Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 14
Overexpression Of hsa-miR-664a-3p Is Associated With Cigarette Smoke-Induced Chronic Obstructive Pulmonary Disease Via Targeting FHL1
Authors Zhong S, Chen C, Liu N, Yang L, Hu Z , Duan P, Shuai D, Zhang Q , Wang Y
Received 25 July 2019
Accepted for publication 12 September 2019
Published 9 October 2019 Volume 2019:14 Pages 2319—2329
DOI https://doi.org/10.2147/COPD.S224763
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Chunxue Bai
Shan Zhong,1,2 Chengshui Chen,3 Naijia Liu,2 Li Yang,3 Zhangli Hu,2 Pengfei Duan,1 Diquan Shuai,2 Qingying Zhang,1 Yun Wang2
1Department of Preventive Medicine, Shantou University Medical College, Shantou, Guangdong 515041, People’s Republic of China; 2Center for Research and Technology of Precision Medicine, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, Guangdong 518055, People’s Republic of China; 3Department of Respiratory, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang 325000, People’s Republic of China
Correspondence: Yun Wang
Center for Research and Technology of Precision Medicine, College of Life Sciences and Oceanography, Shenzhen University (Xili Campus), No. 1066, Xueyuan Ave, Nanshan Distract, Shenzhen, Guangdong 518055, People’s Republic of China
Tel +86 755 2695 8895
Fax +86 755 2653 4274
Email [email protected]
Qingying Zhang
Department of Preventive Medicine, Shantou University Medical College, Shantou, Guangdong 515041, People’s Republic of China
Tel +86 754 8825 9850
Fax +86 754 8856 6774
Email [email protected]
Background: Chronic obstructive pulmonary disease (COPD) is recognized as a chronic lung disease with incomplete reversible airflow limitation, but its pathophysiology was still not clear. This study aimed at investigating regulatory roles of special miRNA–mRNA axis in COPD development.
Methods: Differentially expressed miRNAs and downstream mRNAs were screened from the Gene Expression Omnibus (GEO) dataset by using the LIMMA package in R software. Weighted Gene Co-expression Network Analysis (WGCNA) was used to construct a co-expression network for COPD. The correlation of dysregulated miRNA(s) and COPD was analyzed, and miRNAs with significant differences were validated in peripheral blood mononuclear cells (PBMCs) from COPD patients by real-time PCR. Regulatory roles of candidate miRNAs and targeted mRNAs were investigated in vitro study.
Results: Thirteen modules of co-expressed miRNAs and mRNAs were constructed from a selected cohort with WGCNA. Turquoise module with 12 differentially expressed miRNAs and 120 mRNAs was significantly correlated with COPD. The expression of hsa-miR-664a-3p, an upregulated miRNA in the module, was increased both in lung tissue and PBMCs from COPD patients, whereas that targeted four and a half LIM domains 1 (FHL1) gene was decreased and positively correlated with forced expiratory volume in 1 sec (FEV1)/forced vital capacity (FVC%) (r = 0.59, p < 0.01). In vitro, luciferase activity assay revealed FHL1 as a target of hsa-miR-664a-3p and it could be directly downregulated by overexpression of hsa-miR-664a-3p. Furthermore, cigarette smoke extract could increase hsa-miR-664a-3p level and decrease FHL1 level in Beas-2B cells.
Conclusion: The present study validated significant upregulation of hsa-miR-664a-3p in COPD patients, and its target gene FHL1 was downregulated and positively correlated with FEV1/FVC%; both hsa-miR-664a-3p and FHL1 could be regulated by cigarette smoke extract. Results of bioinformatic analyses and expanded validation suggest that the axis from hsa-miR-664a-3p to FHL1 might play a key role in cigarette smoke-induced COPD, and the exact mechanism should be confirmed in further studies.
Keywords: COPD, miRNA, mRNA, co-expression networks, biomarker
Introduction
Chronic obstructive pulmonary disease (COPD) is recognized as a chronic lung disease with incomplete reversible airflow limitation and predicted to be the third leading cause of death worldwide by 2020,1,2 that resulting in an important problem threatening public health and quality of life. COPD is usually caused by being exposed to noxious particles or gases, cigarette smoking is one of the major risk factors.3 Under complex condition, chronic airway inflammation and emphysema are the two main pathological subtypes of COPD, which cause immunological disorder and destruction of lung tissue.4,5 However, the pathogenesis and effective biomarkers of COPD diagnose and treatment have not yet been elucidated.
