Back to Journals » Veterinary Medicine: Research and Reports » Volume 13
Multidrug-Resistant Microbial Therapy Using Antimicrobial Peptides and the CRISPR/Cas9 System
Authors Getahun YA, Ali DA , Taye BW, Alemayehu YA
Received 16 March 2022
Accepted for publication 27 July 2022
Published 11 August 2022 Volume 2022:13 Pages 173—190
DOI https://doi.org/10.2147/VMRR.S366533
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Young Lyoo
Yared Abate Getahun,1 Destaw Asfaw Ali,2 Bihonegn Wodajnew Taye,3 Yismaw Alemie Alemayehu4
1Livestock and Fishery Research Center, College of Agriculture, Arba Minch University, Arba Minch, Southern Nation Nationalities and Peoples Regional State, Ethiopia; 2Department of Paraclinical Studies, College of Veterinary Medicine, Gondar University, Gondar City, Amhara Regional State, Ethiopia; 3Faculty of Veterinary Medicine, College of Agriculture, Assosa University, Assosa City, Benshangul Gumez Regional State, Ethiopia; 4Department of Animal Science, College of Agriculture, Wollega University, Nekemtie City, Oromia Regional State, Ethiopia
Correspondence: Yared Abate Getahun, Email [email protected]
Abstract: The emergence and spread of multidrug-resistant microbes become a serious threat to animal and human health globally because of their less responsiveness to conventional antimicrobial therapy. Multidrug-resistant microbial infection poses higher morbidity and mortality rate with significant economic losses. Currently, antimicrobial peptides and the CRISPR/Cas9 system are explored as alternative therapy to circumvent the challenges of multidrug-resistant organisms. Antimicrobial peptides are small molecular weight, cationic peptides extracted from all living organisms. It is a promising drug candidate for the treatment of multidrug-resistant microbes by direct microbial killing or indirectly modulating the innate immune system. The CRISPR/Cas9 system is another novel antimicrobial alternative used to manage multidrug-resistant microbial infection. It is a versatile gene-editing tool that uses engineered single guide RNA for targeted gene recognition and the Cas9 enzyme for the destruction of target nucleic acids. Both the CRISPR/Cas9 system and antimicrobial peptides were used to successfully treat nosocomial infections caused by ESKAPE pathogens, which developed resistance to various antimicrobials. Despite, their valuable roles in multidrug-resistant microbial treatments, both the antimicrobial peptides and the CRISPR/Cas systems have various limitations like toxicity, instability, and incurring high manufacturing costs. Thus, this review paper gives detailed explanations of the roles of the CRISPR/Cas9 system and antimicrobial peptides in circumventing the challenges of multidrug-resistant microbial infections, its limitation and prospects in clinical applications.
Keywords: antimicrobial peptides, clinical applications, CRISPR/cas system, multidrug-resistant organism
Introduction
Antimicrobials are the most important and useful therapeutic discovery in the history of medicine. It allows living beings to survive the microbial disease, enhances invasive surgical procedures, ensures animal health, and protects the food chain since 1928 after the discovery of penicillin. However, the indiscriminate use of antibiotics and disinfectants creates selective pressure on the microbes, which leads to the emergence of multidrug-resistant (MDR) microbes persisting as serious global health threats.1
Multidrug-resistant bacteria are defined as acquired non-susceptibility to at least one drug in three or more antimicrobial classes.2 The main mechanisms of action of resistance are outer membrane impermeability in gram-negative bacteria, production of drug degrading enzymes, efflux pumps, and modification of targets are the mechanisms used by bacteria to resist the toxic effect of antimicrobials. The pathogens acquire these resistance mechanisms intrinsically or via horizontal gene transfer or mutations as a consequence of exposure to different drugs.3–5 The common pathogen responsible for nosocomial infections and developing resistance to most of the antimicrobials is known to be “ESKAPE” by its acronym name, which stands for Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.6 Currently, 700, 000 people are dying each year globally from resistant microbial infections, with a projected cost of $100 trillion.7 Research findings propagated that there will be a total loss of effectiveness of all the antibiotics available today, with an estimated death of 10 million people each year from antibiotic-resistant bacteria by 2050 if actions have not been taken.8,9
Most conventional antimicrobials are unable to cure multidrug-resistant microbial infections. It also requires a long-time development process and incurs huge investment. The challenges of using conventional antimicrobials pave the way for the development and use of next-generation antimicrobials like the Clustered regularly interspaced short palindromic repeat (CRISPR/Cas9) and antimicrobial peptides to confront the spread of multidrug-resistant microorganisms (MDROs).10–12
Antimicrobial peptides are bioactive small proteins, produced by all living organisms as essential components of the innate immune system, as the first line of defence against microbial infection in eukaryotes, or produced as a competitive strategy in prokaryotes, to inhibit the growth of other unrelated microbial species.13 Antimicrobial peptides are used as a promising drug candidate to treat ESKAPE pathogens; which are not treated by conventional antimicrobials.14 The majority of the antimicrobial peptides are involved in microbial killing by directly acting on the cell membrane of the microbes, and few of them are involved in microbial killing by acting on the cell wall of the microbes, and others can also kill the microbes by inhibiting the protein and nucleic acid synthesis.13,15,16
The CRISPR/Cas9 system has also a great role in controlling the spread and emergence of multidrug-resistant microbes by knocking out target genes and making the microbes susceptible to conventional antimicrobials or killing the microbes as a result of valuable gene knockout.
This review focuses on the use of antimicrobial peptides and the CRISPR/Cas9 system as alternative antimicrobial therapy to circumvent the challenges of multidrug-resistant microbes and their limitations and prospects in clinical applications.
Antimicrobial Peptides: Multidrug-Resistant Microbial Therapy, Their Limitations and Prospects
Antibiotics are the most important discovery in the history of mankind that saves countless lives of animals and humans, but the effectiveness of antibiotics diminished through time led to the emergence and spread of multidrug-resistant microbes.17,18 Several factors can be considered for the current rise in antimicrobial resistance in hospitals and communities. Indiscriminate use of antibiotics in human and veterinary medicine, lack of proper regulatory mechanisms, and a decline in novel antimicrobial discovery and poor sanitation practices are the major factors responsible for the widespread multidrug-resistant microbes.19,20
The common MDR bacteria that cause nosocomial infection include methicillin-resistant Staphylococcus aureus (MRSA), MDR Pseudomonas aeruginosa, and carbapenem-resistant A. baumannii, Escherichia coli and K. pneumoniae, vancomycin-resistant enterococci (VRE) and extensive drug-resistant (XDR) tubercle bacilli.21 The emergence of multidrug-resistant bacteria has prompted the need to develop new strategies to address the problem of drug resistance.22 The impact of multidrug-resistant organisms is mitigated directly by treating infectious microbes with potent and efficacious new antimicrobial agents like antimicrobial peptides or indirectly by enhancing the immune status of the host through vaccination, feeding prebiotics, probiotics and passive antibody transfer as shown in (Figure 1).23
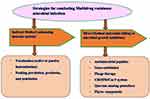 |
Figure 1 Alternative strategies to combat antimicrobial resistance. Data from Sharma et al.23 |
Antimicrobial peptides (AMPs) also known as host defence peptides have both microbicidal and immunomodulatory activities. It has been used as potent next-generation antibiotics since they are bioactive small proteins, naturally produced by all living organisms, and represent the first line of defence against microbial attacks in the eukaryotes or produced as a competitive strategy in prokaryotes, to limit the growth of other microorganisms.19,24 They are potent and broad-spectrum activity against bacteria, yeasts, fungi, viruses and parasites displaying bacteriostatic, microbicidal and cytolytic properties.25,26 AMP developed from synthetic and natural sources is currently used as an alternative therapy for multidrug-resistant microbial infection.27
The antimicrobial peptides drawn from bacteria known as bacteriocins have an important role in inhibiting or killing other bacterial strains with no self-harm.28 Bacteriocins provide important prospects for the development of alternative antibiotic strategies against different bacterial pathogens and are also useful in the food industry as a preservative.21 In 1939, the microbiologist Dubos isolated antimicrobial peptides, from the soil Bacillus strain and named it “gramicidin”, which had been demonstrated for protecting mice from pneumococcal infection.19 Colistin is also an important peptide drug derived from bacteria used for treating several MDR nosocomial pathogens such as methicillin-resistant Staphylococcus aureus (MRSA), multidrug-resistant P. aeruginosa, and carbapenem-resistant pathogens, but it has limited clinical application due to numerous side effects.29,30
Most therapeutically active AMPs are short having 10–50 amino acid length, and it has a positive net charge (ranges from +2 to +11) and an amphipathic structure, as they contain both hydrophobic and hydrophilic residues. Ionic AMPs are less common in therapeutic use and have a negative net charge ranging from (−1 to −7) usually isolated from vertebrates, invertebrates and plants.19 The biosynthesis of AMP takes place in three distinct ways: classical ribosomal synthesis through a ribosomal translation of mRNA in all species of life; non-ribosomal synthesis specifically, in filamentous fungi and bacteria; or proteolytic digestion of proteins produced from vertebrates, invertebrates and plants.19
