Back to Journals » Cancer Management and Research » Volume 12
ONECUT2 Accelerates Tumor Proliferation Through Activating ROCK1 Expression in Gastric Cancer
Authors Chen J, Chen J, Sun B , Wu J, Du C
Received 31 March 2020
Accepted for publication 30 June 2020
Published 21 July 2020 Volume 2020:12 Pages 6113—6121
DOI https://doi.org/10.2147/CMAR.S256316
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Jie Chen,* Jinggui Chen,* Bo Sun, Jianghong Wu, Chunyan Du
Department of Gastric Surgery, Fudan University Shanghai Cancer Center, Fudan University, Shanghai 200032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chunyan Du; Jianghong Wu
Department of Gastric Surgery, Fudan University Shanghai Cancer Center, Fudan University, 1205 Rm., 3# Bldg., 270 Dong an Road, Shanghai 200032, People’s Republic of China
Email [email protected] [email protected]
Background: Transcription factors (TFs) are key regulators which control gene expression during cancer initiation and progression. In the current study, we aimed to explore the proliferative function and clinical significance of TFs in gastric cancer (GC).
Methods: Differential analysis was used to investigate the overall expression difference between normal and tumor tissues of each TF in TCGA-STAD cohort. The quantitative real-time polymerase chain reaction (qRT-PCR) was performed to confirm the mRNA expression of one cut homeobox 2 (ONECUT2) in GC tissues. Western blot analysis was conducted to confirm the protein knockdown efficiency. Cell counting, colony formation, and GC xenograft model assays were performed to confirm the proliferative function of ONECUT2 in GC cells. Gene set enrichment analysis (GESA) and qRT-PCR were conducted to confirm the affected signaling pathways and downstream targets of ONECUT2.
Results: Our data indicated that a TF named ONECUT2 was highly expressed in GC and correlated with patients’ poor prognosis. Importantly, knockdown of ONECUT2 dramatically decreased GC cells proliferation, whereas overexpression of ONECUT2 promoted carcinogenesis in GC. Kyoto encyclopedia of genes and genomes (KEGG) analysis revealed that the upregulating ONECUT2 induced the activation of Wnt signaling pathway and cell cycle regulation pathway. We further identified that ONECUT2 boosted gastric cancer cell proliferation through enhancing ROCK1 (Rho associated coiled-coil containing protein kinase 1) mRNA expression. High level of ROCK1 expression rescued proliferative behavior of ONECUT2-deficient GC cells.
Conclusion: Our findings demonstrated that ONECUT2 promoted GC cells proliferation through activating ROCK1 expression at the DNA level, suggesting that ONECUT2-ROCK1 axis might be a potential therapeutic target in GC.
Keywords: transcriptional factor, ONECUT2, ROCK1, gastric cancer, carcinogenesis
Introduction
Gastric cancer (GC) is one of the most malignant cancers, which is the third leading cause of cancer-related deaths.1 Due to the limited knowledge on pathogenesis and treatment of gastric cancer, the 5-year overall survival in GC patients remains gloomy. The transcriptional dysregulation and oncogene addiction are the hallmarks of cancer.2 Importantly, with the extensive exploration using deep sequencing studies in the area of cancer genome, researchers have identified that the differential expression and recurrent somatic mutations of TFs could influence and control cancer status and transform its malignant properties.3,4 Significantly, targeting these cancer-dependent TFs shows a promising therapeutic effect in many types of tumors.5
Recent evidence has highlighted the crucial roles of TFs in manipulating GC cells proliferation.6,7 For example, the TEA domain family member 4 (TEAD4), a component of Hippo signaling pathway, was highly expressed in GC tissues and positively associated with poor outcomes in GC patients. Knockdown of TEAD4 significantly decreased the growth of GC cells both in vitro and in vivo.8 YAP and TAZ, the key factors of the Hippo signaling pathway, activate gastric cancer initiation through transcriptional and posttranscriptional upregulation of MYC.9 Targeting YAP1 expression with molecular inhibitors reduced Gal-3-mediated aggressive phenotypes in gastric adenocarcinoma cells, which highlighted the potential therapeutic functions of TFs inhibitors.10 However, the full oncogenic roles of TFs in gastric cancer still remain unclear.