MicroRNAs (miRNAs) are a kind of endogenous non-coding RNAs, of 20 to 24 nt, which can participate in regulating physiological and pathological conditions by binding to the 3′ untranslated region (3′ UTR) of target mRNAs.6 Growing evidence has revealed that matched miRNA–mRNA pairs can also play vital roles in the pathogenesis and development of COPD.7–15 For instance, Ezzieet, et al, found that the expression of miR-15b was increased and that of its targeted gene, SMAD7, was decreased in COPD lung tissues, and their co-expression participated in the COPD process via a transforming growth factor β (TGF-β) signal pathway.7 MiR-29b expression was found decreased in both plasma and lung tissue of COPD, and was significantly associated with functional changes of the lung. In addition, miR-29b could regulate cigarette smoke extract (CSE)-induced interleukin 8 (IL-8) expression by targeting bromodomain 4 in bronchial epithelial cells.8 Soeda and colleagues demonstrated that there was a progressive reduction in the plasma miR-106b level in COPD patients with a negative correlation of disease duration, which could be an important clinical indicator for COPD.15 Hence, the relation between miRNAs and target genes can play a vital role in COPD progress, which might provide novel approaches to the management of COPD that have been receiving a great deal of attention.
With high-throughput sequencing technique, abundant expression profiles of miRNAs and mRNAs have been provided to databases for the public. Gene Expression Omnibus (GEO) dataset and bioinformatic analysis have been widely used to search disease-related biomarkers in various medical researches.16–18 Dr. Ma’en and colleagues have found that network based on analyzed data represented a novel approach to discover biomarkers in peripheral blood mononuclear cells (PBMCs) of COPD.19 Based on bioinformatics, a study by Liu et al, was identified that the expression of miR-23a and miR-145 was significantly decreased in COPD patients, compared with healthy controls. Furthermore, miR-23a might be as a promising biomarker of discriminating frequent exacerbators from non-frequent exacerbators of COPD via ROC curve analysis.20 In this study, it was aimed at finding certain miRNAs that can indicate risks of COPD based on co-expression profiles of miRNA to its targeted mRNA, and to further investigate their roles on the pathogenesis and development of COPD.
Materials And Methods
Selection Of Differentially Expressed miRNAs And mRNAs
Data of miRNA and mRNA expression in lung tissues for 25 individuals (17 COPD and 8 normal smokers) were obtained from an NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE38974).7 The whole gene expression matrix was normalized by the quantile method, then transformed with the log2-scale. Finally, miRNAs and mRNAs with differential expression between COPD and normal smokers were further screened by using the LIMMA package in R software, with criteria |fold change| >1.5 and positive false discovery rate <0.05.
Weighted Gene Co-Expression Network Analysis (WGCNA)
A scale-free gene co-expression network, including differentially expressed miRNAs and mRNAs, was constructed by using the WGCNA package in R software.21 First, data from samples were clustered with the “hclust” function for detecting outliers. Then, an appropriate soft threshold power β was chosen with the “pickSoftThreshold” function to build a scale-free topology. The height was set at 0.80, and the β was determined as 14 (Fig. S1). Next, the adjacency matrix was calculated based on the β value, and then transformed into a topological overlap matrix (TOM) and corresponding dissimilarity (1-TOM). Genes with similar expression pattern were clustered, and modules were divided by “cutreeDynamic” functions with default parameters. Because modules identified by the dynamic tree cut algorithm may be similar, they were merged with a height cutoff of 0.15.
Key Co-Expression Modules Of COPD And Functional Enrichment Analysis
Module eigengenes (MEs) were defined as the first principal component in each module and could summarize expression patterns of all genes in a module. To determine the key modules most relevant to COPD, the MEs for each module were calculated with the “moduleEigengenes” function, and correlation with COPD was analyzed with the “Pearson” method. The database for annotation, visualization and integrated discovery (DAVID) tools provided a comprehensive set of functional annotation tools for investigating biological significance with a list of differentially expressed mRNAs. Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis was implemented in DAVID for exploring signal transduction pathways.
