Back to Journals » Neuropsychiatric Disease and Treatment » Volume 19
NR3C1 and NR3C2 Genes Increase the Risk of Suicide Attempt in Psychiatric Disorder Patients with History of Childhood Trauma
Authors Sanabrais-Jiménez MA , Esquivel-López AA , Sotelo-Ramírez CE , Aguilar-García A, Ordoñez-Martínez B, Jiménez-Pavón J, Madrigal-Lara MV, Díaz-Vivanco AJ, Camarena B
Received 18 July 2023
Accepted for publication 11 November 2023
Published 24 November 2023 Volume 2023:19 Pages 2561—2571
DOI https://doi.org/10.2147/NDT.S431176
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Roger Pinder
Marco Antonio Sanabrais-Jiménez,1 Ayerim Alma Esquivel-López,2 Carlo Esteban Sotelo-Ramírez,1 Alejandro Aguilar-García,1 Bruno Ordoñez-Martínez,1 Joanna Jiménez-Pavón,3 María Victoria Madrigal-Lara,3 Alan Jair Díaz-Vivanco,3 Beatriz Camarena1
1Departamento de Farmacogenética, Instituto Nacional de Psiquiatría Ramón de la Fuente Muñiz, Ciudad de Mexico, Mexico; 2Departamento de Genética Molecular, Instituto de Fisiología Celular, Universidad Nacional Autónoma de México, Ciudad de Mexico, Mexico; 3Dirección de Servicios Clínicos, Instituto Nacional de Psiquiatría “Ramón de la Fuente Muñiz”, Ciudad de Mexico, Mexico
Correspondence: Beatriz Camarena, Departamento de Farmacogenética, Instituto Nacional de Psiquiatría Ramón de la Fuente Muñiz, Calz Mexico-Xochimilco 101, Col. San Lorenzo Huipulco, Ciudad de Mexico, 14370, Mexico, Tel +52 (55) 4160-5513, Email [email protected]
Background: Hypothalamic-pituitary-adrenal axis gene variants and childhood trauma (CT) are considered risk factors for suicide attempt (SA). The aim of the present study was analyzed gene x environment (GxE) interaction of NR3C1, NR3C2, and CT, and NR3C1 and NR3C2 gene expression in the development of SA with CT.
Participants and Methods: A total of 516 psychiatric Mexican patients from Instituto Nacional de Psiquiatría Ramón de la Fuente Muñiz. Among them, 274 had SA at least once and 242 had not SA. Genetic variants of NR3C1 and NR3C2 were genotyped in all the patients, of which were obtained the CT information from medical records. Additionally, the gene expression of NR3C1 and NR3C2 was also analyzed for a subsample of 96 patients, obtaining the TC information from Childhood Trauma Questionnaire (CTQ).
Results: The analysis showed a GxE interaction of NR3C1, NR3C2, and CT (OR=2.8, 95% CI [1.9– 3.9], p< 0.0001). Interactions were also observed with neglect (OR=2.1, 95% CI [1.4– 3.1], p< 0.0001), emotional abuse (OR=2.1, 95% CI [1.5– 3], p< 0.0001), and sexual abuse (OR=2.4, 95% CI [1.4– 2.9], p< 0.0001) in the prediction of SA. The analysis of gene expression identified an overexpression of NR3C1 in SA patients with high scores for physical and sexual abuse (p< 0.0001; p< 0.0006, respectively) and emotional neglect (p=0.014). An underexpression was observed of NR3C2, associated with high scores of trauma subtypes (p< 0.0001) except physical neglect. Additionally, we observed an overexpression of NR3C1 gene in patients with SA carriers of A allele of rs6191 (p=0.0015). Also, overexpression of NR3C1 gene in carriers of G allele of rs6198 and underexpression of NR3C2 gene in carriers of G allele of rs5522 (p< 0.0001).
Conclusion: Our findings suggest that genetic variants of NR3C1 and NR3C2 differentially affect expression levels, increasing the susceptibility to SA in psychiatric patients with a history of CT.
Keywords: NR3C1, NR3C2, childhood trauma, suicide attempt, gene x environment interaction, gene expression
Introduction
Suicidal behavior is a major public health issue. An estimated 800,000 deaths annually are attributed to suicide; these show a high prevalence of psychiatric disorders, supported by psychological autopsy studies.1–3 Family, twin, and adoption studies show genetic and environmental components to suicidal behavior, estimating a heritability of 45%, which suggests that genetic factors could be interacting with environmental factors in the risk factors for suicide attempt (SA).4 In particular, childhood trauma (CT) has been found to affect the stress response in individuals with genetic vulnerability to suicidal behavior.5
The hypothalamic-pituitary-adrenal (HPA) axis is considered the main regulator of the stress system stimulating forward and feedback inhibition loops involving the brain, pituitary, and adrenal glands, which regulates cortisol production.6 During acute stress, cortisol levels increase and pulsatility remains intact, which is beneficial for promoting the survival of the fittest as part of the fight-or-flight response.7 However, chronic exposure to stress can result in the reversal of beneficial effects, generating long-term high cortisol levels, which can become a maladaptive stress response, which has been associated with major depressive disorder, bipolar disorder patients with a late age-of-onset, and drug-naïve first-episode patients with psychosis.7–9 Cortisol is released from the adrenal glands and exerts its effects in the hippocampus by binding with the glucocorticoid receptor (GR) and the mineralocorticoid receptor (MR). The GR has a relatively low affinity to cortisol but is gradually occupied by higher levels of the steroid hormone under acute stress conditions. Intracellular MR, however, has ten times its affinity to cortisol, leading to a higher occupancy, even under baseline conditions. These two hippocampal steroid receptors exert opposite actions: the MR mediates the tonic influences of basal cortisol levels, and the GR facilitates negative feedback actions in response to higher cortisol levels to maintain steroid homeostasis.10 The two genes are thus essential for activating and modulating the HPA axis.