ONECUT2 gene belongs to the ONECUT family (ONECUT1, ONECUT2, and ONECUT3), and it was firstly identified as a transcriptional factor that stimulates hnf-3beta gene expression and is required for liver differentiation and metabolism.11 ONECUT2 couples with ONECUT1 to mediate horizontal cell development through the control of Pax6 expression.12 It also regulates pancreas morphogenesis, pancreatic and enteric endocrine differentiation.13 Recent evidence revealed that ONECUT2 is a driver of neuroendocrine prostate cancer, which enhances SMAD3 expression and modulates HIF1α chromatin-binding, and it is an important master regulator of metastatic castration-resistant prostate cancer.14 However, the clinical significance and oncogenic role of ONECUT2 in GC remains to be explored. This study aimed to investigate the relationship between TFs expression and GC cells proliferation. To this end, we examined differentially expressed TF genes in TCGA-STAD cohort and validated their proliferative ability using the CRISPR-Cas9 system in AGS cells.
Materials and Methods
Gastric Cancer Samples
The gastric cancer tissues were acquired from the Department of Gastric Surgery, Fudan University Shanghai Cancer Center, Fudan University (Shanghai, China). All the samples were stored in liquid nitrogen and the informed consent has been obtained from all gastric cancer patients.
Cell Culture
The GC cells MKN-45, AGS, and the human embryonic kidney 293T (HEK-293T) cells were obtained from American Type Culture Collection (ATCC). Human MKN-45 and AGS cells were cultured in RPMI1640 and HEK-293T cells in DMEM at 37°C with 5% CO2. All of the culture medium was supplemented with 10% FBS and 100 µg/mL penicillin and streptomycin.
RNA Interference
To target ONECUT2 expression, small interfering RNAs (siRNAs) were acquired from RiboBio (Guangzhou, China). The transfection with siRNAs were used Lipofectamine RNAiMAX reagent (Invitrogen) in GC cells. After 24–48 hours, cells were collected for RNA and protein extraction and molecular function validation. The siRNA sequences used for targeting ONECUT2 are listed in Supplementary Table S1.
RNA Extraction and Real-Time Quantitative PCR Analysis
RNA was extracted using the RNeasy mini kit (Qiagen). Briefly, cells were lysed with buffer RLT and 70% ethanol was added to the homogenized lysate. Then the sample was transferred to a RNeasy spin column and centrifuged and washed with buffers RW1 and RPE. Next, 50 µL of RNAse-free water was added to the spin column and centrifuged to collect total RNA. The reverse transcription (RT) reactions were performed using a PrimeScript RT Reagent Kit (TaKaRa, Shiga, Japan). The real-time PCR was done using SYBR Premix Ex Taq II (TaKaRa) and detected using a QuantStudio 7 Flex sequence detection system (Thermo Fisher Scientific). The PCR results were normalized to β-actin with the use of the comparative CT method. Primers to test genes mRNA expression are shown in Supplementary Table S1.
Lentivirus Preparation and Infection
The GFP, ONECUT2, Cas9 overexpression plasmid, and the sgRNA targeting ONECUT2 coupled with psPAX2 and pMD2.G were transfected into HEK-293T cells with the usage of Lipofectamine 2000 (Invitrogen). After 48–72 hours, lentivirus-containing supernatant was acquired and used to infect the GC cells MKN-45 and AGS.
Colony Formation Assay
The colony formation assay was performed by seeding 2000 cells per well into 6-well plates, and incubating the plates for 12 days. Then the colonies were fixed with methanol, stained by crystal violet (Sigma-Aldrich), and their number was counted using CCK-8 assay.
