs; Mtb, Mycobacterium tuberculosis; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; oe, overexpression; PBS, phosphate-buffered saline; RIP, RNA immunoprecipitation; RT-qPCR, reverse transcription quantitative polymerase chain reaction; TNF-α, tumor necrosis factor-α; WB, Western blotting.
Data Sharing Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no competing interests.
References
1. Lawn SD, Zumla AI. Tuberculosis. Lancet. 2011;378(9785):57–72. doi:10.1016/S0140-6736(10)62173-3
2. Scott C, Kirking HL, Jeffries C, et al. Tuberculosis trends–United States, 2014. MMWR Morb Mortal Wkly Rep. 2015;64(10):265–269.
3. Lyon SM, Rossman MD. Pulmonary tuberculosis. Microbiol Spectr. 2017;5(1). doi:10.1128/microbiolspec.TNMI7-0032-2016
4. Zumla A, Raviglione M, Hafner R, von Reyn CF. Tuberculosis. N Engl J Med. 2013;368(8):745–755. doi:10.1056/NEJMra1200894
5. Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci. 2008;65(4):545–562. doi:10.1007/s00018-007-7355-9
6. Matsushima K, Isomoto H, Inoue N, et al. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int J Cancer. 2011;128(2):361–370. doi:10.1002/ijc.25348
7. Yang T, Ge B. miRNAs in immune responses to Mycobacterium tuberculosis infection. Cancer Lett. 2018;431:22–30. doi:10.1016/j.canlet.2018.05.028
8. Zheng ML, Zhou NK, Luo CH. MiRNA-155 and miRNA-132 as potential diagnostic biomarkers for pulmonary tuberculosis: a preliminary study. Microb Pathog. 2016;100:78–83. doi:10.1016/j.micpath.2016.09.005
9. Liu F, Chen J, Wang P, et al. MicroRNA-27a controls the intracellular survival of Mycobacterium tuberculosis by regulating calcium-associated autophagy. Nat Commun. 2018;9(1):4295. doi:10.1038/s41467-018-06836-4
10. Ma C, Li Y, Li M, et al. microRNA-124 negatively regulates TLR signaling in alveolar macrophages in response to mycobacterial infection. Mol Immunol. 2014;62(1):150–158. doi:10.1016/j.molimm.2014.06.014
11. Dorhoi A, Iannaccone M, Farinacci M, et al. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J Clin Invest. 2013;123(11):4836–4848. doi:10.1172/JCI67604
12. Li Z, Wong KY, Calin GA, Chng WJ, Chan GC, Chim CS. Epigenetic silencing of miR-340-5p in multiple myeloma: mechanisms and prognostic impact. Clin Epigenetics. 2019;11(1):71. doi:10.1186/s13148-019-0669-2
13. Xu Y, Liu Z, Lv L, et al. MiRNA-340-5p mediates the functional and infiltrative promotion of tumor-infiltrating CD8(+) T lymphocytes in human diffuse large B cell lymphoma. J Exp Clin Cancer Res. 2020;39(1):238. doi:10.1186/s13046-020-01752-2
14. Zhao L, Zhang X, Wu Z, et al. The downregulation of MicroRNA hsa-miR-340-5p in IAV-Infected A549 cells suppresses viral replication by targeting RIG-I and OAS2. Mol Ther Nucleic Acids. 2019;14:509–519. doi:10.1016/j.omtn.2018.12.014
15. Li X, Huang S, Yu T, et al. MiR-140 modulates the inflammatory responses of Mycobacterium tuberculosis-infected macrophages by targeting TRAF6. J Cell Mol Med. 2019;23(8):5642–5653. doi:10.1111/jcmm.14472
16. Bao Y, Zhu Y, He G, et al. Dexmedetomidine attenuates neuroinflammation In LPS-stimulated BV2 microglia cells through upregulation of miR-340. Drug Des Devel Ther. 2019;13:3465–3475. doi:10.2147/DDDT.S210511
17. Li D, Zhou J, Yang B, Yu Y. microRNA-340-5p inhibits hypoxia/reoxygenation-induced apoptosis and oxidative stress in cardiomyocytes by regulating the Act1/NF-kappaB pathway. J Cell Biochem. 2019;120(9):14618–14627. doi:10.1002/jcb.28723