Construction Of Regulatory Network
The bioinformatic prediction algorithm TargetScan was used to predict target genes for each differentially expressed miRNA. The correlation between differentially expressed miRNAs and mRNAs was assayed by Pearson correlation analysis: only the miRNA–mRNA pair with r < −0.5, p < 0.05 was considered significant, and selected as a novel candidate for further investigation. Then, regulatory network was constructed based on differently expressed miRNAs and correlated target mRNAs by using Cytoscape software.
Sample Preparation And Validation
Peripheral blood samples from 48 individuals (24 smokers with COPD and 24 normal smokers) were obtained from The First Affiliated Hospital of Wenzhou Medical University and written informed consent was obtained with all subjects. The experimental procedures were approved by the Medical Ethics Committee of The First Affiliated Hospital of Wenzhou Medical University (approved no.: 2016131). The exclusion criteria were including the history of severe infection, autoimmune disease, solid tumor and other lung diseases. Importantly, the individual who did meet the standard set, the ratio of FEV1 to FVC<0.70 was classified into COPD group after bronchodilator treatment. PBMCs were isolated with human lymphocyte separation medium (Solarbio, China) and stored at −80°C.
Cell Culture
Human bronchial epithelial cells Beas-2B (American Type Culture Collection, ATCC, USA) were cultured in DMEM supplemented with 10% fetal bovine serum in a humidified incubator under 5% CO2 at 37°C. Cells were then transfected with an hsa-miR-664a-3p mimic or non-targeting control (Sangon Biotech, China) with Lipofectamine 2000 reagent (Invitrogen, USA), according to the manufacturer’s protocol, or treated with 2% CSE for 24 hrs. CSE was prepared by bubbling the smoke of two filterless cigarettes through 10 mL DMEM at 2 mins per cigarette for 100% CSE, and this solution was then passed through a 0.22-μM filter for sterilization and stored at −80°C.
Luciferase Activity Assay
The PsiCHECK-2 vector (Promega, USA) harboring the wild-type and mutated FHL1 3ʹ-UTR was co-transfected with an hsa-miR-664a-3p mimic or negative control into HEK293T cells (ATCC). Luciferase activity was detected by using the Dual-Luciferase Reporter Assay System (Promega), according to the manufacturer’s instruction. Firefly luciferase activity was normalized to renilla luciferase activity.
Quantitative Real Time-PCR (qRT-PCR)
Total RNA was extracted from cells by using the M5 HiPer Universal Plus RNA Mini Kit (Mei5 Biotechnology, China). cDNA was synthesized with the cDNA synthesis kit or Mir-X miRNA First-Strand Synthesis Kit (both were obtained from TaKaRa, Japan). Primers for qRT-PCR were designed (listed in Table S1) and synthesized by Sangon Biotech (China), and the primer for U6 and universal reverse primer for miRNAs were supported by Mir-X miRNA First-Strand Synthesis Kit. qRT-PCR amplification involved using SYBR Green PCR Premix Ex TaqTM II reagents (TaKaRa) with the QuantStudio 6 FlexI real-time system (Applied Biosystems, USA). Relative mRNA expression was determined with the 2−ΔCt or 2−ΔΔCt method in comparison to endogenous controls (U6 or GAPDH).
ELISA
Cells were treated with CSE, then levels of IL-6 and IL-8 were determined in supernatant from Beas-2B cells by using commercial ELISA kits (Sino Biological, China), according to the manufacturer’s instructions.
Western Blot Analysis
Total protein was extracted from Beas-2B cells and lysed, then the concentration was determined by using a BCA kit (Thermo, USA). Equal amounts of proteins from each sample were separated by 12% SDS-PAGE and transferred to a nitrocellulose membrane (Millipore Co, USA). After blocking with non-fat milk, the membrane was incubated with specific primary antibody at 4°C overnight. After washing with TBST, membranes were incubated with secondary antibody at room temperature for 1 hr. Primary antibodies for FHL1 and GAPDH were from Abcam and Cell Signaling Technology (both in USA). Immunoreactive signals were quantified by using Image Lab (Bio-Rad, USA).