Gastric Cancer Xenograft Model
Animals used in this study were treated humanely using the Guide for the Care and Use of Laboratory Animals of Fudan University and all animal experiments were approved by the Committee on the Ethics and Welfare of Laboratory Animal Science of Fudan University. ONECUT2 knockdown GC AGS cells and control cells (2×106 cells in 200 µL of RPMI1640 medium) were subcutaneously injected in the lower back of mice (n=6 each group). Tumor size was measured every 5 days. At the end of 25 days, mice were sacrificed and tumor weight was determined.
Dual-Luciferase Assay
Dual-Luciferase assay was performed using Dual-Luciferase Reporter Assay (Promega). In brief, GC cells were transfected with either ONECUT2 mixed siRNAs or negative control siRNA coupled with luciferase plasmids and renilla plasmids in a 24-well plate. After 24 hours, growth media were removed and cells were lysed with PLB buffer. Lysate were transferred into a 96-well plate, the luciferase and renilla substrate were added, and the luminescence was measured.
ChIP Assay
GC cells were cross-linked, quenched, and sonicated using Bioruptor UCD-200 (Diagenode, Liege, Belgium). Solubilized chromatin was immunoprecipitated with Flag antibody (Sigma) and Dynabeads® Protein G (Thermo Fisher Scientific). The beads were washed (500 mM NaCl, 50 mM Tris 7.5, 1% NP40, 0.02% SDS, proteinase inhibitor) and the DNA was collected using MinElute Reaction Cleanup Kit (Qiagen, Hilden, Germany). The sequences of the primers used are listed in Supplementary Table S1.
Western Blot Analysis
Western blot analysis was done as previously described.15 The nitrocellulose membrane was incubated with anti-ONECUT2 antibody and then with horseradish peroxidase-conjugated secondary antibodies. The detailed list of antibodies used is provided in Supplementary Table S2.
Statistical Analysis
We used the R package limma to investigate the overall statistical difference between each TFs in TCGA-STAD cohort, The P-value under multiple different analysis was adjusted using Benjamini & Hochberg false discovery rate (FDR) methods. The different analysis of quantified variates was conducted using Student’s t-test after the normal inspection test. We used the Log rank test to compare the difference between the Kaplan–Meier curves for each TF gene. The univariate and multi-variate prognosis analysis were estimated by uni- and multi-variate Cox proportional hazards regression model. The statistical differences between each of the two qualitative variables were analyzed by Pearson’s Chi-squared Test. The R software (https://www.r-project.org/) was used to conduct statistical analysis. The two-side statistical analysis with a P-value<0.05 was deemed as statistically significant.
Results
Transcription Factors are Differentially Expressed in GC Samples
To explore the differential expression and potential clinical significances of TFs in gastric cancer development and progression, we systematically examined the mRNA expression heterogeneity of TFs in the TCGA-STAD cohort by limma package. We discovered that 72 TF genes were highly-expressed in gastric cancer, inversely the 27 TFs were down-regulated in tumor tissues compared with normal tissues (FDR<0.2, fold change>2; Figure 1A). Moreover, we tested the potential prognosis of TFs using uni- and multi-variate Cox regression analysis. The results identified that 11 down-regulated TF and 34 upregulated TF were significantly correlated with patient overall survival (P<0.05; Figure 1A). Furthermore, we have tested the fold-change in expression levels of candidate-dysregulated TFs using hazard ratio of Cox results, and correlation coefficients of TFs and tumor stage (Figure 1B). Importantly, we investigated a possible correlation between cell proliferation rate and upregulation of TFs in the AGS cell line. The high ONECUT2 mRNA expression levels in the tissue of gastric cancer patients was significantly correlated with patient prognosis and carcinogenesis (Figure 1C). These results demonstrated that a group of TFs was dysregulated in gastric cancer, which was associated with possible biological functions. Among these TF genes, those belonging to the ONECUT family serve as potential oncogenic regulators in gastric cancer tissues.