Classification of Antimicrobial Peptides
Antimicrobial peptides are classified into various groups based on their source of extraction, activity on the microbes or cancer cells, structural characteristics, and amino acid-rich residues.24,32
Antimicrobial Peptide Classification Based on Source
Antimicrobial peptides are universal and essential components of the immune system in all life forms. Several AMPs have been discovered from both prokaryotic and eukaryotic organisms as promising antimicrobial agents against multidrug-resistant microbes.14,33 In eukaryotic organisms, antimicrobial peptide synthesis takes place in lymph, epithelial cells of the gastrointestinal and genitourinary systems, phagocyte and lymphocyte cells of the immune system. In higher organisms, AMPs are localized on the skin and mucosal epithelia exposed to the pathogen.19
Based on the source of extraction and discovery, AMPs are classified into mammalians, amphibians, microorganisms, and insects according to statistical data in the antimicrobial peptide database (APD 3) (Figure 2).19,32 The AMPs from amphibians, insects, mammals and fishes account for about 75.65% of therapeutically useful AMPs, whereas plants and microorganisms consist of the remaining 25% of the total AMP sources.34,35
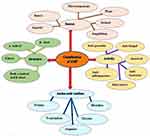 |
Figure 2 Classification of antimicrobial peptides. Adapted from Huan Y, Kong Q, Mou H, Yi H. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front Microbiol. 2020;11:582779.32 |
The antimicrobial peptides discovered in humans, sheep, cattle, and vertebrates mainly consist of cathelicidins and defensins. The defensin AMPs may be divided into α, β and θ defensins based on the position of disulfide bonds. Cathelicidin LL-37, a well-known AMP derived from the human body, is generally detected in the skin of infants, while human beta-defensin 2 (hBD-2) is frequently expressed in elder humans.32 Milk is a full source of AMPs, produced by enzymatic hydrolysis of milk. Several AMPs were recognized from α -lactalbumin, β -lactoglobulin, lactoferrin, and casein fractions.32,33 The common AMP extracted from amphibians is esculentins, brevinins, ranacyclins, ranatuerins, nigrocin-2, magainins, dermaseptins, bombinins, temporins, and japonicins-1 and −2, and palustrin-2.36 Antimicrobial peptides extracted from the frog skin have potent antimicrobial activity against antibiotic-resistant bacteria, protozoa, yeasts, and fungi through permeating and destroying the plasma membrane and inactivating intracellular targets. The Magainin is the famous amphibian AMP extracted from the skin secretions of frogs, which exhibits potent tumoricidal and broad-spectrum antimicrobial activities against bacteria, fungi, and protozoa by disrupting the pathogen membrane.21
Peptides extracted from marine sources have different functions and structures, as compared to peptides isolated from terrestrial sources, considering the different adaptive pressure on the organisms through the evolutionary process.37 The Cancrin AMP is the first AMP isolated from the sea amphibian Rana cancrivora.32 Cathelicidins (CATH BRALE and codCath1) derived from fish show potential antibacterial activity to a broad spectrum of Gram-positive and Gram-negative bacteria.38
Classification of Antimicrobial Peptides Based on Their Activity
Based on their activity, AMPs are divided into 18 categories: antibacterial, antiviral, antifungal, antiparasitic, anti-human immunodeficiency virus (HIV), and anti-tumour peptides as shown in (Figure 2). AMPs have antimicrobial activity in a huge variety of microorganisms via membrane disruption or non-membranolytic destruction of microorganisms.39 AMPs like nisin, cecropins and defensins have proven antimicrobial activity for both Gram-positive and Gram-negative microorganisms.19,32,33,40
Antifungal peptides (AFPs) are excellent alternative drug candidates for combating pathogenic fungi, including Candida albicans, Cryptococcus neoformans, Aspergillus fumigatus, Histoplasma capsulatum and molds. Many of the AFPs can also inhibit the growth of A. flavus, which is the causative agent of aflatoxins.32,41,42
Antiparasitic peptides are used for the treatment of numerous protozoa parasites including Leishmania spp., Plasmodium spp., Trypanosomes spp., and Schistosoma species. Antiprotozoal peptides are chemotherapeutics compounds, commonly originated from insects, and destabilize the protozoan homeostasis.43
Antiviral peptides (AVPs) display a robust killing effect on viruses by inhibiting viruses at different stages of their life cycle, such as entry, attachment, replication, transcription, translation, maturation, and release. AVPs act via three main mechanisms: some inhibit virus attachment and its fusion to the cell membrane; some disrupt the envelope of target viruses, and a few others interact with viral polymerase causing the inhibition of virus duplication.43
Anticancer peptides show anticancer activity by a mechanism of recruiting immune cells (including dendritic cells) to kill cancer cells, inducing necrosis or apoptosis, inhibiting angiogenesis to prevent tumour metastasis, and activating regulatory useful proteins to intervene with the gene transcription and translation of tumour cells.32
Classification of Antimicrobial Peptides Based on Their Structure
Based on their secondary structure, antimicrobial peptides are categorized into four groups: helical peptides, β-strand/sheet peptides, mixed helical/sheet peptides and prolonged non-helical/sheet peptides (Figure 2). Most AMPs undergo a conformational change from a flexible unstructured solution to a rigid conformation when interacting with membranes. The presence of disulfide bridges is required for structural stabilization and serves the biological function of the peptide. Salt bridge and head-to-tail cyclization are additional factors that support the overall stability of the peptide’s secondary structure.18,44
The α -helix AMPs are abundant in nature and can be isolated from several species such as plants, insects, amphibians, fish and mammals. The LL37, produced by neutrophils and epithelial cells, and the human lactoferrin, produced by the iron-binding glycoprotein lactoferrin and found in milk and exocrine secretions, are the two most common AMPs within the α-helical groups.19,45
Beta-sheet AMPs consist of at least two β-strands, and many linear systems employ β-hairpin-like conformations. Most members of this AMP contain conserved cysteine residues organized to form disulfide bonds and form amphipathic molecules. Beta-sheet AMP has a more stable structure and does not undergo large conformational changes when interacting with phospholipid membranes.46 Defensin AMP is the main component of β-sheet AMP. Alpha-defensins are predominantly present in neutrophils, while β-defensins are predominantly secreted by epithelial cells of various tissues.19,33,44,47
The α-helix/β-sheet mixed structure stabilizes three or four disulfide bridges. The cysteine-stabilized α/β (CSαβ) structural motif is composed of a single α-helix and β-sheets with two or three antiparallel chains and is first recognized by the antibacterial insect defensin and scorpion neurotoxin.47
Physicochemical Properties of Therapeutically Used Antimicrobial Peptides
In the design and preparation of AMPs, individuals should look at characteristics like the secondary structure, charge, hydrophobicity, hydrophobic moment, amphipathicity, polar angle, and peptide length that are responsible for modulating the antibiotic activity of AMPs.39 The AMPs are usually less than 100 amino acid residues long, having a positive net charge (+2 to +11), with high lysine and arginine content, and a significant number of hydrophobic residues are preferable in antimicrobial therapeutics.24 The cationic nature can be attributed to the presence of lysine and arginine and sometimes histidine residues.46,48
Hydrophobicity is also a key characteristic of all AMPs and it is at least 50% of hydrophobic residues consisting of valine, leucine, isoleucine, alanine, methionine, phenylalanine, tyrosine and tryptophan integrated within the antimicrobial peptide sequence.19,49 Hydrophobicity governs the quantity to which the water-soluble AMPs might be capable of partitioning into the membrane lipid bilayer. Hydrophobic movement and polar angle are also responsible for the interactions among the peptide and the membrane after the peptide adheres to the membrane surface. It is needed for membrane permeabilization; however, immoderate ranges of hydrophobicity can result in mammalian cell toxicity and lack of antimicrobial selectivity.7,14,46 The amphipathicity of AMPs should also be considered, because of the capacity to function in a solution under hydrophobic and hydrophilic conditions. Amphipathicity allows for enhancing the inhibitory capacity of AMPs against microorganisms.39
Antimicrobial Peptides Mechanisms of Action to Combat Multidrug-Resistant Microbes
The primary function of antimicrobial peptides is the killing of invading pathogens; however, they also act as modulators of the innate immune system. Antimicrobial peptides (HDPs) are active against numerous microorganisms like viruses, fungi and parasites through disruption of the integrity of the membrane or modulating the immune response of the host to kill cancer cells and intracellular pathogens. Antimicrobial peptides also have good antimicrobial activities against biofilm-forming bacterial pathogens.9,19,31,50
Most of the AMPs work by disrupting the integrity of the cell membrane, while a few AMPs can kill microbes by inhibiting the intracellular process including protein synthesis, nucleic acid synthesis, enzyme activity or cell wall synthesis.43,45,51,52 The gramicidin, daptomycin, oritavancin, telavancin, and colistin AMPs are approved by the American Food and Drug Administration (FDA) for the treatment of various microbes, and they are membrane-active peptides.53,54
Antimicrobial peptides have interaction with phospholipids of the microbial cell membrane through the process of biochemical or biophysical affinity (electrostatic or hydrophobic interactions), structural adjustment of the target membrane, accumulation as much as an active stoichiometric level disrupting the target membrane through permeabilization or depolarization or inflicting some other direct/indirect abnormality in its function that may be transitory or permanently, and finally, having access to the membranes.19,33,52,55