Glucocorticoid (NR3C1) and mineralocorticoid (NR3C2) receptor genes codify to proteins containing a variable amino-terminal transactivation domain, a small well-conserved DNA-binding domain that comprises two repeats of a zinc finger protein motif, and a relatively conserved carboxy-terminal domain responsible for hormone binding.11 NR3C1 is located at 5q31-32 and the NR3C2 gene at 4q31.1–31.2. NR3C1 is expressed in two main isoforms due to alternative splicing of exon 9, generating the GRα and GRβ proteins. GRα diverges from GRβ only at the carboxyl-terminal end, with 15 nonhomologous amino acids. This difference impairs GRβ from binding to steroids such as cortisol. The GRα isoform is thus considered a biologically active receptor isoform.12
The NR3C1 gene contains a functional polymorphism, rs6198 (A → G), which alters the rates of GRα/GRβ expression, where the G allele favors the expression of the GRβ isoform. G allele carriers have also shown significantly higher cortisol responses following the psychological stressor and dexamethasone suppression test.11 Another genetic variant is the rs6191, located in exon 9β in the 3’UTR region, in linkage disequilibrium with rs6198, a block related to mRNA stability and increased protein expression of the receptor.13 Moreover, rs33388 is in the region of intron 2, which has been suggested as a target sequence involved in regulating the NR3C1 gene or as a binding site for specific miRNAs regulating the network of genes involved in the regulation of the HPA axis.13
The expression and activity of the MR have also been associated with functional genetic variants of NR3C2. One of them is rs5522, a G → A substitution in exon 2, resulting in an amino acid change (Ile180Val), where a carrier of the Val allele was associated with a loss of cortisol function when used as a ligand, and with increased saliva and plasmatic cortisol responses to psychosocial stressors and the dexamethasone suppression test.11,14 The single-nucleotide polymorphism (SNP) rs2070951 (C/G) of the NR3C1 gene has also been associated with modulation of MR activity through the altering of expression, transactivation, and activity. In vitro studies have demonstrated that the C allele is associated with increased expression of MR.10
Despite to the limited literature, there are some studies about NR3C1, NR3C2, CT, and SA. It was observed an interaction between NR3C1 and physical neglect increasing the risk of dependence of crack/cocaine.15 Another study shows that NR3C1 and emotional trauma (abuse or neglect) in childhood interact inducing reductions in left hippocampal volumes suggesting vulnerability to mood disorders in adolescent girls.16 It is important to mention that substance use disorder and mood disorders are highly implicated with SA.17 For other hand, it was observed significant association between NR3C1 and SA in patients with schizophrenia.18 Two studies failed to find an association between suicide behavior and the NR3C2 gene.19,20 To date, few studies analyzing NR3C1, NR3C2, and CT have been conducted on patients with SA. A gene x environment (GxE) study found no interaction between 19 gene variants involved in the HPA axis, including NR3C1, and early childhood abuse in patients with SA and bipolar disorder.21 A previous study failed to identify interaction of NR3C1, NR3C2, and CT in the development of SA in bipolar disorder patients.20 The only gene expression study has been of NR3C1, showing an under-expression (UE) of GRα in patients with suicidal behavior and childhood abuse.22 To date, the expression of NR3C2 has not been evaluated in SA. Findings concerning the interaction of NR3C1 and NR3C2 with CT in the development of SA have not been conclusive.
Aim
The aim of this study was to analyze GxE interaction of NR3C1, NR3C2, and CT, and the gene expression of NR3C1 and NR3C2 in the development of SA in patients with CT.
Hypothesis
Genetic variants of the NR3C1 and NR3C2 genes are associated with differential expression levels, which interact with a history of childhood trauma, contributing to an increase of at least one SA.
Materials and Methods
Study Design
The study design was comparative, observational, and transversal.