ONECUT2 Predicts Poor Patient Prognosis in GC
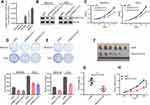
We further explored the ONECUT2 differently expressed status in gastric cancer tissues and normal tissues in our selected samples using qRT-PCR on tissues obtained from the patients in our gastric cancer cohort (n=116). Consistently, the ONECUT2 was overexpressed in GC samples compared with nontumor tissues (NTs) at the mRNA levels (P<0.001; Figure 2A). The results of Kaplan–Meier analysis showed that GC patients with high ONECUT2 expression levels had poor survival outcomes, as shown by overall survival (OS, P=0.0009; Figure 2B) and disease-free survival (DFS, P=0.0002; Figure 2C). Additionally, we investigated a possible correlation between clinicopathological features and tumor ONECUT2 expression in gastric cancer patients (Figure 2D). To further investigate whether high expression level of ONECUT2 was an independent prognostic factor in our internal gastric cancer patient cohort (n=116), we conducted a multi-variate analysis using the Cox proportional hazard regression model comparing ONECUT2 expression values with other clinical characteristics as covariables (age, gender, tumor stage, number of distant metastatic status, and tumor size). Gastric cancer patients with high expression level of ONECUT2 in tumors presented a 3.65-fold high risk of death (P<0.05, 95% confidence interval [CI]=1.94–6.90: Figure 2E). Thus, these results comprehensively indicated that ONECUT2 can serve as a promising prognostic biomarker in gastric cancer patients.
ONECUT2 Enhances GC Cells Proliferation
To verify the proliferative function of ONECUT2 in gastric cancer cells, we tested the ONECUT2 mRNA levels in GC cell lines and found ONECUT2 was highly expressed in AGS, MKN-45, and SNU668 cells, but relatively low to not expressed in SNU484 and MKN-74 cells (Figure 3A). We then employed two small interfering RNAs (siRNAs) that target to ONECUT2 mRNA in MKN-45 and AGS cells. Western blot analysis showed a strong knockdown efficacy of ONECUT2 expression (Figure 3B). CCK-8 and colony formation assays revealed that knockdown of ONECUT2 mRNA levels using siRNAs decreased the proliferation of gastric cancer cells (Figure 3C and D). We also used a lenti-clustered regularly interspaced short palindromic repeats (CRISPR) deletion system to knockdown ONECUT2 and found similar functional results validated with colony formation assay (Figure 3E). Considering our in vitro results, we tested the in vivo proliferative property of ONECUT2 in AGS gastric cancer cells, which were infected with ONECUT2 knockdown sgRNA lentivirus using the CRISPR deletion system and subcutaneously injected into the flanks of 6-week-old nude mice. Strikingly, the knockdown of ONECUT2 significantly abolished tumorigenicity (Figure 3F), as determined by analyzing tumor weight (Figure 3G) and size (Figure 3H). These findings indicated that ONECUT2 enhanced gastric cancer cell proliferation both in vitro and in vivo.
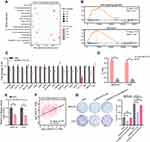
ONECUT2 Controls ROCK1 Expression at the DNA Level
To further understand the details of the oncogenic mechanism of ONECUT2 in gastric cancer, we performed RNA-sequencing using ONECUT2 overexpression in AGS cells and found that high expression of ONECUT2 was positively correlated with the activation of cell cycle regulation and Wnt signaling pathways (Figure 4A and B). To determine the direct downstream targets of ONECUT2, we analyzed genes that were correlated the most (top 20), which were enriched in the cell cycle pathway and Wnt signaling pathway, and found that knockdown of ONECUT2 could decrease sterile alpha motif domain containing 3 (SAMD3) and protein kinase C alpha (PRCKA) mRNA levels. Of note, the inhibition of ONECUT2 could significantly abolish Rho associated coiled-coil containing protein kinase 1 (ROCK1) mRNA levels (Figure 4C). ROCK1 encodes a protein serine/threonine kinase, and is activated by the GTP-bound form of Rho to regulate the Wnt signaling pathway. Recent studies revealed that ROCK1 acts as an oncogene to enhance GC carcinogenesis.16,17 To further determine the transcriptional regulation of ROCK1 by ONECUT2, we performed ChIP-qPCR and found ONECUT2 protein was enriched on ROCK1 promoter region (Figure 4D). Dual-luciferase assay also identified knockdown of ONECUT2 decreased ROCK1 promoter activity (Figure 4E). Furthermore, we found that the expression of ONECUT2 mRNA levels was positively correlated with the expression of ROCK1 mRNA levels in TCGA-STAD cohort (Figure 4F). These findings suggested that ROCK1 was the direct downstream target of ONECUT2. Importantly, overexpression of ROCK1 could rescue the proliferative behavior of ONECUT2-deficient gastric cancer cells (Figure 4G). Taken together, these studies showed that ONECUT2 exerted its proliferative function through transcriptional regulation of ROCK1 in gastric cancer.