The cytoplasmic membranes of both the Gram-positive and Gram-negative microorganisms are reached in phospholipids, phosphatidylglycerol, cardiolipin, and phosphatidylserine, which have negatively charged head groups, which are highly attractive for positively charged AMPs. The outer layer of mammalian cell membranes is neutral in charge, made up of phosphatidylethanolamine, phosphatidylcholine and sphingomyelin;24 such a membrane composition distinction between the microbes and the host enhances the selective toxicity of antimicrobial peptides towards microbes.56–58
The non-membrane target AMPs are divided into two categories: the ones that concentrate on the bacterial cell wall and the other is intracellular targets. Similar to conventional antibiotics, AMPs can inhibit bacterial cell wall synthesis. Most traditional antibiotics bind to unique proteins involved in the synthesis of the cell wall components, whereas AMPs regularly interact with numerous precursor molecules, which can be required for cell wall synthesis.46 Antimicrobial peptides like vancomycin, oritavancin, dalbavancin, and telavancin are among FDA authorized cell wall synthesis inhibitor peptides.53
Antimicrobial peptides can also traverse the cell barrier through direct penetration or endocytosis. Endocytosis follows either micropinocytosis or receptor-mediated pathways. The edeine, tuberactinomycins and dityromycin are peptides with this kind of action. The edeine antimicrobial pentapeptide binds to the (P-site) of each 30S subunits of the 70S ribosomes of the prokaryotes, thus, inhibiting protein synthesis.14,51,53 Anionic AMPs bind with other cationic AMPs to form cationic salt bridges, and then they interact with negatively charged microbial membranes, permitting their penetration into the cell. Once they reach the cytoplasm, they will attach to the ribosomes or inhibit ribonuclease activity.19
Antimicrobial peptides also have immunomodulatory activities. The immunomodulatory activities displayed through AMPs include activation of immune cells, enhanced chemotaxis of immune cell differentiation including dendritic cell maturation and initiation of adaptive immunity. Antimicrobial peptides stimulate angiogenesis, improve wound healing, and decrease scar formation through the repression of cytokine-mediated and Toll-like receptor (TLR) mediated release of proinflammatory cytokines and reactive oxygen species, induction of anti-inflammatory cytokines, and scavenging of bacterial endotoxins.43,53,59
Synergistic therapy of traditional antibiotics and antimicrobial peptides is a promising and cost-effective alternative to manage the problems of multidrug-resistant microbial infections and to solve the inadequacies of antimicrobial monotherapy.22 The microorganism is much less possibly developing resistance to antimicrobial cocktails. The synergy of AMPs with antibiotics allows bacterial pores open for longer durations, prevents pore repair, increases perturbation of bacterial intracellular functions and helps other bacterial killing mechanisms and decreases host toxicity; low concentrations of each antimicrobial are needed to carry out a large antimicrobial effect.60
The Advantage of Antimicrobial Peptides Over Conventional Antibiotics
A group of life-threatening nosocomial pathogens commonly known by their acronym name ESKAPE” for Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter are less susceptible to the majority of conventional antimicrobial treatments.61 The rapid emergence of multidrug-resistant pathogens and the reduced effectiveness of traditional antimicrobial agents have caused serious problems in the health sector. Multidrug-resistant microbes develop various virulence factors, which enhance the colonization, pathogenesis and escape from the antibiotic attack and immune system mounting. Thus, antimicrobial peptides (AMPs) offer excellent potential strategies for combating the various impacts of multidrug-resistant microorganisms.62
Compared to conventional antimicrobials, AMPs are less prone to resistance selection and show rapid killing effects, broad-spectrum activity, and good clinical effects on MDR microbial therapy.21 Another important advantage of antimicrobial peptides over traditional antimicrobial agents is the activation of the innate immune system to enhance the body’s defences against multidrug-resistant microbes.63 Besides their poor efficacy in treating nosocomial infections, conventional antibiotics disrupt the normal microflora, but antimicrobial peptides are specific to the pathogenic microbe.64 Antimicrobial peptides are synergistically administered with conventional antimicrobials; with little dose can efficiently treat multidrug-resistant microbes. For example, the combination of butaprotegulin 1 and human β-defensin, LL37 with intracellular targeted antibiotics (gentamicin, ofloxacin, and rifampicin) have a good effect against MRSA, Micrococcus luteus, A. baumannii, K. pneumoniae, P. aeruginosa, and E. coli. Some AMPs are currently being evaluated in clinical trials as new antimicrobial agents. Different scientific investigations are undergoing to develop effective MDR microbial therapy, and some of them are now used clinically for the treatment of multidrug-resistant microbial infection as indicated in (Table 1).65
 |
Table 1 FDA Approved Cell Wall and Cell Membrane Acting Antimicrobial Peptides |
Delivery Techniques of Antimicrobial Peptides
Similar to other proteins, AMPs are affected by proteolytic and gastric degradation, which makes oral AMP delivery challenging. Appropriate delivery vehicles are important to enhance AMP bioavailability and safety, keeping it from chemical and enzymatic degradation, stopping aggregation and improving controlled release.66
Delivery vehicles are regularly harnessed for drug administration to enhance the biological properties of therapeutic AMPs. Inorganic nanoparticles such as gold, silver, and silica nanoparticles; polymeric nanoparticles like chitosan and hyaluronic acid; lipid nanoparticles especially liposomes, solid lipid nanoparticles, and nanostructured lipid carriers can be used for the delivery of AMP.45,67 Currently, silver nanoparticles (AgNPs) and gold nanoparticles (AuNPs) are frequently used in the delivery of AMPs due to their efficient delivery as well as synergistic antimicrobial activity. In addition to these, silicon derivatives nanoparticles have paramount significance to deliver AMP against MDR microbes.22,29,67,68
Antimicrobial Peptides Clinical Applications in Veterinary Practices
Antimicrobial peptides used for livestock treatments are classified into two broad families, particularly cathelicidins and defensins. The cathelicidin family of AMPs incorporates small cationic AMPs which are stored in neutrophils and macrophages.69 They are a part of the innate immune system and are normally proteolytically active proteins having a conserved cathelicidin domain. The defensin groups of AMPs consist of six cysteine residues that form the three disulfide bonds.70
Among the different applications of AMPs used to counteract the spread of multidrug-resistant microbes in animals, some AMPs had shown promising therapeutic efficacy for the treatment of animal diseases. The Swine intestinal antimicrobial peptides (SIAMP) have a good antimicrobial effect for the treatment of infectious bronchitis virus (IBV) in chickens (Table 2). The research confirmed that broiler chickens have got an increase in body weight gain and feed performance in continuous heat stress conditions.71
 |
Table 2 Antimicrobial Peptides Extracted from Animals and Effective Against Multidrug Resistant-Microbial Therapy |
The Frog caerin, European sea bass dicentracin and NK-lysine peptides (NKLPs) have good inhibitory outcomes on Nodavirus, Septicaemic haemorrhagic virus, Infectious pancreatic necrosis virus and Spring viremia carp virus, which can be devastating to fish farming. The AMP in soybean meal fermented via Bacillus subtilis E20 successfully inhibits Vibrio parahaemolyticus and Vibrio alginolyticus.32 Liver expressed AMP extracted from Topmouth culter fish (LEAP-2) showed inhibitory activity on penicillin-resistant Aeromonas hydrophila.72
The ovine pulmonary surfactant-related anion peptide (SAAP), the primary discovery of anionic AMP with 5–7 aspartate residues, had antimicrobial activity to Mannheimia haemolytica of ovine, with the presence of Zn ions.38 The panteth cells secrete α-defensins in reaction to bacterial antigens, which include LPS and muramyl dipeptide to inhibit intestinal bacterial infections.19
AMPs like cathelicidin and temporin-SHd display inhibition activity towards parasite infections. Epi-1, a marine synthetic AMP, can remarkably inhibit Trichomonas vaginalis via way of destroying its membrane.32,73 Indolicidin belongs to the family of cathelicidin AMPs, found from bovine neutrophils as a tridecapeptide and confirmed a bactericidal activity on Staphylococcus aureus.74 The human- β defensin 3 (hBD-3), the sheep myeloid peptide (AMP-29), the rat cathelin related antimicrobial peptide (rCRAMP), and bovine myeloid antimicrobial peptide (BMAP-27) demonstrate robust microbicidal activity against pathogenic E. coli, Pseudomonas aeruginosa, MRSA, and Acinetobacter baumannii, inhibit biofilm formation and modulate the immune system of the host.21
Antimicrobial peptides like Bombinins H2 and H4 extracted from the skin secretions of frog species Bombina variegata inhibit Leishmaniasis disease. Antimicrobial peptides extracted from numerous Bacillus strains have proven inhibitory activity towards Shigella, Salmonella, E. coli, Staphylococcus aureus, and pneumonia. Antimicrobial peptides extracted from Pseudomonas display good antimicrobial activity against Shigella, Salmonella, E. coli, and Staphylococcus aureus.33
Antimicrobial Peptides Limitations
Despite their effective therapeutic use in the treatment of multidrug-resistant microbes, AMP also has unwanted traits like toxicity to eukaryotic cells, which could result in hemolysis, nephrotoxicity, immunogenicity, and neurotoxicity. Susceptibility to proteolysis by bacterial proteases; high production cost, lack of an appropriate delivery system to the target site, and undefined pharmacokinetic profile of AMPs are the other drawbacks of using AMPs for therapeutic purposes.14,75 In addition, their cationic and amphiphilic nature results in excessive binding to serum proteins after parenteral administration, with consequent rapid removal from blood circulation and accumulation within the reticuloendothelial system, thus, ensuing in toxic effects and decreased therapeutic activity.54,67,76