Sample
We included a sample of 516 Mexican patients (404 female and 112 male), who met DSM-5 criteria for bipolar disorder (n = 231), major depressive disorder (n = 221), borderline personality disorder (n = 34), and other psychiatric disorders (n = 30). They were recruited from the affective disorder’s clinic, the outpatient service, the emergency department, and the inpatient population of the Instituto Nacional de Psiquiatría Ramón de la Fuente Muñiz from October 2016 to November 2021. The sample included 274 patients (53%) who had made at least one suicide attempt and 242 (47%) who had not attempted suicide. SA was defined as self-destructive behavior with the intent to end one’s life, regardless of the resulting damage. The information about CT was systematically obtained by psychiatrists in the clinical interview and registered in the patients’ medical records. The CT was evaluated by a psychiatrist who reviewed the clinical information and classified the different subtypes of CT according to the criteria of Bernstein and Fink (1998) for neglect (emotional and physical) and abuse (emotional, physical, and sexual).23
In addition, we analyzed the gene expression in a subsample of 96 patients (86 female and 10 male) that included 63 patients with major depressive disorder (65.6%), 24 with borderline personality disorder (25%), and nine with other psychiatric disorders (9.4%). The assessment of CT for these patients was assessed with the Childhood Trauma Questionnaire (CTQ) short form, which evaluates five types of traumas: sexual, physical, and emotional abuse, and physical and emotional neglect.23
Inclusion criteria were psychiatric patients diagnosed with DSM-5 with and without SA; female and male patients over 18 years of age; Mexican Mestizos with a family background of three generations born in México; and capacity to provide written informed consent. Additionally, the patients implicated in the gene expression analyzes did not presented medical conditions that generate an imbalance in the HPA axis. Whereas exclusion criteria were psychiatric patients not competent to consent to the study, and insufficient sample to genetic analysis.
Ethical Approval
This study was conducted in agreement with the Declaration of Helsinki standards. The study protocol was approved by the Ethics Committee of the Instituto Nacional de Psiquiatría Ramón de la Fuente Muñiz (Approval No. CEI/C/015/2019). All participants in the study provided written informed consent.
Genotyping
DNA was extracted from peripherical leukocytes using the Flexigene DNA kit (Qiagen; Minneapolis, MN). NR3C1 (rs6198, rs6191, and rs33388) and NR3C2 (rs5522 and rs2070951) polymorphisms were selected, given that they had been previously studied in people with SA and CT.20,21 Genotyping was analyzed using TaqMan assays for rs6198 (C_8951023_10), rs6191 (C_3234245_20), rs33388 (C_1046426_10), rs5522 (C_12007869_20), and rs2070951 (C_1594392_10). Genotypes were identified with the TaqMan allele-specific assay method using the ABI Prism®7500 sequence detection system according to the manufacturer´s protocols (Applied Biosystems Inc.; Foster City, CA). The final volume of the reaction was 7μL, consisting of 100ng of genomic DNA, 1X TaqMan Universal Master Mix, and 0.71X SNP Genotyping Assay Mix (Applied Biosystems Inc.). After denaturing at 95°C for 10 min, 40 cycles of PCR were performed under the following conditions: denaturing at 95°C for 15s and annealing at 60°C for 1 min. The genotyping was performed without knowing the clinical status of the subjects.
Gene Expression
Total RNA was isolated from the blood samples collected within the first 24 hours after hospitalization (7:00–9:00 am), using the reagent TRIzol (Roche; Germany). The first-strand cDNA was synthesized with SuperScript™ IV VILO™ Master Mix (Thermofisher Scientific) using 500 ng of total RNA. Gene expression was analyzed with custom TaqMan assays for NR3C1 (Hs00353740_m1) and NR3C2 (Hs01031809_m1), with Glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Hs02786624_g1) as a reference gene transcript, using the ABI Prism®7500 Sequence Detection System according to the manufacturer´s protocols (Applied Biosystems Inc.; Foster City, CA). Real-time PCR was performed using a final reaction volume of 10μL, consisting of 62.5 ng of resulting cDNA, 1X TaqMan Universal Master Mix, and 1X Gene Expression Assay Mix (Applied Biosystems Inc.). cDNA was amplified for 40 cycles at 95°C for 15s and 60°C for 1 min. All samples were performed in duplicate and information about the study sample blinded.
Statistical Analyses
Demographic and clinical characteristics were analyzed using chi-square and Student’s t-tests. The Shapiro–Wilk test was used to analyze the normality of the total and subscale scores of the CTQ short form. Total CTQ, emotional neglect, and emotional abuse scores were divided into low and high trauma groups defined by the upper and lower 27% of the total scores, due to their normal distribution, based on previous grouping methods.24,25 Physical neglect, physical abuse, and sexual abuse were not normally distributed; a median-split approach was therefore used to dichotomize these scores between high and low trauma severity.26
The statistical analyses were performed using the program RStudio version 4.2.0.27 The power analysis was performed with the R package “gap” version 1.2.3–6,28 showing a sample power of 0.99, assuming an additive genetic model, a risk allele frequency of 0.09, a population prevalence of SA of 2.7%, an α level of 0.05, and a portion of cases of 0.53 in a sample of 516 patients. Analysis of genotype and allele distributions in patients with and without SA were performed using chi-square tests with Epidat version 3.1.29 Bonferroni correction for multiple testing was applied (for five polymorphisms corrected at p < 0.01). A linkage disequilibrium (LD) matrix based on the patients’ D’ parameter was estimated using Haploview version 4.2.30 THESIAS software was used to analyze haplotype effects. The results were expressed as a haplotypic odds ratio in reference to the most frequent haplotype.31
We also analyzed the GxE interaction using Multifactor Dimensionality Reduction (MDR) software version 3.0.2 and MDR permutation testing software version 1.0 beta.32,33 The MDR method is used for detecting high-order nonlinear or non-additive interactions in case control studies with small sample size to improve the effect of multiple genetic loci and environment in the development of a disease.33,34 MDR collapses multi-locus and environment data into a single-dimensional variable with two levels: high and low risk.32 The interaction models are evaluated using testing balanced accuracy (TBA), a measure of how often individuals are correctly classified with respect to their case/control status, and cross-validation consistency (CVC), which evaluates the consistency with which individuals are classified. The best final model is defined as one with values of TBA between 0.55 and 0.69 and a CVC of 10, which is considered strong evidence of a multifactor interaction, and a significant p-value derived from 1000 permutations.34,35
The gene expression analyses were performed using GraphPad Prism software version 5.0.0.36 All of the results were normalized to GAPDH. Cycle threshold values were averaged and relative NR3C1 and NR3C2 expression levels were determined using the comparative cycle threshold method.37 Differences between NR3C1 and NR3C2 expression of patients with and without SA were calculated using a t-test. Changes in gene expression in relationship to low or high total CT score and the subscale scores were analyzed using ANOVA and Tukey’s multiple comparison post-hoc test or the Kruskal–Wallis test and Dunn’s multiple comparison post-hoc test.