Discussion
In this study, we found that a transcriptional factor ONECUT2 was overexpressed in the tissues of GC patients, which predicted poor patient prognosis. Strikingly, the knockdown of ONECUT2 significantly abolished GC cells proliferation in vitro and in vivo. We further revealed that high expression of ONECUT2 was positively correlated with Wnt signaling and cell cycle pathways activation and enhanced ROCK1 mRNA expression. Importantly, the overexpression of ROCK1 could rescue ONECUT2-deficient GC cells proliferation, suggesting that the ONECUT2-ROCK1 axis may represents a potential therapeutic target for GC patients.
Transcriptional factors are the key factors that manipulate gene expression and remodel cancer cell homeostasis, thus maintaining malignant properties of tumors. To investigate the mechanisms of such transcriptional regulation in GC, we firstly analyzed the differential expression of TFs, evaluated their proliferative functions, and defined the oncogenic role for ONECUT2 in GC. Our results showed that ONECUT2 was highly upregulated in GC tissues compared with nontumorous tissues. Consistently, GC patients with highly expressed ONECUT2 exhibited worse overall survival. These findings suggested that ONECUT2 may play a crucial role in gastric cancer progression. We further revealed that ONECUT2 enhanced GC cell proliferation both in vitro and in vivo. Importantly, ONECUT2 could induce ROCK1 expression at the DNA level by binding to its promoter and activating the Wnt signaling pathway. Moreover, overexpression of ROCK1 could rescue the proliferation of ONECUT2-deficient GC cells. Our studies suggested that the ONECUT2–ROCK1 axis may serve as a potential therapeutic target for gastric cancer patients.
ONECUT2 has been identified as an oncogene to accelerate tumor progression in many cancers;18,20 for example, ONECUT2 plays a role as a diver gene that activates SMAD3, regulates hypoxia signaling, and leads to higher degrees of hypoxia in neuroendocrine prostate cancer.14 ONECUT2 also acts as a master regulator of androgen receptor networks in metastatic castration-resistant prostate cancer. Importantly, inhibition of ONECUT2 by a small molecule compound, suppressed prostate cancer metastasis, which shows that ONECUT2 is a potential drug target in the metastatic phase of aggressive prostate cancer.21 These studies highlighted the key roles of ONECUT2 in cancer initiation and development.