Statistical Analysis
Statistical analysis is involved in using GraphPad Prism 6.0 (GraphPad Software Inc., San Diego, CA, USA). Student’s t-test and Mann–Whitney test were used for analyzing two groups with or without normal distribution, and Spearman correlation analysis was used for correlation analysis. p < 0.05 was considered statistically significant.
Results
Construction Of Weighted Gene Co-Expression Network
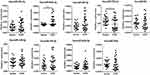
WGCNA was used to analyze 2098 differentially expressed mRNAs and 80 miRNAs (Fig. S2) from 25 individuals with miRNA and mRNA expression profiles. After merging modules with high similarity, 13 co-expression modules were identified. The correlation between MEs and COPD is shown in Figure 1A. Among the 13 modules, the green-yellow and turquoise modules were the most significant modules relevant to COPD, with Pearson r = 0.95 (p = 6e−13) and −0.95 (p = 8e−13), respectively. Functional enrichment analysis was performed to infer the potential functions of mRNAs and miRNAs in the two modules. The results from DAVID tools showed 4 miRNAs and 80 mRNAs in the green-yellow module and 15 miRNAs and 354 mRNAs in the turquoise module (Table S2). KEGG pathway analysis revealed that those target genes in the turquoise module were involved in TGF-β signaling, NF-kB signaling and apoptosis (Figure 1B); genes in the green-yellow module were involved in protein binding-bridging, copper ion binding and BMP binding (Figure 1C). Thus, target genes in the turquoise module had more potential roles for COPD progression than that in green-yellow module.
Regulatory Networks Of Genes In The Turquoise Module
Regulatory networks in the turquoise module were constructed on the basis of inverse correlation between miRNA and target genes by using Cytoscape. A total of 12 differentially expressed miRNAs and matched inversely correlated 120 mRNAs were included in the regulatory networks. Ten miRNAs (hsa-miR-574-3p, hsa-miR-339-5p, hsa-miR-664a-3p, hsa-miR-186-5p, hsa-miR-454-3p, hsa-miR-642a-5p, hsa-miR-766-3p, hsa-miR-518b, hsa-miR-634 and hsa-miR-625-3p) were upregulated and two miRNAs (hsa-miR-24-3p and hsa-miR-508-5p) were downregulated in COPD lung tissue vs normal smoker lung tissue (Figure 2). In the networks, hsa-miR-664a-3p had the most regulatory target genes (n=63), whereas hsa-miR-766-3p and hsa-miR-625-3p had 35 and 33 differentially expressed target genes (Table S3). Among differentially expressed genes, PTAR1 and DDR2 were regulated by 5 miRNAs, and hsa-miR-186-5p, hsa-miR-642a-5p, hsa-miR-766-3p and hsa-miR-664a-3p were predicted to have binding sites for both genes.
Validation Of Upregulated miRNAs In PBMCs From COPD Patients
The 10 upregulated miRNAs in the turquoise module were recorded by fluorescent intensity of miRNA relative expression and analyzed by transformation to a log2-scale. Except for hsa-miR-625-3p, 9 miRNAs showed significantly increased expression (p < 0.05 or p < 0.01) (Figure 3). Those 9 miRNAs in PBMCs from COPD patients vs normal smokers (24 for each group) were investigated. Their characteristics were listed in Table 1. Only the expression of hsa-miR-664a-3p was significantly higher in PBMCs from COPD patients than normal smokers (p < 0.01). The expression of hsa-miR-766-3p in PBMCs was significantly decreased (Figure 4), which was inconsistent with its expression in lung tissue of COPD patients (Figure 3).