The Prospects of Antimicrobial Peptides in Clinical Applications
The ability to chemically modify the structure of antimicrobial peptides opens up almost unlimited possibilities for therapeutic applications. A variety of chemical modification strategies are developed to address the hurdles related to AMP therapeutics. Cyclization, the substitution of genetically encoded amino acids by noncanonical residues and N-/C-terminal adjustments facilitate keeping AMPs away from proteolytic degradation, increasing permeability and metabolic stability.69,77 For example, AMP Polybia-MPI, from the venom of the social wasp Polybia paulista has broad-spectrum activity and can destroy both Gram-positive and Gram-negative microorganisms and fungi. To prevent proteolytic degradation of polybia-MPI through the enzymes, the D-lysine’s substituted analogue (D-lys-MPI) and the D-enantiomer of polybia-MPI created and showed proven therapeutic effects (D-MPI).54 Another example of chemically modified AMP used for efficient therapeutics is S-thanatin produced by substitution of the 15th amino acid threonine with serine in the insect-derived thanatin, demonstrating high microorganism selectivity and broad antimicrobial activity.78
The topical application of AMPs promotes the migration of keratinocytes and fibroblasts and contributes considerably to an accelerated wound healing process. Parenteral administration of AMPs by using nanotechnological approaches avoids the main disadvantages of AMPs, such as instability and toxicity, and provides a controlled delivery profile with prolonged activity.54,78 Synergistic therapeutic applications of AMP with conventional antibiotics reduce the number of peptides needed for effective treatment, costs and extend the lifetime of conventional antibiotics.79
Applying CRISPR-Cas System as an Alternative Antimicrobials
The CRISPR/Cas system is an adaptive immune system of bacteria and archaea to protect themselves from invasive nucleic acids. The CRISPR motifs located in archaea and bacteria are not found in viral or eukaryotic genomes. Among the numerous bacterial and archaeal populations, around 36% of the bacteria and 75% of the archaea incorporate the CRISPR-Cas system in their genome. In 1987, Nakata et al first reported a set of highly homologous sequences in the 3’ end of E. coli iap (alkaline phosphatase) gene.8,80–82 In 2012, Doudna, Charpentier et al first reported that the CRISPR-associated protein 9 can be tailored to genome editing with a customized CRISPR RNA (crRNA) together with a transactivating CRISPR RNA (tracrRNA) or an artificial single guide RNA (sgRNA) which becomes a chimeric crRNA-tracrRNA hybrid.83 The genetic loci of the CRISPR-Cas system incorporate the CRISPR array, which is comprised of repeated sequences and smaller sized flanking sequences (spacers). The spacers of CRISPR arrays are known as protospacers, which are derived from the DNA of invading phages or plasmids. Cas proteins are key elements of the CRISPR system, which are encoded upstream of the CRISPR array to determine the system activity.84–86 The CRISPR is different from other repetitive DNA sequences in that the repeats are 21 bp to 37 bp length and are interspaced by similar-sized non-repetitive DNA, and they may be clustered in a single or numerous loci at the chromosome.87,88
Types of the CRISPR/Cas System
Based on the differences in the Cas protein content, the CRISPR/Cas systems are divided into two classes, six types (I–VI) and 33 subtypes. In this classification, class-1 consists of types I, III, and IV, together with sixteen subtypes that incorporate multiple Cas proteins as effector modules, which determine crRNA-binding complexes and mediate together in pre-crRNA processing and interference. Class 2 consists of type II, V, and VI, together with 17 subtypes that incorporate a single, large, multidomain crRNA-binding protein, which is involved in all activities required for interference in all variants, and in pre-crRNA processing in some variants.8,85,89,90
Each kind of CRISPR/Cas system possesses a special protein composition for expression, interference, and adaptation modules. They are differentiated by the presence of unique proteins they contain: Cas3 for type I, Cas9 for type II, Cas10 for type III, Csf1 for type IV and Cas12a (Cpf1) for Type V. The class-1 CRISPR/Cas system undertakes interference through the use of multi-Cas effector protein complex, while the Class 2 system uses a single effector protein for interference.80,91 Among the type II CRISPR/Cas systems, CRISPR/Cas9 usually isolated from Streptococcus pyogenes are extensively studied and carried out for gene edition, because of its simplicity, versatility, efficiency and specificity.8,84,85,92
Mechanisms of Action of the CRISPR/Cas9
The CRISPR–Cas9 system is designed to perform as a precision antimicrobial, able to eliminate drug-resistant microbes. The CRISPR–Cas9 selectively removes target genes involved in antibiotic resistance, biofilm formation, and a virulence factor.8,16
The mechanism of action of the CRISPR-Cas system has 3 stages: adaptation, expression, and interference. During the adaptation stage, about 30 bp of the invading foreign DNA integrates into the leader side of the CRISPR locus, and protospacer adjacent motifs (PAMs) had been decided on from spacer sequences of the host genome. During the expression stage, RNA is transcribed from the spacers of the CRISPR locus.8,93
The Cas9 is a dual RNA-guided DNA endonuclease required for interference and immunity in the type II CRISPR/Cas system. In addition to crRNA, Cas9 requires trans-activating crRNA (tracrRNA), a small RNA that bears complementarity to the repeat regions of crRNA. Engineered single-guide RNA (sgRNA) chimaera of tracrRNA: crRNA has been developed for genome engineering applications. Cas9 identifies target DNA via PAM popularity and subsequent base pairing of the guide RNA. If the target DNA shows enough complementarity to the RNA guide, Cas9 generates a blunt, double-strand break three bp upstream of the PAM.91,94,95 The single guide RNAs can be designed to target antibiotic resistance, virulence, or critical genes specific to the pathogens. Once Cas9 binds to its guide RNA, the complex is ready to search for complementary target DNA. Target search and recognition require both complementary base pairing among the 20-nucleotide spacer sequence and a protospacer in the target DNA, in addition to the presence of a conserved PAM sequence adjacent to the target site, which is important for precise recognition of the target genes. The PAM sequence is important for the discrimination between self- and non-self-sequence for selective toxicity.96
Depending on experimental design, targeting specific outcomes such as cell death or growth inhibition of the target bacterium, specific deletion of genes from pathogens, elimination of antibiotic resistance plasmids, or transcriptional repression of a target gene.85,96 The Cas9 use two nuclease domains: a well-conserved RuvC domain including the three split RuvC motifs and an HNH domain that is living in the centre of the protein. The RuvC domain cleaves non-complementary DNA strands, while the HNH domain cleaves complementary DNA strands at a specific site three bp from the NGG PAM sequence to produce a predominantly blunt-ended double-strand break.85,96,97
Use of the CRISPR/Cas9 System in the Treatment of Multidrug-Resistant Microbes
The power of nucleic acid destruction by RNA makes the CRISPR/Cas system a promising candidate for the development of next-generation antimicrobial agents to combat infectious diseases, especially those caused by AMR pathogens.88,98 The Cas9 nuclease in the CRISPR/Cas II system uses a guide RNA to identify the target DNA via Watson–Crick base pairing. The sequences present in the CRISPR-guided RNA are specific to the invader sequence so that it can easily replace this sequence with the sequence of interest to retarget the CRISPR/Cas9 nuclease.99
Depending on where the target gene is located, CRISPR/Cas system can be used as an antibacterial agent in two different approaches, a pathogen-focused approach and a gene-focused approach. Targeting specific regions of the bacterial chromosome is a pathogen-focused approach. This approach results in bacterial cell death and targets pathogenic strain killing. A gene-focused approach involves targeting plasmids that carry the AMR gene. This approach removes the plasmid and causes bacteria sensitive to antibiotics.100 Pathogen-focused approach can help treat well-defined infections or target-specific strains of mixed microbial communities, while gene-focused approaches are undefined. It can be used to treat bacterial infections and generally reduce the abundance of the AMR gene in microbial communities.99
Unlike conventional antimicrobials, the CRISPR/Cas system can distinguish between symbiotic and pathogenic species by highly specific sequence targeting. Scientists transformed E. coli and Staphylococcus aureus with a plasmid encoding a cas9-driven RNA that accurately degrades antibiotic resistance genes.99 Currently, this technology is mostly in the preclinical phase, and the main goal is to provide precision antimicrobials for multidrug-resistant microbial therapy. Among several clinical studies of the CRISPR/Cas9 antimicrobial therapy, only two of them have antimicrobial efficacy as compared to traditional antibiotics. For example, Manipulated cas9 and crRNA of the methicillin resistance gene (mecA) of a phagemid (pDB121:: mecA) for treating a clinical isolate of Staphylococcus aureus showed a reduction of the disease by 49.6% after treatment.8,83,84 Studies using a mouse skin colonization model showed that CRISPR/Cas9 in S. aureus significantly reduced S. aureus skin colonization compared to other treatment conditions.101 Another study by Rodrigues, McBride, Hullahalli, Palmer, Duerkop, chemotherapy102 showed that CRISPR/Cas9, which targets the erythromycin resistance gene ermB, significantly reduced the overall development of erythromycin-resistant E. faecalis in the intestine after treatment.