18. Ou M, Zhang C, Chen J, Zhao S, Cui S, Tu J. Overexpression of MicroRNA-340-5p inhibits pulmonary arterial hypertension induced by APE by downregulating IL-1beta and IL-6. Mol Ther Nucleic Acids. 2020;21:542–554. doi:10.1016/j.omtn.2020.05.022
19. Brodtkorb M, Lingjaerde OC, Huse K, et al. Whole-genome integrative analysis reveals expression signatures predicting transformation in follicular lymphoma. Blood. 2014;123(7):1051–1054. doi:10.1182/blood-2013-07-512392
20. Singer-Leshinsky S. Pulmonary tuberculosis: improving diagnosis and management. JAAPA. 2016;29(2):20–25. doi:10.1097/01.JAA.0000476207.96819.a7
21. Iannaccone M, Dorhoi A, Kaufmann SH. Host-directed therapy of tuberculosis: what is in it for microRNA? Expert Opin Ther Targets. 2014;18(5):491–494. doi:10.1517/14728222.2014.897696
22. Kang R, Yao DF, Xu GZ, Zhou YH. The knockdown of SNHG3 inhibits the progression of laryngeal squamous cell carcinoma by miR-340-5p/YAP1 axis and Wnt/beta-catenin pathway. Neoplasma. 2020;67(5):1094–1105. doi:10.4149/neo_2020_191022N1073
23. Liu D, Lin L, Wang Y, et al. PNO1, which is negatively regulated by miR-340-5p, promotes lung adenocarcinoma progression through Notch signaling pathway. Oncogenesis. 2020;9(5):58. doi:10.1038/s41389-020-0241-0
24. Zhang Z, Liang X, Ren L, et al. LINC00662 promotes cell viability and metastasis in esophageal squamous cell carcinoma by sponging miR-340-5p and upregulating HOXB2. Thorac Cancer. 2020.
25. Tan J, Wang W, Song B, Song Y, Meng Z. Integrative analysis of three novel competing endogenous RNA biomarkers with a prognostic value in lung adenocarcinoma. Biomed Res Int. 2020;2020:2837906. doi:10.1155/2020/2837906
26. Xiong Q, Wu S, Wang J, et al. Hepatitis B virus promotes cancer cell migration by downregulating miR-340-5p expression to induce STAT3 overexpression. Cell Biosci. 2017;7:16. doi:10.1186/s13578-017-0144-8
27. Song F, Wei M, Wang J, et al. Hepatitis B virus-regulated growth of liver cancer cells occurs through the microRNA-340-5p-activating transcription factor 7-heat shock protein A member 1B axis. Cancer Sci. 2019;110(5):1633–1643. doi:10.1111/cas.14004
28. Tobin DM, Roca FJ, Oh SF, et al. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell. 2012;148(3):434–446. doi:10.1016/j.cell.2011.12.023
29. Scriba TJ, Penn-Nicholson A, Shankar S, et al. Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease. PLoS Pathog. 2017;13(11):e1006687. doi:10.1371/journal.ppat.1006687
30. Djoba Siawaya JF, Chegou NN, van den Heuvel MM, et al. Differential cytokine/chemokines and KL-6 profiles in patients with different forms of tuberculosis. Cytokine. 2009;47(2):132–136. doi:10.1016/j.cyto.2009.05.016
31. Gao L, Pu X, Huang Y, Huang J. MicroRNA-340-5p relieved chronic constriction injury-induced neuropathic pain by targeting Rap1A in rat model. Genes Genomics. 2019;41(6):713–721. doi:10.1007/s13258-019-00802-0
32. Liaunardy-Jopeace A, Bryant CE, Gay NJ. The COP II adaptor protein TMED7 is required to initiate and mediate the delivery of TLR4 to the plasma membrane. Sci Signal. 2014;7(336):ra70. doi:10.1126/scisignal.2005275
33. Wang W, Nag SA, Zhang R. Targeting the NFkappaB signaling pathways for breast cancer prevention and therapy. Curr Med Chem. 2015;22(2):264–289. doi:10.2174/0929867321666141106124315
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.