Results
Analysis of Demographic and Clinical Characteristics
Analysis of demographic characteristics revealed significant age and sex differences between patients with and without SA (Table 1). There were significant differences in primary diagnoses, with a high level of major depressive disorder and borderline personality disorder in patients with SA (Table 1). The SA patients showed greater comorbidity with borderline personality disorder, eating disorder, and post-traumatic stress disorder than those without SA (Table 1). There were differences in all CT subtypes, except for physical neglect, between patients with and without SA.
 |
Table 1 Demographic and Clinical Characteristics of the Sample |
Association Analysis
Genotype and allele distributions of NR3C1 (rs6198, rs6191, and rs33388) and NR3C2 (rs5522 and rs2070951) gene polymorphisms are shown in Table 2. There were no differences in NR3C1 or NR3C2 gene polymorphisms between patients with and without SA (Table 2).
 |
Table 2 Genotype and Allele Frequencies of NR3C1 and NR3C2 SNPs in Patients with and without Suicide Attempt |
Haplotype Analysis
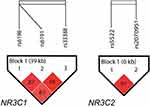
The NR3C1 and NR3C2 haplotype structures are shown in Figure 1. NR3C1 showed a block composed of rs6198, rs6191, and rs33388 (D’ = 0.83, r2 = 0.14), and NR3C2 showed a block composed of rs5522 and rs2070951 (D’ = 0.91, r2 = 0.35). The haplotype analysis did not show any statistical differences between subjects with and without SA (data not shown).
Gene Expression
There were no significant differences in NR3C1 and NR3C2 gene expression levels between patients with and without SA (t = 0.7, p = 0.48; t = 1.4, p = 0.15, respectively). Analysis of the gene expression in relationship to CT and its subtypes showed overexpression (OE) of NR3C1 associated with high scores for physical abuse (p < 0.0001), sexual abuse (p = 0.0006), and emotional neglect (p = 0.014) in patients with SA. It also detected NR3C1 OE in subjects with high scores for total CTQ (p = 0.0012) and physical neglect (p < 0.0001) compared with subjects with low scores. No gene expression changes were detected between the groups after the post-hoc test (p > 0.05) for emotional abuse. The analysis of NR3C2 gene expression and CT showed UE in psychiatric patients with high scores for total CTQ (p < 0.0001), emotional abuse (p < 0.0001), physical abuse (p < 0.0001), sexual abuse (p < 0.0001), and emotional neglect (p < 0.0001) compared with patients that presented low scores.
Analysis of Association Between SNPs and Gene Expression
Patients were subgrouped into carriers and non-carriers of alleles related to previous reports of altered function or differential levels of gene expression of NR3C1 and NR3C2;10,11,13 the NR3C1/rs6198 were subgrouped into AA and AG+GG carriers, the rs6191 into CC and CA+AA carriers, and rs33388 into AA y AT/TT, the NR3C2/rs5522 into AA and AG+GG carriers, and the rs2070951 into CC and GG+CG carriers. An OE of NR3C1 was found in patients with SA who were A allele carriers of rs6191 (p = 0.0015); also, an OE of NR3C1 in carriers of the G allele of rs6198 (p < 0.0001) and an UE of NR3C2 in carriers of the G allele of rs5522 (p < 0.0001) in the sample as a whole. No differences in gene expression were found between those with and without SA in NR3C1/rs33388 (p = 0.43) or NR3C2/rs2070951 (p = 0.2).
Gene x Environment Interaction
Finally, we analyzed the GxE interaction for the risk of developing a SA using the different subgroups of carriers or non-carriers of alleles related to differential levels of function. In the analysis of NR3C1, NR3C2, and CT, we found a prediction of SA risk (TBA = 0.6027) with an OR of 2.76 (1.93–3.94). In addition, the analysis of CT subtypes showed three best interaction models in relation to neglect (TBA = 0.5621; OR 2.12; 95% CI [1.45–3.11]), emotional abuse (TBA = 0.5616; OR 2.13; 95% CI [1.48–3.05]), and sexual abuse (TBA = 0.5673; OR 2.38; 95% CI [1.56–3.64]) for the risk of developing SA. All GxE interaction models showed a CVC of 10 out of 10 and p < 0.0001 (Table 3).