 |
Table 1 The Demographic And Clinical Characteristics Of The Recruited Subjects |
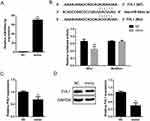
Validation Of FHL1 Expression In PBMCs From COPD Patients
The expression of hsa-miR-664a-3p was significantly increased in both lung tissues and PBMCs from COPD patients. In the regulation network of miRNA and mRNA pairs, hsa-miR-664a-3p could bind to 63 targeted regulatory genes. Four and a half LIM domains 1 (FHL1) was predicted as one of the target genes of hsa-miR-664a-3p, and has been reported to have a downregulated trend in COPD.40 Validation experiments showed that FHL1 expression was significantly decreased in both lung tissues (Figure 5A) and PBMCs from COPD patients (Figure 5C) (both p < 0.01). The expression of FHL1 was inversely correlated with that of has-miR-664a-3p in lung tissues (rs = −0.79, p < 0.01; Figure 5B). FEV1/FVC% was positively correlated with FHL1 expression (rs = 0.59, p < 0.01; Figure 5D) and inversely with hsa-miR-664a-3p expression (rs = −0.21, p = 0.1587; Figure 5E) in this disease.
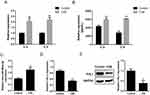
Hsa-miR-664a-3p Directly Targeted FHL1
From the above results, TargetScan was used to predict FHL1 3′-UTR binding site(s) for hsa-miR-664a-3p. Dual luciferase reporter assay was used to explore whether FHL1 was a direct target of hsa-miR-664a-3p. The expression of hsa-miR-664a-3p was significantly increased in HEK293T cells after transfection with the hsa-miR-664a-3p mimic as compared with controls (Figure 6A). Relative luciferase activity of the 3′-UTR-FHL1 wild type was significantly suppressed with hsa-miR-664a-3p mimic transfection (p < 0.01), but it was unchanged with co-transfection of the 3′-UTR-FHL1 mutant and hsa-miR-664a-3p mimic as compared with negative controls (Figure 6B). In addition, FHL1 mRNA and protein expression were negatively regulated by hsa-miR-664a-3p in Beas-2B cells (Figure 6C and D).
Expression Of hsa-miR-664a-3p And FHL1 In CSE-Induced Beas-2B Cells
To further study the roles of hsa-miR-664a-3p and FHL1 expression in CSE-induced Beas-2B cells, Beas-2B cells were treated with 2% CSE. As compared with controls, after treated with 2% CSE, IL-6, IL-8 and hsa-miR-664a-3p mRNA expression were increased 2.06-, 2.19- and 1.74-fold, respectively (Figure 7A and C), and FHL1 expression was significantly decreased down to 51% (Figure 7D). The secretion levels of IL-6 and IL-8 were increased with CSE treatment vs control treatment (Figure 7B). FHL1 protein expression was also downregulated by 62% with CSE treatment, compared with controls (Figure 7E).
Discussion
As a leading cause of death worldwide, COPD has a considerable impact on public health and quality of life.2 To find novel biomarkers of COPD, this study screened COPD-related miRNAs with targeted mRNAs from the GEO dataset GSE38974, which contained both miRNA and mRNA expression profiles in lung tissues from both normal smokers and smokers with COPD. With bioinformatics analysis, one module showed strong associations with COPD, and 9 miRNAs in this module were significantly upregulated in lung tissues from COPD patients. In COPD patients, unlike in patients with solid tumor, obtaining tissues or epithelial cells from lungs is difficult, so the expression of those 9 miRNAs in PBMCs from COPD patients was validated. Among the 9 significantly expressed miRNAs, only hsa-miR-664a-3p was significantly upregulated, which showed a similar trend as in lung tissue.
A few reports showed that hsa-miR-664a-3p has different functions in several diseases. Yoneda and coworkers analyzed serum miRNA profiles in samples from participants with and without chronic periodontitis by using microarray and real-time PCR; the expression of hsa-miR-664a-3p was higher in individuals with periodontitis than controls and was a candidate serum biomarker for chronic periodontitis.22 Modak et al, revealed differential expression of hsa-miR-664a-3p in patients with cardioembolic stroke as compared with controls.23 Aberrant expression of hsa-miR-664a-3p was found in various malignancies, such as gastric cancer, breast cancer, cervical cancer and osteosarcoma.24–27 However, the precise role and underlying mechanism of hsa-miR-664a-3p in chronic respiratory disease have not been elucidated.