CRISPR-Cas9 Delivery Technique
CRISPR/Cas9 system delivery requires finding a golden means that combines safe delivery and powerful gene editing. The Cas9 and guide RNAs can be introduced in the form of DNA, RNA/mRNA, or ribonucleoprotein.85,92,103 The delivery methods are usually divided into physical (electroporation and microinjection), viral (lentiviral, adenoviral, and AAV vectors), and non-viral (plasmids, lipid and polymeric nanoparticles, and extracellular vesicles).104–106
Physical methods, such as microinjection and electroporation, are more applicable to ex vivo CRISPR-Cas9-based genome editing therapy. They offer a high yield of transfected cells. However, the harmfulness of those strategies restricts their suitability to change zygotes or for ex vivo experiments.107,108 Electroporation is an extensively used technique to deliver proteins and nucleic acids into mammalian cells.108 The permeability of the cell membrane is temporarily increased during electroporation, permitting proteins or nucleic acids to enter the cells. Electroporation is appropriate for all sorts of CRISPR-Cas9 systems, such as plasmid-based CRISPR-Cas9 systems, to deliver the mixture of Cas9 and the sgRNA or the Cas9/sgRNA RNP. The hurdles behind CRISPR/Cas9 delivery using electroporation are that plasmid DNA is only integrated into about 0.01% of the target cells as shown in (Table 3). In addition, electroporation induces a large number of cell death.97,109,110
 |
Table 3 The Pros and Cons of Using the Different CRISPR-Cas9 Delivery Techniques |
Both viral vectors and plasmids can deliver the Cas9 gene and gRNA concurrently or separately. Adeno-associated viruses (AAVs) are very good vectors for in vivo gene delivery and gene editing. They can infect each dividing and non-dividing cell in various tissues, mediate long time periods and robust transgene expression, induce minimum immunogenicity, and no longer induce disease by themselves.107
Lentivirus is another commonly used viral vector for gene therapy. In addition to its slight immunogenicity and long-term expression of transduced genes, lentivirus has high infection efficiency, even in non-dividing cells. This is important for gene modification in tissues including the liver, brain, and muscle.97,107,111
Direct delivery of the Cas9 protein complexed with sgRNA is the most widely studied method in recent years. The purified Cas9 protein is positively charged and can efficiently form a complex with sgRNA, which is referred to as Cas9/sgRNA ribonucleoprotein complexes (RNPs). Direct delivery of RNPs has several advantages, including rapid action, low off-target effect, high gene editing efficiency, no requirement of codon optimization and promote selection and reduced toxicity and immune responses. In addition, RNPs do now no longer rely on transcriptional or translational cellular machinery for particular enzymatic gene editing activity.97,105,106
CRISPR-Cas9 Clinical Applications
Gene therapy is a technique of converting the genetic makeup of the target host to produce disease-resistant or highly productive organisms or it can be done by knocking out microbial genetic material through specific gene editing techniques to kill or make it susceptible to antimicrobials. Therapeutic genes are introduced into the target cells of a patient by directly targeting a specific harmful gene to treat a disease. If we take tumour gene therapy as an example, numerous approaches including tumour-suppressor gene therapy, antisense gene therapy, drug-resistant gene therapy, and immune gene therapy have been implemented.112 Several oncogenes (including KRAS, 45 PI3K, IDH2, etc.) and tumour suppressor genes (p53, RB1, and VHL) display numerous mutations in most cancer affected patients.113,114
CRISPR/ Cas9 Genome Editing for the Treatment of Animal Disease
Research on genome editing in animals is focused on improving animal productivity performance (eg, meat, milk, and egg), health, and welfare. CRISPR-Cas has the capacity for use as an antimicrobial for the target cleavage of antibiotic-resistant genes located on plasmids. The CRISPR/Cas9 nickase mediated exogenous knock-in of natural resistance-associated macrophage protein-1 (NRAMP1) in bovine fetal fibroblasts (BFFs) is performed to create tuberculosis-resistant genetically modified cattle.50,95
The gene encodes for the major prion protein PrP (protease-resistant protein). Protein molecules can exist in multiple isoforms: the normal or cellular form symbolized as PrPC type and protease-resistant abnormal form which is symbolized as PrPSc(scrapie). PrPSc is an infectious protein that causes fatal disorders in bovines and humans such as bovine spongiform encephalopathy, Creutzfeldt–Jakob disease, and a chronic wasting disease has been knockout PRNP coding exon 3 using CRISPR/Cas9.50,115,116 Interleukin-10 receptor alpha gene (IL10RA) is a proinflammatory cytokine associated with Mycobacterium avium subspecies paratuberculosis (MAP) infection and mastitis in dairy cattle. The CRISPR/Cas9 gene-editing tool was used to knock out IL10RA gene in mammary alveolar-T cells (MAC-T cells).116 The removal of a virulent gene, virB10 or RpolA that cause Brucella infection has got a promising effect after transduction with the CRISPR/Cas9 carrying vector against bacterial RpolA. The study done by Rodrigues, McBride, Hullahalli, Palmer, and Duerkop102 engineered conjugative plasmid pPD1 with a complete, constitutively expressed CRISPR-Cas9 targeting cassette that efficiently transfers to Enterococcus faecalis for the selective elimination of ermB gene (encoding erythromycin resistance) and tetM gene (encoding tetracycline resistance).85,102
The Limitation and Prospects of the CRISPR/Cas9 System
The Limitations of the CRISPR-Cas9 Technology
Despite the numerous advantages and advancements in the CRISPR system, there are several limitations to the implementation of a successful gene-editing tool. The off-target effects are the most important situation for in vivo therapeutic applications in eukaryotic organisms. The target specificity depends on the gRNA of Cas9 and PAM sequences and the off-target cleavage in the genome; this ends in precise knockout of valuable alleles like disease resistance and heat tolerance, as well as haplotypes in local domestically well-adapted cattle breed genome.116–118
Another drawback of using the CRISPR/Cas9 is the requirement for a PAM close to the target site. The Cas9 from the bacteria Streptococcus pyogenes (SpCas9) is one of the most substantially used Cas9s with a relatively short established PAM recognition site. On the contrary, SpCas9 is quite big and hard to pack into AAV vectors.119 The CRISPR induced double-strand break (DSBs) frequently cause apoptosis regardless of the intended gene edit which requires safety concerns revealed during usage of this tool in human pluripotent stem cells (hPSCs) which demonstrated that tumor suppressor protein p53 activation in response to the toxic DSBs introduced by CRISPR system.118 Even though CRISPR has a significant therapeutic application, it can only be effective in all cell types which can carry out active mitosis. Homologous double-strand repair (HDR) to maintain gene knockout by the CRISPR/cas9 system is difficult to achieve in cells that are not mitotically dividing, like neuron cells.113
Prospects of Using the CRISPR/Cas9 System
To overcome the challenges associated with the CRISPR/Cas gene edition, scientists developed exceptional online gene-editing platforms and efficiently utilized them.118 Nowadays, researchers have also developed Cas9 variants that are particular to be engineered to reduce the OTEs and maintain editing efficacy.119
Direct delivery of the CRISPR/Cas9 system has a ribonucleoprotein (RNP) complex consisting of Cas9 protein and single-guide RNA (sgRNA) has emerged as an effective and widespread method for genome editing because of its benefits of transient genome editing and decreased off-target effects and cleaves the target sequences directly after delivery and, contrary to stable transfection, are degraded afterwards.80,87,120 To avoid the hurdles of HDR in low mitotically dividing cells, using non-homologous or microhomology mediated integration of cassettes has a promising effect.113
Conclusion and Recommendations
Antimicrobials have a great impact on the improvement of life. However, abusive use of antimicrobials in the health care and agricultural sector has led to the emergence and spread of multidrug-resistant microbes globally; which are difficult to treat with conventional antimicrobials. Currently, alternative antimicrobials like antimicrobial peptides and the CRISPR/Cas9 are explored and used as promising drug candidates to circumvent the trouble of multidrug-resistant microbial infections. Despite, their role to tackle the spread and emergence of multidrug-resistant microbes, antimicrobial peptides are prone to proteolytic enzymes, cytotoxic, lack efficient delivery systems and require high manufacturing costs. The off-target effect in the CRISPR/Cas system is the major problem observed in the gene-editing function of the CRISPR/Cas9 system.