 |
Table 3 Best Models for the Risk of Suicide Attempt Between NR3C1, NR3C2 and Childhood Trauma |
Discussion
This study investigated the effect of two candidate genes involved in the activation and modulation of the HPA axis and CT in the development of SA in a sample of Mexican patients with psychiatric disorders. Consistent with previous studies, we observed a high proportion of women, primary diagnosis of major depressive disorder, and comorbidity with borderline personality disorder.38–40 Our CT assessment was carried out by psychiatrists using clinical interviews and medical records with the operative definition of neglect and abuse proposed by Bernstein and Fink.23 CT frequencies were found to be similar to those reported in previous studies using the CTQ in SA patients.41–43 This study did not used a healthy control group and to our knowledge, there are not Mexican studies that analyzed the CT in a healthy group. A study in healthy Colombian population obtained the information of the subtypes of CT, showing a similar frequency of sexual abuse in our patients without SA (21.74–19%); also, observed similar frequencies of emotional (36.84–42.3%) and physical abuse (29.63–29.9%) and emotional neglect (28.57–31.7%) with our patients with SA, and higher frequency of physical neglect (30.56%) compared with the groups with (14.2%) and without SA (10.7%).44 Could be interesting to analyze the frequency of the CT in a healthy Mexican population to know more directly their impact in the development of SA in psychiatric patients.
We did not find an association between NR3C1 and NR3C2 gene polymorphisms and SA. Yin et al45 likewise found no association of an NR3C1 SNP in patients with SA.
SA is a multifactorial behavior caused by the interaction of various genes and environmental factors. Our study showed that the patients with SA with a history of emotional neglect or physical or sexual abuse presented OE of the NR3C1 gene. A previous study observed an UE of NR3C1 in a sample of patients with suicidal behavior and a background of childhood abuse, as compared with a control group.22 The difference in the results may be explained by the inclusion in their sample of patients with SA and suicidal ideation, and the measurement of gene expression measure of 9β isoform of NR3C1. In contrast, our study included only patients with SA and measured the total gene expression of NR3C1. We also analyzed the gene expression of NR3C2, observing that both genes presented changes in the level of gene expression related to high scores for neglect and abuse. Our findings suggest that a history of CT in psychiatric patients affects the gene expression of both steroid hormone receptors. Several studies in psychiatric disorder patients with CT reported changes in the methylation patterns altering the expression levels of NR3C1.46 Regarding NR3C2, a study showed an under-expression of NR3C2 in hippocampus of suicide victims compared with non- suicide subjects.47 To our knowledge, the effects of CT in the NR3C2 expression levels have not been investigated in SA. Therefore, it is important to replicate our findings in an independent sample.
We also observed GxE interaction of NR3C1, NR3C2, and CT in psychiatric patients with SA. In contrast, two previous studies did not show interaction. The first study showed no interaction of NR3C1, NR3C2, and CT in Caucasian bipolar disorder patients with SA.20 The second analyzed 235 SNPs of genes involved in the HPA axis and reported no interaction between NR3C1, childhood physical or sexual abuse, and SA in patients of European-American ancestry with bipolar disorder.21
These SNPs may alter the binding of transcription factors, creating a new protein-binding motif or causing alterations in the binding affinity of transcription factors. Alteration of the nucleotide sequence in a protein-binding motif may alter the binding of DNA transcription factors, affecting their regulatory function.48 To find out how rs6198 and rs6191 of NR3C1 and rs5522 of NR3C2 could affect the regulation of gene expression by altering the dynamics of transcription factors, we searched in JASPAR49 for DNA-binding motifs. We found that rs6198/NR3C1 is located near predictive cis-regulatory element (CRE) regions, which could be altering the expression of the neighboring genes. CREs are DNase hypersensitive sites that are commonly shared with histone modifications such as H4K4me3 and H3K27ac, or CTCF sites, where their function is to regulate the transcription of target genes. It is important to note that this SNP is in a region of DNA binding motifs, such as FOXD3, HOXB, and SOX2, that play an important role in the development of the nervous system.50 In addition, we observed that rs6198 correlates with the DNA binding motif of the factor associated with RNApolII ZNF354A, with a possible role in the regulation of the transcription of its target genes.51 ZNF354A belongs to the family of zinc finger proteins. The binding motifs must remain intact for correct binding to DNA, and deregulation in the anchoring of these proteins could impact the transcription of target genes, particularly ZNF354A, because of its association with RNAPolII.51 The rs6191 SNP of NR3C1 is in the consensus sequence of binding of the KLF9 transcription factor regulating the transcription of genes; we hypothesize that alteration of a base pair can affect the binding of the transcription factor KLF9 to DNA, increasing transcription levels.52 Finally, rs5522 of NR3C2 belongs to the consensus sequence described for the binding of the transcription factor HOXD11, which plays a key role in cell differentiation, the maintenance of cell identity, and the DNA-binding motif of the TFAP4 transcription factor, whose function is the positive regulation of its target genes.53,54 We suggest that a change in its binding motif can be associated with inhibition in the transcription of NR3C2 through modifications in the binding strength of the protein to DNA.
Based on all of these observations, our hypothesis is that the genetic variants in NR3C1 and NR3C2 are implicated in changes in gene expression due to anomalies in the binding of transcription factors to DNA and in the structure of the binding motif. These generate changes in the density of GR and MR in the hippocampus, which in turn produce deficient activation and modulation of the HPA axis in response to stress, increasing the susceptibility to SA in psychiatric patients with a history of CT.