Here, we demonstrated that hsa-miR-664a-3p was significantly upregulated in COPD. However, the expression of hsa-miR-766-3p was inconsistent in lung tissue and PBMCs of COPD patients, that was upregulated in lung tissues and downregulated in PBMCs. An opposite trend of miRNA expression between tissues and blood samples was reported in other disease studies.28,29 An explanation is that miRNAs passive release may occur during tissue injury, or its expression may occur through microvesicles or exosome-mediated transfer, which release more miRNAs into the blood stream and then into hemocytes. The potential mechanisms are needed to be further studied.
The detailed role and underlying mechanism of hsa-miR-664a-3p in COPD disease were demanded to elucidate. We found hsa-miR-664a-3p expression negatively correlated with FHL1 expression, as a direct target gene in the pathogenesis of COPD. FHL1 encodes a member of FHL protein family, functions as a transcription factor and is involved in many cellular processes, especially in the regulation of muscle diseases, accompanied by early respiratory failure.30,31 FHL1 expression is also found significantly upregulated in pulmonary hypertension, particularly in the pulmonary vasculature.32 Furthermore, FHL1 has an important role in CSE-induced proliferation of pulmonary arterial smooth muscle cells and hypoxia-induced pulmonary hypertension.33,34 However, numerous studies implied that FHL1 may have a tumor suppressor activity and play significant roles in tumorigenesis and progression. Its expression was found downregulated in many cancers,35–39 such as gastric, lung, prostate and breast cancer, suggesting that FHL1 has the various physiological and pathological functions. In COPD, FHL1 was detected in type II pneumocytes, macrophages, bronchi and capillaries of lung tissue from COPD patients and was found downregulated in CSE-induced primary human lung cells.40
Here we found FHL1 expression downregulated in both lung tissues and PBMCs from COPD patients as compared with normal smokers, and the expression was negatively correlated with hsa-miR-664a-3p level in lung tissue. In addition, FHL1 expression was positively correlated with FEV1/FVC%. Previous studies demonstrated that cigarette smoke exposure is the crucial risk factor for COPD pathogenesis, accompanied by release of IL-6 and IL-8 in airway epithelial cells.3,41 In this study, CSE exposure significantly increased the expression of IL-6 and IL-8 in Beas-2B cells, for consistent results with previous studies. Simultaneously, it was identified that increased hsa-miR-664a-3p expression and decreased FHL expression in CSE-induced Beas-2B cells, which showed a significant negative correlation in vitro study of COPD.
Conclusions
The present study revealed increased expression of hsa-miR-664a-3p in both lung tissues and PBMCs from COPD as compared with normal smokers, and the miRNA could regulate pulmonary function by negatively regulating its target, FHL1. Moreover, CSE treatment could significantly increase the expression of hsa-miR-664a-3p and decrease that of FHL1 in Beas-2B cells, and with higher inflammtion factors. Thus, hsa-miR-664a-3p and its targeted FHL1 may play a pivotal role in CSE-induced COPD, but the exact mechanisms are needed by further study.