To further reveal the characteristic of ONECUT2 in GC, we analyzed its expression in TCGA database and found that ONECUT2 was highly expressed in pancreatic adenocarcinoma, esophageal carcinoma, hepatocellular carcinoma, and extremely highly expressed in GC. The GEPIA database also showed similar results (http://gepia.cancer-pku.cn/). Meanwhile, we also assessed the expression of ONECUT1 and ONECUT3; however, in both genes, the average expression level was lower than that of ONECUT2. By analyzing the clinical information available on GC patients in our cohort, we firstly found that GC patients with a high level of ONECUT2expression exhibited worse overall survival and disease-free survival, which was consistent with the data of TCGA-STAD cohort (data of TCGA-STAD was not shown). Therefore, ONECUT2 may serve as a potential oncogene in GC. Using siRNA and CRISPR deletion system, we found that knockdown of ONECUT2 significantly inhibited GC cell proliferation, which is in agreement with another recent study by Seo et al.22 Our works firstly identified that ONECUT2 controlled the Wnt signaling pathway and regulated ROCK1 expression. ROCK1 gene encodes a serine/threonine kinase protein and is activated when bound to the GTP-bound form of RhoA protein. ROCK1 is one of the key regulators of the Wnt/β-catenin pathway and it has been deemed as an oncogene in many cancers, including the GC, which accelerates cancer proliferation and migration. The overactivation of the Wnt signaling pathway in GC has also been validated by many previous reports. Our findings supported the newly identified importance of the ONECUT2-ROCK1 axis in GC. Our next works will be focused on evaluating the therapeutic value of the molecular inhibitors that specifically target ONECUT2 in GC.
Our findings also identified other potential proliferative TFs in GC such as RAD51 recombinase (RAD51) and zinc finger protein 367 (ZNF367). Liu et al23 found RAD51 mediated resistance of cancer stem cells to PARP inhibition in triple-negative breast cancer. Importantly, He et al24 reported that RAD51 potentiated synergistic effects of chemotherapy with PCI-24,781 and Cis-Diamminedichloroplatinum in GC. In GC, the clinical significance and detailed molecular function of RAD51 also need to be explored further.
Conclusion
Our works highlighted that ONECUT2 enhances GC cells proliferation through the transcriptional activation of ROCK1 by binding to its promoter and the subsequent upregulation of the Wnt signaling pathway. We further suggest that the ONECUT2-ROCK1 axis could be a promising therapeutic target in gastric cancer.
Abbreviations
TFs, transcription factors; ONECUT2, one cut homeobox 2; TCGA, the cancer genome atlas; GC, gastric cancer; NTs, nontumor tissues; STAD, stomach adenocarcinoma; GSEA, gene set enrichment analysis; ROCK1, Rho associated coiled-coil containing protein kinase 1; OS, overall survival; DFS, disease-free survival; KEGG, Kyoto encyclopedia of genes and genomes; qRT-PCR, quantitative real-time polymerase chain reaction; FDR, false discovery rate.
Acknowledgments
We are grateful for Dr Didier Trono’s gifts of the psPAX2 and pMD2.G lentivirus plasmids.
Author Contributions
Chunyan Du and Jianghong Wu designed the study; Jie Chen and Jinggui Chen acquired the data; Chunyan Du and Bo Sun performed the analysis of data. Jie Chen and Jinggui Chen wrote the paper with comments from all authors. All authors contributed to data analysis, drafting, or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
2. Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi:10.1016/j.cell.2016.12.013
3. Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nat Rev Genet. 2013;14:703–718. doi:10.1038/nrg3539
4. Bhagwat AS, Vakoc CR. Targeting transcription factors in cancer. Trends Cancer. 2015;1:53–65. doi:10.1016/j.trecan.2015.07.001
5. Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi:10.1016/j.cell.2011.08.017