We acknowledge some strengths of this study reside in that the operational definition of CT was defining by consensus of the group of psychiatrists based on the definition of Bernstein and Fink.23 In addition, the use of MDR program for the GxE interaction minimized the false positive results due to multiple testing, avoid problems developed with the use of parametric statistics in the analysis of high-order interactions, and it is not assumed a genetic model.34
There are several limitations to this study. First, we did not include the CTQ to assess CT in the entire sample, so we cannot perform the GxE interaction dichotomizing between high and low trauma. However, previous studies consider it valid to obtain the information from retrospective reports.55,56 Second, we analyzed the total expression of the NR3C1 and NR3C2 genes. However, there are many isoforms for GR and MR, and it would have been useful to analyze the gene expression in these different isoforms. Third, we did not include the assessment of personality traits involved in the development of SA, such as impulsivity, aggressiveness, and anxiety.
Conclusion
In summary, our study supports the evidence of interaction between NR3C1, NR3C2, and a history of CT in the development of a SA. Our findings suggest that genetic variants of these two steroid hormone receptors are related to differential expression levels, increasing the risk of a SA in psychiatric patients with a history of neglect, emotional abuse, and sexual abuse. However, these findings should be further analyzed in a larger sample of psychiatric patients with SA, taking into consideration clinical evaluations related to personality traits and the expression profiles of genes involved in the stress regulator system to elucidate the role of the HPA axis and CT in the etiology of SA.
Abbreviations
SA, suicide attempt; CT, childhood trauma; HPA, hypothalamic-pituitary-adrenal; GR, glucocorticoid receptor; MR, mineralocorticoid receptor; NR3C1, glucocorticoid receptor gene; NR3C1, mineralocorticoid receptor gene; SNP, single-nucleotide polymorphism; GxE, gene x environment; UE, under-expression; CTQ, Childhood Trauma Questionnaire; GAPDH, Glyceraldehyde-3-phosphate dehydrogenase gene; LD, linkage disequilibrium; MDR, Multifactor Dimensionality Reduction; TBA, testing balanced accuracy; CVC, cross-validation consistency; OE, overexpression; CRE, cis-regulatory element.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
We thank the patients for their participation in this study. The study was presented at the World Congress of Psychiatric Genetics 2023 as a poster presentation with interim findings, and published in “Conference Abstract” in European Neuropsychopharmacology: doi.org/10.1016/j.euroneuro.2023.08.260
Author Contributions
All authors made significant contributions to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all this areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The study was funded by Instituto Nacional de Psiquiatria Ramón de la Fuente Muñiz.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ludwig B, Kienesberger K, Carlberg L, et al. Influence of CRHR1 polymorphisms and childhood abuse on suicide attempts in affective disorders: a GxE approach. Frontiers in Psychiatry. 2018;9:165. doi:10.3389/fpsyt.2018.00165
2. Moitra M, Santomauro D, Degenhard L, et al. Estimating the risk of suicide associated with mental disorders: a systematic review and meta-regression analysis. J Psychiatr Res. 2021;137:242–249. doi:10.1016/j.jpsychires.2021.02.053
3. Sutar R, Kumar A, Yadav V. Suicide and prevalence of mental disorders: a systematic review and meta-analysis of world data on case-control psychological autopsy studies. Psych Res. 2023;329:115492. doi:10.1016/j.psychres.2023.115492
4. Mirkovic B, Laurent C, Podlipski MA, Frebourg T, Cohen D, Gerardin P. Genetic association studies of suicidal behavior: a review of the past 10 years, progress, limitations, and future directions. Frontiers in Psychiatry. 2016;7:158. doi:10.3389/fpsyt.2016.00158
5. González-Castro TB, Juárez-Rojop IE, Tovilla-Zárate CA, et al. Gene-environment interaction between HPA-axis genes and trauma exposure in the suicide behavior: a systematic review. J Psychiatr Res. 2023;164:162–170. doi:10.1016/j.jpsychires.2023.06.011
6. Keller J, Gomez R, Williams G, et al. HPA axis in major depression: cortisol, clinical symptomatology and genetic variation predict cognition. Mol Psychiatry. 2017;22(4):527–536. doi:10.1038/mp.2016.120
7. Russell G, Lightman S. The human stress response. Nature reviews. Endocrinology. 2019;15(9):525–534. doi:10.1038/s41574-019-0228-0
8. Staufenbiel SM, Penninx BW, Spijker AT, Elzinga BM, van Rossum EF. Hair cortisol, stress exposure, and mental health in humans: a systematic review. Psychoneuroendocrinology. 2013;38(8):1220–1235. doi:10.1016/j.psyneuen.2012.11.015
9. Koumantarou Malisiova E, Mourikis I, Darviri C, et al. Hair cortisol concentrations in mental disorders: a systematic review. Physiol Behav. 2021;229:113244. doi:10.1016/j.physbeh.2020.113244
10. Terock J, Van der Auwera S, Janowitz D, Wittfeld K, Teumer A, Grabe HJ. Functional polymorphisms of the mineralocorticoid receptor gene NR3C2 are associated with diminished memory decline: results from a longitudinal general-population study. Mol Genet Genomic Med. 2020;8(9):e1345. doi:10.1002/mgg3.1345
11. Rovaris DL, Mota NR, de Azeredo LA, et al. MR and GR functional SNPs may modulate tobacco smoking susceptibility. J Neural Transm. 2013;120(10):1499–1505. doi:10.1007/s00702-013-1012-2
12. Pujols L, Mullol J, Picado C. Alpha and beta glucocorticoid receptors: relevance in airway diseases. Curr Allergy Asthma Rep. 2007;7(2):93–99. doi:10.1007/s11882-007-0005-3
13. Szczepankiewicz A, Leszczyńska-Rodziewicz A, Pawlak J, et al. Glucocorticoid receptor polymorphism is associated with major depression and predominance of depression in the course of bipolar disorder. J Affect Disord. 2011;134(1–3):138–144. doi:10.1016/j.jad.2011.06.020
14. Derijk RH, Schaaf MJ, Turner G, et al. A human glucocorticoid receptor gene variant that increases the stability of the glucocorticoid receptor beta-isoform mRNA is associated with rheumatoid arthritis. J Rheumatol. 2001;28(11):2383–2388.