Ethics Approval
The experimental procedures were approved by the Medical Ethics Committee of The First Affiliated Hospital of Wenzhou Medical University (approved no.: 2016131).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Reid DJ, Pham NT. Emerging therapeutic options for the management of COPD. Clin Med Insights Circ Respir Pulm Med. 2013;7:7–15. doi:10.4137/CCRPM.S8140
2. Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. lancet. 2012;380(9859):2095–2128. doi:10.1016/S0140-6736(12)61728-0
3. Hogg JC, Timens W. The pathology of chronic obstructive pulmonary disease. Annu Rev Pathol-Mech Dis. 2009;4:435–459. doi:10.1146/annurev.pathol.4.110807.092145
4. Decramer M, Janssens W, Miravitlles M. Chronic obstructive pulmonary disease. Lancet. 2012;379:1341–1351. doi:10.1016/S0140-6736(11)60968-9
5. Singh D, Agusti A, Anzueto A, et al. Global strategy for the diagnosis,management, and prevention of chronic obstructive lung disease: the GOLD science committee report 2019. Eur Respir J. 2019;53(5):1900164. doi:10.1183/13993003.01184-2018
6. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
7. Ezzie ME, Crawford M, Cho JH, et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2012;67(2):122–131. doi:10.1136/thoraxjnl-2011-200089
8. Tang K, Zhao J, Xie J, et al. Decreased miR-29b expression is associated with airway inflammation in chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol. 2019;316(4):621–629. doi:10.1152/ajplung.00436.2018
9. Pottelberge GRV, Mestdagh P, Bracke KR, et al. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2011;183(7):898–906. doi:10.1164/rccm.201002-0304OC
10. Ebrahimi A, Sadroddiny E. MicroRNAs in lung diseases: recent findings and their pathophysiological implications. Pulm Pharmacol Ther. 2015;34:55–63. doi:10.1016/j.pupt.2015.08.007
11. Xue H, Li MX. MicroRNA-150 protects against cigarette smoke-induced lung inflammation and airway epithelial cell apoptosis through repressing p53: microRNA-150 in CS-induced lung inflammation. Hum Exp Toxicol. 2018;37(9):920–928. doi:10.1177/0960327117741749
12. Shen W, Liu J, Zhao G, et al. Repression of Toll-like receptor-4 by microRNA-149-3p is associated with smoking-related COPD. Int J Chron Obstruct Pulmon Dis. 2017;12:705–715. doi:10.2147/COPD.S128031
13. Conickx G, Mestdagh P, Avila Cobos F, et al. MicroRNA profiling reveals a role for microRNA-218-5p in the pathogenesis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2017;195(1):43–56. doi:10.1164/rccm.201506-1182OC
14. Xu H, Sun Q, Lu L, et al. MicroRNA-218 acts by repressing TNFR1-mediated activation of NF-κB, which is involved in MUC5AC hyper-production and inflammation in smoking-induced bronchiolitis of COPD. Toxicol Lett. 2017;280:171–180. doi:10.1016/j.toxlet.2017.08.079
15. Soeda S, Ohyashiki JH, Ohtsuki K, et al. Clinical relevance of plasma miR-106b levels in patients with chronic obstructive pulmonary disease. Int J Mol Med. 2013;31(3):533–539. doi:10.3892/ijmm.2013.1251
16. Huang Y, Zhu J, Li W, et al. Serum microRNA panel excavated by machine learning as a potential biomarker for the detection of gastric cancer. Oncol Rep. 2018;39(3):1338–1346. doi:10.3892/or.2017.6163
17. Shangguan H, Tan SY, Zhang JR. Bioinformatics analysis of gene expression profiles in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2015;19(11):2054–2061.
18. Tian X, Zhu X, Yan T, et al. Differentially expressed lncRNAs in gastric cancer patients: a potential biomarker for gastric cancer prognosis. J Cancer. 2017;8(13):2575–2586. doi:10.7150/jca.19980
19. Nie Y, Chen V, Shannon CP, et al. Network-based analysis reveals novel gene signatures in peripheral blood of patients with chronic obstructive pulmonary disease. Respir Res. 2017;18(1):72–80. doi:10.1186/s12931-017-0558-1
20. Liu X, Qu J, Xue W, et al. Bioinformatics-based identification of potential microRNA biomarkers in frequent and non-frequent exacerbators of COPD. Int J Chron Obstruct Pulmon Dis. 2018;13:1217–1228. doi:10.2147/COPD.S163459
21. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9(1):559–572. doi:10.1186/1471-2105-9-559
22. Yoneda T, Tomofuji T, Ekuni D, et al. Serum microRNAs and chronic periodontitis: a case-control study. Arch Oral Biol. 2019;101:57–63. doi:10.1016/j.archoralbio.2019.03.009
23. Modak JM, Roy-O’Reilly M, Zhu L, et al. Differential microribonucleic acid expression in cardioembolic stroke. J Stroke Cerebrovasc Dis. 2019;28(1):121–124. doi:10.1016/j.jstrokecerebrovasdis.2018.09.018
24. Wang L, Li B, Zhang L, et al. miR‐664a‐3p functions as an oncogene by targeting Hippo pathway in the development of gastric cancer. Cell Prolif. 2019;52:12567–12580. doi:10.1111/cpr.12567
25. Wu L, Li Y, Li J, et al. MicroRNA-664 targets insulin receptor substrate 1 to suppress cell proliferation and invasion in breast cancer. Oncol Res Featuring Preclinical Clin Cancer Ther. 2019;27:459–467. doi:10.3727/096504018X15193500663936
26. Yang Y, Liu H, Wang X, et al. Up-regulation of microRNA-664 inhibits cell growth and increases cisplatin sensitivity in cervical cancer. Int J Clin Exp Med. 2015;8(10):
27. Bao Y, Chen B, Wu Q, et al. Overexpression of miR-664 is associated with enhanced osteosarcoma cell migration and invasion ability via targeting SOX7. Clin Exp Med. 2017;17(1):51–58. doi:10.1007/s10238-015-0398-6
28. Zhu J, Zheng Z, Wang J, et al. Different miRNA expression profiles between human breast cancer tumors and serum. Front Genet. 2014;5:149–155. doi:10.3389/fgene.2014.00149
29. Brase JC, Wuttig D, Kuner R, et al. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer. 2010;9(1):306–314. doi:10.1186/1476-4598-9-306
30. Shathasivam T, Kislinger T, Gramolini AO. Genes, proteins and complexes: the multifaceted nature of FHL family proteins in diverse tissues. J Cell Mol Med. 2010;14(12):2702–2720. doi:10.1111/j.1582-4934.2010.01176.x
31. Ding L, Wang Z, Yan J, et al. Human four-and-a-half LIM family members suppress tumor cell growth through a TGF-β-like signaling pathway. J Clin Invest. 2009;119(2):349–361. doi:10.1172/JCI35930
32. Kwapiszewska G, Wygrecka M, Marsh LM, et al. Fhl-1, a new key protein in pulmonary hypertension. Circulation. 2008;118(11):1183–1194. doi:10.1161/CIRCULATIONAHA.107.761916
33. Yue J, Guan J, Wang X, et al. MicroRNA-206 is involved in hypoxia-induced pulmonary hypertension through targeting of the HIF-1α/Fhl-1 pathway. Lab Invest. 2013;93(7):748–759. doi:10.1038/labinvest.2013.63
34. Li Y, Pu G, Chen C, et al. Inhibition of FHL1 inhibits cigarette smoke extract-induced proliferation in pulmonary arterial smooth muscle cells. Mol Med Rep. 2015;12(3):3801–3808. doi:10.3892/mmr.2015.3787
35. Asada K, Ando T, Niwa T, et al. FHL1 on chromosome X is a single-hit gastrointestinal tumor-suppressor gene and contributes to the formation of an epigenetic field defect. Oncogene. 2013;32(17):2140–2149. doi:10.1038/onc.2012.228
36. Sakashita K, Mimori K, Tanaka F, et al. Clinical significance of loss of Fhl1 expression in human gastric cancer. Ann Surg Oncol. 2008;15(8):2293–2300. doi:10.1245/s10434-008-9904-3
37. Niu C, Liang C, Guo J, et al. Downregulation and growth inhibitory role of FHL1 in lung cancer. Int J Cancer. 2012;130(11):2549–2556. doi:10.1002/ijc.26259
38. Cao W, Liu J, Xia R, et al. X-linked FHL1 as a novel therapeutic target for head and neck squamous cell carcinoma. Oncotarget. 2016;7(12):14537–14550. doi:10.18632/oncotarget.7478
39. Li X, Jia Z, Shen Y, et al. Coordinate suppression of Sdpr and Fhl1 expression in tumors of the breast, kidney, and prostate. Cancer Sci. 2008;99(7):1326–1333. doi:10.1111/j.1349-7006.2008.00816.x
40. Heinbockel L, Marwitz S, Schromm AB, et al. Identification of novel target genes in human lung tissue involved in chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2018;13:2255–2259. doi:10.2147/COPD.S161958
41. Comer DM, Kidney JC, Ennis M, et al. Airway epithelial cell apoptosis and inflammation in COPD, smokers and nonsmokers. Eur Respir J. 2013;41(5):1058–1067. doi:10.1183/09031936.00063112
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.