6. Xu TP, Liu XX, Xia R, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648–5661. doi:10.1038/onc.2015.18
7. Wu J, Qin W, Wang Y, et al. SPDEF is overexpressed in gastric cancer and triggers cell proliferation by forming a positive regulation loop with FoxM1. J Cell Biochem. 2018;119:9042–9054. doi:10.1002/jcb.27161
8. Shuai Y, Ma Z, Liu W, et al. TEAD4 modulated LncRNA MNX1-AS1 contributes to gastric cancer progression partly through suppressing BTG2 and activating BCL2. Mol Cancer. 2020;19:6. doi:10.1186/s12943-019-1104-1
9. Choi W, Kim J, Park J, et al. YAP/TAZ initiates gastric tumorigenesis via upregulation of MYC. Cancer Res. 2018;78:3306–3320. doi:10.1158/0008-5472.CAN-17-3487
10. Ajani JA, Estrella JS, Chen Q, et al. Galectin-3 expression is prognostic in diffuse type gastric adenocarcinoma, confers aggressive phenotype, and can be targeted by YAP1/BET inhibitors. Br J Cancer. 2018;118:52–61. doi:10.1038/bjc.2017.388
11. Jacquemin P, Lannoy VJ, Rousseau GG, Lemaigre FP. OC-2, a novel mammalian member of the ONECUT class of homeodomain transcription factors whose function in liver partially overlaps with that of hepatocyte nuclear factor-6. J Biol Chem. 1999;274:2665–2671. doi:10.1074/jbc.274.5.2665
12. Klimova L, Antosova B, Kuzelova A, Strnad H, Kozmik Z. ONECUT1 and ONECUT2 transcription factors operate downstream of Pax6 to regulate horizontal cell development. Dev Biol. 2015;402:48–60. doi:10.1016/j.ydbio.2015.02.023
13. Vanhorenbeeck V, Jenny M, Cornut JF, et al. Role of the ONECUT transcription factors in pancreas morphogenesis and in pancreatic and enteric endocrine differentiation. Dev Biol. 2007;305:685–694. doi:10.1016/j.ydbio.2007.02.027
14. Guo H, Ci X, Ahmed M, et al. ONECUT2 is a driver of neuroendocrine prostate cancer. Nat Commun. 2019;10:278. doi:10.1038/s41467-018-08133-6
15. Chen D, Wang H, Chen J, et al. MicroRNA-129-5p regulates glycolysis and cell proliferation by targeting the glucose transporter SLC2A3 in gastric cancer cells. Front Pharmacol. 2018;9:502. doi:10.3389/fphar.2018.00502
16. Yu X, Ma C, Fu L, Dong J, Ying J. MicroRNA-139 inhibits the proliferation, migration and invasion of gastric cancer cells by directly targeting rho-associated protein kinase 1. Oncol Lett. 2018;15:5977–5982. doi:10.3892/ol.2018.8038
17. Hu CB, Li QL, Hu JF, Zhang Q, Xie JP, Deng L. miR-124 inhibits growth and invasion of gastric cancer by targeting ROCK1. Asian Pac J Cancer Prev. 2014;15:6543–6546. doi:10.7314/APJCP.2014.15.16.6543
18. Sun Y, Shen S, Liu X, et al. MiR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting ONECUT2 in colorectal carcinoma. Mol Cell Biochem. 2014;390:19–30. doi:10.1007/s11010-013-1950-x
19. Lu T, Wu B, Yu Y, et al. Blockade of ONECUT2 expression in ovarian cancer inhibited tumor cell proliferation, migration, invasion and angiogenesis. Cancer Sci. 2018;109:2221–2234. doi:10.1111/cas.13633
20. Ma Q, Wu K, Li H, et al. ONECUT2 overexpression promotes RAS-driven lung adenocarcinoma progression. Sci Rep. 2019;9:20021. doi:10.1038/s41598-019-56277-2
21. Rotinen M, You S, Yang J, et al. ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nat Med. 2018;24:1887–1898. doi:10.1038/s41591-018-0241-1
22. Seo EH, Kim HJ, Kim JH, et al. ONECUT2 upregulation is associated with CpG hypomethylation at promoter-proximal DNA in gastric cancer and triggers ACSL5. Int J Cancer. 2020;146:3354–3368. doi:10.1002/ijc.32946
23. Liu Y, Burness ML, Martin-Trevino R, et al. RAD51 mediates resistance of cancer stem cells to PARP inhibition in triple-negative breast cancer. Clin Cancer Res. 2017;23:514–522. doi:10.1158/1078-0432.CCR-15-1348
24. He WL, Li YH, Hou WJ, et al. RAD51 potentiates synergistic effects of chemotherapy with PCI-24781 and cis-diamminedichloroplatinum on gastric cancer. World J Gastroenterol. 2014;20:10094–10107. doi:10.3748/wjg.v20.i29.10094
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.