15. Rovaris DL, Mota NR, Bertuzzi GP, et al. Corticosteroid receptor genes and childhood neglect influence susceptibility to crack/cocaine addiction and response to detoxification treatment. J Psychiatr Res. 2015;68:83–90. doi:10.1016/j.jpsychires.2015.06.008
16. Malhi GS, Das P, Outhred T, et al. Effect of stress gene-by-environment interactions on hippocampal volumes and cortisol secretion in adolescent girls. Aust N Z J Psychiatry. 2019;53(4):316–325. doi:10.1177/0004867419827649
17. Hoertel N, Franco S, Wall MM, et al. Mental disorders and risk of suicide attempt: a national prospective study. Mol Psychiatry. 2015;20(6):718–726. doi:10.1038/mp.2015.19
18. De Luca V, Tharmalingam S, Zai C, et al. Association of HPA axis genes with suicidal behaviour in schizophrenia. J Psychopharmacol. 2010;24(5):677–682. doi:10.1177/0269881108097817
19. Supriyanto I, Sasada T, Fukutake M, et al. Association of FKBP5 gene haplotypes with completed suicide in the Japanese population. Prog Neuro Psychopharmacol Biol Psychiatry. 2011;35(1):252–256. doi:10.1016/j.pnpbp.2010.11.019
20. Segura AG, Mitjans M, Jiménez E, et al. Association of childhood trauma and genetic variability of CRH-BP and FKBP5 genes with suicidal behavior in bipolar patients. J Affect Disord. 2019;255:15–22. doi:10.1016/j.jad.2019.05.014
21. Breen ME, Seifuddin F, Zandi PP, Potash JB, Willour VL. Investigating the role of early childhood abuse and HPA axis genes in suicide attempters with bipolar disorder. Psychiatricgenetics. 2015;25(3):106–111. doi:10.1097/YPG.0000000000000082
22. Melhem NM, Munroe S, Marsland A, et al. Blunted HPA axis activity prior to suicide attempt and increased inflammation in attempters. Psychoneuroendocrinology. 2017;77:284–294. doi:10.1016/j.psyneuen.2017.01.001
23. Bernstein D, Fink L. Childhood Trauma Questionnarie. A Retrospective Self-Report Manual. San Antonio: The Psychological Corporation; 1998.
24. Burke T, Pinto-Grau M, Lonergan K, et al. Measurement of social cognition in amyotrophic lateral sclerosis: a population based study. PLoS One. 2016;11(8):e0160850. doi:10.1371/journal.pone.0160850
25. Zhang Y, Lü W. Effect of childhood maltreatment on cardiovascular response habitation to repeated psychosocial stress. Int J Psychophysiol. 2022;172:10–16. doi:10.1016/j.ijpsycho.2021.12.005
26. Asmal L, Kilian S, du Plessis S, et al. Childhood trauma associated white matter abnormalities in first-episode schizophrenia. Schizophrenia bulletin. 2019;45(2):369–376. doi:10.1093/schbul/sby062
27. R Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2018. Available from: https://www.R-project.org/.
28. Zhao JH. A genetic analysis package with R. Cambridge, UK; 2020. Available from: https://cran.r-project.org/web/packages/gap/vignettes/gap.html.
29. Hervada Vidal X, Santiago Pérez M, Vázquez Fernández E, Castillo Salgado C, Loyola Elizondo E, Silva Ayçaguer LC. Program for epidemiological analysis of tabulated data Epidat 3.0. Rev Esp Salud Publica. 2004;78(2):277–280. doi:10.1590/S1135-57272004000200013
30. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi:10.1093/bioinformatics/bth457
31. Tregouet DA, Garelle V. A new JAVA interface implementation of THESIAS: testing haplotype effects in association studies. Bioinformatics. 2007;23(8):1038–1039. doi:10.1093/bioinformatics/btm058
32. Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19(3):376–382. doi:10.1093/bioinformatics/btf869
33. Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24(2):150–157. doi:10.1002/gepi.10218
34. Ritchie MD, Hahn LW, Roodi N, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69(1):138–147. doi:10.1086/321276
35. Moore JH. MDR 101—part 4—results. Epistasis blog from the computational genetics laboratory at the university of Pennsylvania; 2019. Available from: http://www.epist_asisb_log.org/2006/12/.