It has been recommended to apply global communications on the adoption of alternative antimicrobials, reducing the toxicity and enhancing the therapeutic efficacy of antimicrobials using bioinformatics tools during the selection and formulation of antimicrobial peptides and the CRISPR/Cas9 system. Using appropriate drug delivery materials like nanoparticles is also recommended to administer alternative antimicrobials as well as conventional drugs.
Abbreviations
AAV, adeno-associated vector; AFP, anti-fungal peptides; AMP, antimicrobial peptides; APD, antimicrobial peptide database; CRISPR, cluster regularly interspaced palindromic repeat; FDA, Food and Drug Administration; MDRO, multidrug-resistant organisms; MRSA, methicillin-resistant Staphylococcus aureus; OTEs, off target effects; PAM, protospacer adjacent motif; RNP, ribonuclear protein.
Author Contributions
All authors made significant contributions to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval for the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The authors did not receive funds for manuscript preparations.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Abo-Shama UH, El-Gendy H, Mousa WS, et al. Synergistic and antagonistic effects of metal nanoparticles in combination with antibiotics against some reference strains of pathogenic microorganisms. Infect Drug Resist. 2020;13:351–362. doi:10.2147/IDR.S234425
2. Dakal TC, Kumar A, Majumdar RS, Yadav V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front Microbiol. 2016;7:1831. doi:10.3389/fmicb.2016.01831
3. Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4(3):482–501. doi:10.3934/microbiol.2018.3.482
4. Peterson E, Kaur P. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front Microbiol. 2018;9:2928. doi:10.3389/fmicb.2018.02928
5. Morrison L, Zembower TR. Antimicrobial Resistance. Gastrointest Endosc Clin N Am. 2020;30(4):619–635. doi:10.1016/j.giec.2020.06.004
6. Leon-Buitimea A, Garza-Cardenas CR, Garza-Cervantes JA, Lerma-Escalera JA, Morones-Ramirez JR. The demand for new antibiotics: antimicrobial peptides, nanoparticles, and combinatorial therapies as future strategies in antibacterial agent design. Front Microbiol. 2020;11:1669. doi:10.3389/fmicb.2020.01669
7. Gan BH, Gaynord J, Rowe SM, Deingruber T, Spring DR. The multifaceted nature of antimicrobial peptides: current synthetic chemistry approaches and future directions. Chem Soc Rev. 2021;50(13):7820–7880. doi:10.1039/d0cs00729c
8. Gholizadeh P, Köse Ş, Dao S, et al. How CRISPR-Cas system could be used to combat antimicrobial resistance. Infect Drug Resist. 2020;13:1111. doi:10.2147/IDR.S247271
9. Bechinger B, Gorr SU. Antimicrobial peptides: mechanisms of action and resistance. J Dent Res. 2017;96(3):254–260. doi:10.1177/0022034516679973
10. Eleraky NE, Allam A, Hassan SB, Omar MM. Nanomedicine fight against antibacterial resistance: an overview of the recent pharmaceutical innovations. Pharmaceutics. 2020;12(2):142. doi:10.3390/pharmaceutics12020142
11. Vivas R, Barbosa AAT, Dolabela SS, Jain S. Multidrug-resistant bacteria and alternative methods to control them: an overview. J Microbial Drug Resist. 2019;25(6):890–908. doi:10.1089/mdr.2018.0319
12. Tucker AT, Leonard SP, DuBois CD, et al. Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. J Cell. 2018;172(3):618–628 e613. doi:10.1016/j.cell.2017.12.009
13. Rima M, Rima M, Fajloun Z, Sabatier J-M, Bechinger B, Naas TJA. Antimicrobial peptides: a potent alternative to antibiotics. Antibiotics. 2021;10(9):1095. doi:10.3390/antibiotics10091095
14. Biswaro LS, da Costa Sousa MG, Rezende TMB, Dias SC, Franco OL. Antimicrobial peptides and nanotechnology, recent advances and challenges. Front Microbiol. 2018;9:855. doi:10.3389/fmicb.2018.00855
15. Taati Moghadam M, Amirmozafari N, Shariati A, et al. How phages overcome the challenges of drug resistant bacteria in clinical infections. Infect Drug Resist. 2020;13:45–61. doi:10.2147/IDR.S234353
16. Lima R, Del Fiol FS, Balcao VM. Prospects for the use of new technologies to combat multidrug-resistant bacteria. Front Pharmacol. 2019;10:692. doi:10.3389/fphar.2019.00692
17. Yang X, Ye W, Qi Y, Ying Y, Xia Z. Overcoming multidrug resistance in bacteria through antibiotics delivery in surface-engineered nano-cargos: recent developments for future nano-antibiotics. Front Bioeng Biotechnol. 2021;9:696514. doi:10.3389/fbioe.2021.696514
18. Lee TH, Hall KN, Aguilar MI. Antimicrobial peptide structure and mechanism of action: a focus on the role of membrane structure. Curr Top Med Chem. 2016;16(1):25–39. doi:10.2174/1568026615666150703121700
19. Moretta A, Scieuzo C, Petrone AM, et al. Antimicrobial peptides: a new hope in biomedical and pharmaceutical fields. Front Cell Infect Microbiol. 2021;11:668632. doi:10.3389/fcimb.2021.668632
20. Ashraf M, Mustafa B-E, Rehman S-U, Khalid Bashir M, Adnan Ashraf M. Emergence of antimicrobial resistance, causes, molecular mechanisms, and prevention strategies: a bovine perspective. Bovine Science A Key to Sustainable Development; 2019:45.
21. Mwangi J, Hao X, Lai R, Zhang ZY. Antimicrobial peptides: new hope in the war against multidrug resistance. Zool Res. 2019;40(6):488–505. doi:10.24272/j.issn.2095-8137.2019.062
22. Sheard DE, O’Brien-Simpson NM, Wade JD, Separovic F. Combating bacterial resistance by combination of antibiotics with antimicrobial peptides. J Pure Applied Chem. 2019;91(2):199–209. doi:10.1515/pac-2018-0707
23. Sharma C, Rokana N, Chandra M, et al. Antimicrobial resistance: its surveillance, impact, and alternative management strategies in dairy animals. Front Vet Sci. 2017;4:237. doi:10.3389/fvets.2017.00237
24. Amerikova M, Pencheva El-Tibi I, Maslarska V, Bozhanov S, Tachkov K. Antimicrobial activity, mechanism of action, and methods for stabilisation of defensins as new therapeutic agents. Biotechnol Biotechnol Equip. 2019;33(1):671–682. doi:10.1080/13102818.2019.1611385
25. Pacios O, Blasco L, Bleriot I, et al. Strategies to combat multidrug-resistant and persistent infectious diseases. Antibiotics. 2020;9(2):65. doi:10.3390/antibiotics9020065
26. Mahlapuu M, Hakansson J, Ringstad L, Bjorn C. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol. 2016;6:194. doi:10.3389/fcimb.2016.00194
27. Kang HK, Kim C, Seo CH, Park Y. The therapeutic applications of antimicrobial peptides (AMPs): a patent review. J Microbiol. 2017;55(1):1–12. doi:10.1007/s12275-017-6452-1
28. Meade E, Slattery MA, Garvey M. Bacteriocins, potent antimicrobial peptides and the fight against multi drug resistant species: resistance is futile? Antibiotics. 2020;9(1):32. doi:10.3390/antibiotics9010032
29. Chung CR, Jhong JH, Wang Z, et al. Characterization and identification of natural antimicrobial peptides on different organisms. Int J Mol Sci. 2020;21(3):986. doi:10.3390/ijms21030986
30. Soltani S, Hammami R, Cotter PD, et al. Bacteriocins as a new generation of antimicrobials: toxicity aspects and regulations. J FEMS Microbiol Rev. 2021;45(1):fuaa039.
31. Raheem N, Straus SK. Mechanisms of action for antimicrobial peptides with antibacterial and antibiofilm functions. Front Microbiol. 2019;10:2866. doi:10.3389/fmicb.2019.02866
32. Huan Y, Kong Q, Mou H, Yi H. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front Microbiol. 2020;11:582779. doi:10.3389/fmicb.2020.582779
33. Boparai JK, Sharma PK. Mini review on antimicrobial peptides, sources, mechanism and recent applications. Protein Pept Lett. 2020;27(1):4–16. doi:10.2174/0929866526666190822165812
34. Erdem Büyükkiraz M, Kesmen Z. Antimicrobial peptides (AMPs): a promising class of antimicrobial compounds. J Appl Microbiol. 2021;132:1–24.