36. GraphPad Prism (Version 5). La Jolla, CA: GraphPad Software, Inc. Available from: https://www.graphpad.com/scientific-software/prism/.
37. Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 2013;3(3):71–85.
38. Zeng R, Cohen LJ, Tanis T, et al. Assessing the contribution of borderline personality disorder and features to suicide risk in psychiatric inpatients with bipolar disorder, major depression and schizoaffective disorder. Psych Res. 2015;226(1):361–367. doi:10.1016/j.psychres.2015.01.020
39. Borges G, Orozco R, Villatoro J, Medina-Mora ME, Fleiz C, Díaz-Salazar J. [Suicide ideation and behavior in Mexico: encodat 2016]. Ideación e intento de suicidio en México: encodat 2016. Salud Publica Mex. 2019;61(1):6–15. Spanish. doi:10.21149/9351
40. Dong M, Zeng LN, Lu L, et al. Prevalence of suicide attempt in individuals with major depressive disorder: a meta-analysis of observational surveys. Psychol Med. 2019;49(10):1691–1704. doi:10.1017/S0033291718002301
41. Guillaume S, Perroud N, Jollant F, et al. HPA axis genes may modulate the effect of childhood adversities on decision making in suicide attempters. J Psychiatr Res. 2013;47(2):259–265. doi:10.1016/j.jpsychires.2012.10.014
42. Smith CE, Pisetsky EM, Wonderlich SA, et al. Is childhood trauma associated with lifetime suicide attempts in women with bulimia nervosa. Eat Weight Disord. 2016;21(2):199–204. doi:10.1007/s40519-015-0226-8
43. Yin H, Guo J, Xin Q, et al. Influence of the GABA receptor subunit gene polymorphism and childhood sexual abuse on processing speed in major depression and suicide attempt. Frontiers in Psychiatry. 2021;12:712231. doi:10.3389/fpsyt.2021.712231
44. Guillen-Burgos H, Moreno-Lopez S, Acevedo-Vergara K, Pérez-Florez M, Pachón-Garcia C, Gálvez-Flórez JF. Risk of childhood trauma exposure and severity of bipolar disorder in Colombia. Inter J Bipolar Disord. 2023;11(1):7. doi:10.1186/s40345-023-00289-5
45. Yin H, Galfalvy H, Pantazatos SP, et al. Glucocorticoid receptor-related genes: genotype and brain gene expression relationships to suicide and major depressive disorder. Depression Anxiety. 2016;33(6):531–540. doi:10.1002/da.22499
46. Watkeys OJ, Kremerskothen K, Quidé Y, Fullerton JM, Green MJ. Glucocorticoid receptor gene (NR3C1) DNA methylation in association with trauma, psychopathology, transcript expression, or genotypic variation: a systematic review. Neurosci Biobehav Rev. 2018;95:85–122. doi:10.1016/j.neubiorev.2018.08.017
47. López JF, Chalmers DT, Little KY, Watson SJ. Bennett Research Award. Regulation of serotonin1A, glucocorticoid, and mineralocorticoid receptor in rat and human hippocampus: implications for the neurobiology of depression. Biol. Psychiatry. 1998;43(8):547–573. doi:10.1016/s0006-3223(97)00484-8
48. Degtyareva AO, Antontseva EV, Merkulova TI. Regulatory SNPs: altered transcription factor binding sites implicated in complex traits and diseases. Int J Mol Sci. 2021;22(12):6454. doi:10.3390/ijms22126454
49. Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022;50(D1):D165–D173. doi:10.1093/nar/gkab1113
50. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi:10.1016/j.cell.2007.11.019
51. Cho NH, Cheveralls KC, Brunner AD, et al. OpenCell: endogenous tagging for the cartography of human cellular organization. Science. 2022;375(6585):eabi6983. doi:10.1126/science.abi6983
52. Spörl F, Korge S, Jürchott K, et al. Krüppel-like factor 9 is a circadian transcription factor in human epidermis that controls proliferation of keratinocytes. Proc Natl Acad Sci USA. 2012;109(27):10903–10908. doi:10.1073/pnas.1118641109
53. Liu JN, Kong XS, Sun P, Wang R, Li W, Chen QF. An integrated pan-cancer analysis of TFAP4 aberrations and the potential clinical implications for cancer immunity. J Cell & Mol Med. 2021;25(4):2082–2097. doi:10.1111/jcmm.16147
54. Taketani T, Taki T, Shibuya N, et al. The HOXD11 gene is fused to the NUP98 gene in acute myeloid leukemia with t(2;11)(q31;p15). Cancer Res. 2002;62(1):33–37.
55. Kelleher I, Harley M, Lynch F, Arseneault L, Fitzpatrick C, Cannon M. Associations between childhood trauma, bullying and psychotic symptoms among a school-based adolescent sample. Br J Psychiatry. 2008;193(5):378–382. doi:10.1192/bjp.bp.108.049536
56. De Venter M, Van Den Eede F, Pattyn T, et al. Impact of childhood trauma on course of panic disorder: contribution of clinical and personality characteristics. Acta Psychiatr Scand. 2017;135(6):554–563. doi:10.1111/acps.12726
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.