35. Hazam PK, Goyal R, Ramakrishnan V. Peptide based antimicrobials: design strategies and therapeutic potential. Prog Biophys Mol Biol. 2019;142:10–22. doi:10.1016/j.pbiomolbio.2018.08.006
36. Patocka J, Nepovimova E, Klimova B, Wu Q, Kuca K. Antimicrobial peptides: amphibian host defense peptides. J Current Med Chem. 2019;26(32):5924–5946. doi:10.2174/0929867325666180713125314
37. Macedo MWFS, Cunha N, Carneiro JA, et al. Marine organisms as a rich source of biologically active peptides. Front Marine Sci. 2021;8:889. doi:10.3389/fmars.2021.667764
38. Zhang QY, Yan Z-B, Meng Y-M, et al. Antimicrobial peptides: mechanism of action, activity and clinical potential. Military Med Res. 2021;8(1):48. doi:10.1186/s40779-021-00343-2
39. Sultana A, Luo H, Ramakrishna S. Antimicrobial peptides and their applications in biomedical sector. Antibiotics. 2021;10:9. doi:10.3390/antibiotics10091094
40. Gonzalez-Garcia M, Morales-Vicente F, Pico ED, et al. Antimicrobial activity of cyclic-monomeric and dimeric derivatives of the snail-derived peptide Cm-p5 against viral and multidrug-resistant bacterial strains. Biomolecules. 2021;11(5):5. doi:10.3390/biom11050745
41. Thery T, Lynch KM, Arendt EK. Natural antifungal peptides/proteins as model for novel food preservatives. Compr Rev Food Sci Food Safety. 2019;18(5):1327–1360. doi:10.1111/1541-4337.12480
42. Fernandez de Ullivarri M, Arbulu S, Garcia-Gutierrez E, Cotter PD. Antifungal peptides as therapeutic agents. Front Cell Infect Microbiol. 2020;10(105). doi:10.3389/fcimb.2020.00105
43. Moravej H, Moravej Z, Yazdanparast M, et al. Antimicrobial peptides: features, action, and their resistance mechanisms in bacteria. Microb Drug Resist. 2018;24(6):747–767. doi:10.1089/mdr.2017.0392
44. Bin Hafeez A, Jiang X, Bergen PJ, Zhu Y. Antimicrobial peptides: an update on classifications and databases. Int J Mol Sci. 2021;22(21):21. doi:10.3390/ijms222111691
45. Thapa RK, Diep DB, Tønnesen HH. Nanomedicine-based antimicrobial peptide delivery for bacterial infections: recent Advances and Future Prospects. J Pharm Investig. 2021;51(4):377–398. doi:10.1007/s40005-021-00525-z
46. Kumar P, Kizhakkedathu JN, Straus SK. Antimicrobial peptides: diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules. 2018;8(1):4. doi:10.3390/biom8010004
47. Erdem Büyükkiraz M, Kesmen ZJJ. Antimicrobial peptides (AMPs): a promising class of antimicrobial compounds. J Appli Microbiol. 2022;132(3):1573–1596. doi:10.1111/jam.15314
48. Pfalzgraff A, Brandenburg K, Weindl G. Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. J Front Pharmacol. 2018;9:281. doi:10.3389/fphar.2018.00281
49. Aronica PGA, Reid LM, Desai N, et al. Computational methods and tools in antimicrobial peptide research. J Chem Inf Model. 2021;61(7):3172–3196. doi:10.1021/acs.jcim.1c00175
50. Zharkova MS, Orlov DS, Golubeva OY, et al. Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics—a novel way to combat antibiotic resistance? J Front Cellular Infect Microbiol. 2019;9:128. doi:10.3389/fcimb.2019.00128
51. Le CF, Fang CM, Sekaran SD. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob Agents Chemother. 2017;61(4):e02340–02316. doi:10.1128/AAC.02340-16
52. Corrêa JAF, Evangelista AG, De Melo Nazareth T, Luciano FB. Fundamentals on the molecular mechanism of action of antimicrobial peptides. Materialia. 2019;8:100494. doi:10.1016/j.mtla.2019.100494
53. Chen CH, Lu TK. Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics. 2020;9(1):24. doi:10.3390/antibiotics9010024
54. Divyashree M, Mani MK, Reddy D, et al. Clinical applications of antimicrobial peptides (AMPs): where do we stand now? Protein Pept Lett. 2020;27(2):120–134. doi:10.2174/0929866526666190925152957
55. Benfield AH, Henriques ST. Mode-of-action of antimicrobial peptides: membrane disruption versus intracellular mechanisms. Front Med Technol. 2020;2:20. doi:10.3389/fmedt.2020.610997
56. Carratala JV, Serna N, Villaverde A, Vazquez E, Ferrer-Miralles N. Nanostructured antimicrobial peptides: the last push towards clinics. Biotechnol Adv. 2020;44:107603. doi:10.1016/j.biotechadv.2020.107603
57. Mishra B, Reiling S, Zarena D, Wang G. Host defense antimicrobial peptides as antibiotics: design and application strategies. Curr Opin Chem Biol. 2017;38:87–96. doi:10.1016/j.cbpa.2017.03.014
58. Lei J, Sun L, Huang S, et al. The antimicrobial peptides and their potential clinical applications. Am J Transl Res. 2019;11(7):3919–3931.
59. Mahlapuu M, Bjorn C, Ekblom J. Antimicrobial peptides as therapeutic agents: opportunities and challenges. Crit Rev Biotechnol. 2020;40(7):978–992. doi:10.1080/07388551.2020.1796576
60. Duong L, Gross SP, Siryaporn A. Developing antimicrobial synergy with AMPs. Front Med Technol. 2021;3:9. doi:10.3389/fmedt.2021.640981
61. Pandey R, Mishra SK, Shrestha A. Characterisation of ESKAPE pathogens with special reference to multidrug resistance and biofilm production in a Nepalese hospital. Infect Drug Resist. 2021;14:2201–2212. doi:10.2147/IDR.S306688
62. Park SC, Park Y, Hahm KS. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int J Mol Sci. 2011;12(9):5971–5992. doi:10.3390/ijms12095971
63. Rima M, Rima M, Fajloun Z, Sabatier JM, Bechinger B, Naas T. Antimicrobial peptides: a potent alternative to antibiotics. Antibiotics. 2021;10(9):1095.
64. Koeninger L, Osbelt L, Berscheid A, et al. Curbing gastrointestinal infections by defensin fragment modifications without harming commensal microbiota. Commun Biol. 2021;4(1):47. doi:10.1038/s42003-020-01582-0
65. Magana M, Pushpanathan M, Santos AL, et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect Dis. 2020;20(9):e216–e230. doi:10.1016/S1473-3099(20)30327-3
66. Drayton M, Kizhakkedathu JN, Straus SK. Towards robust delivery of antimicrobial peptides to combat bacterial resistance. Molecules. 2020;25(13):3048. doi:10.3390/molecules25133048
67. Teixeira MC, Carbone C, Sousa MC, et al. Nanomedicines for the Delivery of Antimicrobial Peptides (AMPs). Nanomaterials. 2020;10(3):560. doi:10.3390/nano10030560
68. Criel B, Taelman S, Van Criekinge W, Stock M, Briers Y. PhaLP: a database for the study of phage lytic proteins and their evolution. J Viruses. 2021;13(7):1240. doi:10.3390/v13071240
69. Browne K, Chakraborty S, Chen R, et al. A new era of antibiotics: the clinical potential of antimicrobial peptides. Int J Mol Sci. 2020;21(19):7047. doi:10.3390/ijms21197047
70. Kumar R, Ali SA, Singh SK, et al. Antimicrobial peptides in farm animals: an updated review on its diversity, function, modes of action and therapeutic prospects. Vet Sci. 2020;7(4):206. doi:10.3390/vetsci7040206
71. Shi S, Shen T, Liu Y, Chen L, Wang C, Liao C. Porcine myeloid antimicrobial peptides: a review of the activity and latest advances. Front Vet Sci. 2021;8:664139. doi:10.3389/fvets.2021.664139
72. Chen Y, Wu J, Cheng H, et al. Anti-infective effects of a fish-derived antimicrobial peptide against drug-resistant bacteria and its synergistic effects with antibiotic. Front Microbiol. 2020;11:602412. doi:10.3389/fmicb.2020.602412
73. Neshani A, Zare H, Akbari Eidgahi MR, Khaledi A, Ghazvini K. Epinecidin-1, a highly potent marine antimicrobial peptide with anticancer and immunomodulatory activities. BMC Pharmacol Toxicol. 2019;20(1):33. doi:10.1186/s40360-019-0309-7
74. Guo Y, Xun M, Han J. A bovine myeloid antimicrobial peptide (BMAP-28) and its analogs kill pan-drug-resistant Acinetobacter baumannii by interacting with outer membrane protein A (OmpA). Medicine. 2018;97(42):e12832. doi:10.1097/MD.0000000000012832
75. Sarkar T, Chetia M, Chatterjee S. Antimicrobial peptides and proteins: from nature’s reservoir to the laboratory and beyond. J Fronti Chemi. 2021;9:432.
76. Pervin Z, Hassan MM. Synergistic therapeutic actions of antimicrobial peptides to treat multidrug-resistant bacterial infection. J Revi Med Microbiol. 2021;32(2):83–89. doi:10.1097/MRM.0000000000000239
77. Costa F, Teixeira C, Gomes P, Martins MCL. Clinical application of AMPs. Adv Exp Med Biol. 2019;1117:281–298.
78. Gera S, Kankuri E, Kogermann K. Antimicrobial peptides - Unleashing their therapeutic potential using nanotechnology. Pharmacol Ther. 2021;232:107990. doi:10.1016/j.pharmthera.2021.107990
79. Dijksteel GS, Ulrich MMW, Middelkoop E, Boekema B. Review: lessons learned from clinical trials using Antimicrobial Peptides (AMPs). Front Microbiol. 2021;12:616979. doi:10.3389/fmicb.2021.616979
80. Zhang S, Shen J, Li D, Cheng Y. Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing. Theranostics. 2021;11(2):614–648. doi:10.7150/thno.47007
81. Roy S, Naha S, Rao A, Basu S. CRISPR-Cas system, antibiotic resistance and virulence in bacteria: through a common lens. J Prog Mol Biol Transl Sci. 2021;178:123–174.
82. Goren M, Yosef I, Qimron U. Sensitizing pathogens to antibiotics using the CRISPR-Cas system. Drug Resist Updat. 2017;30:1–6. doi:10.1016/j.drup.2016.11.001
83. Palacios Araya D, Palmer KL, Duerkop BA. CRISPR-based antimicrobials to obstruct antibiotic-resistant and pathogenic bacteria. PLoS Pathog. 2021;17(7):e1009672. doi:10.1371/journal.ppat.1009672
84. Aslam B, Rasool M, Idris A, et al. CRISPR-Cas system: a potential alternative tool to cope antibiotic resistance. Antimicrob Resist Infect Control. 2020;9(1):131. doi:10.1186/s13756-020-00795-6
85. Wan F, Draz MS, Gu M, Yu W, Ruan Z, Luo Q. Novel strategy to combat antibiotic resistance: a sight into the combination of CRISPR/Cas9 and nanoparticles. J Pharmaceutics. 2021;13(3):352. doi:10.3390/pharmaceutics13030352
86. Zohra T, Numan M, Ikram A, et al. Cracking the Challenge of Antimicrobial Drug Resistance with CRISPR/Cas9, nanotechnology and other strategies in ESKAPE pathogens. Microorganisms. 2021;9(5):954. doi:10.3390/microorganisms9050954
87. Horodecka K, Duchler M. CRISPR/Cas9: principle, applications, and delivery through extracellular vesicles. Int J Mol Sci. 2021;22(11):6072. doi:10.3390/ijms22116072
88. Duan C, Cao H, Zhang LH, Xu Z. Harnessing the CRISPR-Cas systems to combat antimicrobial resistance. Front Microbiol. 2021;12:716064. doi:10.3389/fmicb.2021.716064
89. Bikard D, Barrangou R. Using CRISPR-Cas systems as antimicrobials. Curr Opin Microbiol. 2017;37:155–160. doi:10.1016/j.mib.2017.08.005
90. Khadempar S, Familghadakchi S, Motlagh RA, et al. CRISPR-Cas9 in genome editing: its function and medical applications. J Cell Physiol. 2019;234(5):5751–5761. doi:10.1002/jcp.27476
91. Hille F, Richter H, Wong SP, Bratovic M, Ressel S, Charpentier E. The biology of CRISPR-cas: backward and forward. Cell. 2018;172(6):1239–1259. doi:10.1016/j.cell.2017.11.032
92. Loureiro A, Da Silva GJ. CRISPR-Cas: converting a bacterial defence mechanism into A State-of-the-art genetic manipulation tool. Antibiotics. 2019;8(1):18. doi:10.3390/antibiotics8010018
93. Kamruzzaman M, Iredell JR. CRISPR-cas system in antibiotic resistance plasmids in Klebsiella pneumoniae. Front Microbiol. 2019;10:2934. doi:10.3389/fmicb.2019.02934
94. Hussain W, Mahmood T, Hussain J, et al. CRISPR/Cas system: a game changing genome editing technology, to treat human genetic diseases. Gene. 2019;685:70–75. doi:10.1016/j.gene.2018.10.072
95. de la Fuente-Núñez C, Lu TK. CRISPR-Cas9 technology: applications in genome engineering, development of sequence-specific antimicrobials, and future prospects. J Integrative Biol. 2017;9(2):109–122. doi:10.1039/c6ib00140h
96. Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi:10.1146/annurev-biophys-062215-010822
97. Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17–26. doi:10.1016/j.jconrel.2017.09.012
98. Serajian S, Ahmadpour E, Oliveira SMR, Pereira M, Heidarzadeh S. CRISPR-cas technology: emerging applications in clinical microbiology and infectious diseases. J Pharmaceuticals. 2021;14(11):1171. doi:10.3390/ph14111171
99. Shabbir MAB, Shabbir MZ, Wu Q, et al. CRISPR-cas system: biological function in microbes and its use to treat antimicrobial resistant pathogens. Ann Clin Microbiol Antimicrob. 2019;18(1):21. doi:10.1186/s12941-019-0317-x
100. Ekwebelem OC, Aleke J, Ofielu E, Nnorom-Dike O. CRISPR-Cas9 system: a revolutionary tool in the fight against antimicrobial resistance. Infect Microb Dis. 2021;3(2):51–56. doi:10.1097/IM9.0000000000000049
101. Wang P, He D, Li B, et al. Eliminating mcr-1-harbouring plasmids in clinical isolates using the CRISPR/Cas9 system. J Antimicrob Chemother. 2019;74(9):2559–2565. doi:10.1093/jac/dkz246
102. Rodrigues M, McBride SW, Hullahalli K, Palmer KL, Duerkop BA. Conjugative delivery of CRISPR-Cas9 for the selective depletion of antibiotic-resistant enterococci. J Antimicrobial Agents Chemother. 2019;63(11):e01454–01419.
103. Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018;25(1):1234–1257. doi:10.1080/10717544.2018.1474964
104. Elaswad A, Khalil K, Cline D, et al.Microinjection of CRISPR/Cas9 protein into channel catfish, Ictalurus punctatus, embryos for gene editing. J Vis Exp. 2018;(131):56275. doi:10.3791/56275
105. Campbell LA, Richie CT, Maggirwar NS, Harvey BK. Cas9 ribonucleoprotein complex delivery: methods and applications for neuroinflammation. J Neuroimmune Pharmacol. 2019;14(4):565–577. doi:10.1007/s11481-019-09856-z
106. Chen G, Abdeen AA, Wang Y, et al. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat Nanotechnol. 2019;14(10):974–980. doi:10.1038/s41565-019-0539-2
107. Behr M, Zhou J, Xu B, Zhang H. In vivo delivery of CRISPR-Cas9 therapeutics: progress and challenges. Acta Pharm Sin B. 2021;11(8):2150–2171. doi:10.1016/j.apsb.2021.05.020
108. Lin JC, Van Eenennaam AL. Electroporation-mediated genome editing of livestock zygotes. Front Genet. 2021;12:648482. doi:10.3389/fgene.2021.648482
109. Alghadban S, Bouchareb A, Hinch R, et al. Electroporation and genetic supply of Cas9 increase the generation efficiency of CRISPR/Cas9 knock-in alleles in C57BL/6J mouse zygotes. Sci Rep. 2020;10(1):17912. doi:10.1038/s41598-020-74960-7
110. Le QA, Tanihara F, Wittayarat M, et al. Comparison of the effects of introducing the CRISPR/Cas9 system by microinjection and electroporation into porcine embryos at different stages. BMC Res Notes. 2021;14(1):7. doi:10.1186/s13104-020-05412-8
111. van Hees M, Slott S, Hansen AH, Kim HS, Ji HP, Astakhova K. New approaches to moderate CRISPR-Cas9 activity: addressing issues of cellular uptake and endosomal escape. Mol Ther. 2021;30(1):32–46. doi:10.1016/j.ymthe.2021.06.003
112. Jiang C, Lin X, Zhao Z. Applications of CRISPR/Cas9 technology in the treatment of lung cancer. Trends Mol Med. 2019;25(11):1039–1049. doi:10.1016/j.molmed.2019.07.007
113. Randhawa S. CRISPR-Cas9 in cancer therapeutics. Prog Mol Biol Transl Sci. 2021;181:129–163.
114. Zhang H, Qin C, An C, et al. Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol Cancer. 2021;20(1):126. doi:10.1186/s12943-021-01431-6
115. Luther DC, Lee YW, Nagaraj H, Scaletti F, Rotello VM. Delivery approaches for CRISPR/Cas9 therapeutics in vivo: advances and challenges. Expert Opin Drug Deliv. 2018;15(9):905–913. doi:10.1080/17425247.2018.1517746
116. Singh P, Ali SA. Impact of CRISPR-Cas9-based genome engineering in farm animals. Vet Sci. 2021;8(7):122. doi:10.3390/vetsci8070122
117. Omodamilola OI, Ibrahim AU. CRISPR technology: advantages, limitations and future direction. J Biomed Pharm Sci. 2018;1(2):115.
118. Yang Y, Xu J, Ge S, Lai L. CRISPR/Cas: advances, limitations, and applications for precision cancer research. Front Med. 2021;8:649896. doi:10.3389/fmed.2021.649896
119. Uddin F, Rudin CM, Sen T. CRISPR gene therapy: applications, limitations, and implications for the future. Front Oncol. 2020;10:1387. doi:10.3389/fonc.2020.01387
120. Cai W, Luo T, Mao L, Wang M. Spatiotemporal delivery of CRISPR/Cas9 genome editing machinery using stimuli-responsive vehicles. Angew Chem Int Ed Engl. 2021;60(16):8596–8606. doi:10.1002/anie.202005644
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
