Back to Journals » Journal of Inflammation Research » Volume 15
Multi-Omics Analysis of Western-style Diet Increased Susceptibility to Experimental Colitis in Mice
Authors Lin L, Li Y, Zhou G, Wang Y, Li L, Han J, Chen M, He Y, Zhang S
Received 3 February 2022
Accepted for publication 8 April 2022
Published 21 April 2022 Volume 2022:15 Pages 2523—2537
DOI https://doi.org/10.2147/JIR.S361039
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Lihui Lin,1,* Ying Li,1,2,* Gaoshi Zhou,1 Ying Wang,1 Li Li,1 Jing Han,1 Minhu Chen,1 Yao He,1 Shenghong Zhang1
1Division of Gastroenterology, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China; 2Division of Gastroenterology, The Seventh Affiliated Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shenghong Zhang; Yao He, Division of Gastroenterology, The First Affiliated Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China, Email [email protected]; [email protected]
Background: Western-style diet (WSD) is associated with inflammatory bowel disease (IBD) prevalence. However, the impact of WSD on IBD development and its underlying mechanism remain unclear. Transcriptomics and metabolomics could be beneficial for identifying key factors in WSD-related experimental IBD susceptibility. However, no such study has been conducted yet. We aimed to analyze the implications of WSD for experimental colitis susceptibility in mice and its underlying mechanism using these high-throughput technologies.
Methods: We fed experimental mice a WSD and a control diet from weaning. After 9 weeks, the mice were treated with 2,4,6 trinitrobenzene sulfonic acid to induce colitis, and the control group was treated with 50% ethanol (commonly used IBD animal model). Genome-wide microarray and liquid chromatography-tandem mass spectrometry were used to identify the differential transcripts and metabolites of experimental colitis with and without pre-illness WSD.
Results: WSD induced more severe inflammation in experimental colitis than the control diet. We found 2540 up-regulated genes and 2737 down-regulated genes in experimental colitis with WSD compared with those for the control diet. In addition, levels of 41 colonic tissue metabolites and 56 serum metabolites showed significant differences. Integrating transcriptomic and metabolomic data, we found major co-expression networks through which WSD promoted experimental IBD susceptibility, including enzymes of biotransformation, glycan synthesis and metabolism, steroid hormone metabolites.
Conclusion: Pre-illness WSD increased experimental colitis susceptibility. Our results could provide important evidences for the potential mechanisms and assist dietary recommendations to better manage IBD.
Keywords: Western-style diet, susceptibility, experimental colitis, inflammatory bowel disease, transcriptomics, metabolomics
Introduction
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), is considered an immune disorder that develops in genetically susceptible individuals exposed to certain environmental factors.1 Environmental exposure, especially through diet, causes great concern owing to its tremendous variability. Diet consumption affects many pathophysiologic processes, such as intestinal barrier regulation, immune responses, gut microecology, and intestinal hormone synthesis and release.2
IBD is common in northern Europe, North America, and Asian industrialized countries and regions,3,4 where Western-style diet (WSD) is popular. WSD includes refined carbohydrates, red meat, carbonated drinks, processed foods and a low proportion of vegetables, fruits, beans and whole grains.5 Studies have identified that WSD contributes to the risk of IBD. For example, imbalance between fatty acid and vegetable and fruit intake has been linked to increased CD risk among children.6 A meta-analysis indicated that pre-illness WSD consumption increases the risk of IBD.7 The ω-6 polyunsaturated fatty acid in WSD triggers glutathione peroxidase 4-restricted intestinal inflammation in CD.8 Anderson et al9 proposed that WSD may affect anti-tumor necrosis factor (TNF) drug response in IBD. However, the impact of WSD on IBD development and its underlying mechanism remain unclear.
High-throughput technologies have helped to establish a more profound understanding of IBD at the molecular level.10 Transcriptomics, the analysis of the output of genetic code, promises to change our understanding of the molecular basis of disease as well as identifying core biological signatures and signaling pathways.11 The metabolome is the downstream product of the transcriptome as well as the collection of the final biochemical reaction products in the life process of an organism. Metabolomics can detect metabolites in a certain period with high-throughput and help analyze them qualitatively and quantitatively. It is advantageous for investigating key biological mechanisms.12
Integrating transcriptomic and metabolomic data is beneficial for exploring major pathways and potential key factors of WSD-related IBD susceptibility. However, no such studies have been conducted yet. Controlling participant diet and collecting a large number of samples are necessary steps to this end. Therefore, choosing an IBD animal model as the research subject is the most appropriate. The chemically induced intestinal inflammation model is the most commonly used animal model of IBD.13,14 Colitis induced using 2,4,6-trinitro-benzene sulfonic acid (TNBS) represents an acute colitis model of IBD. TNBS-induced colitis impairs the intestinal barrier and highly activates CD4+ T-cell-dependent immunity as well as innate immunity.15 In this study, we used genome-wide microarray and liquid chromatography-tandem mass spectrometry (LC-MS/MS) and integrated transcriptomic and metabolomic profiling to investigate the effect of pre-illness WSD on TNBS-induced colitis. Our study provides a unique perspective into the molecular mechanism underlying the effect of pre-illness WSD in experimental colitis susceptibility in mice and reveals potentially critical factors.
Materials and Methods
Study Design
Male Balb/c mice (2 weeks old) weighing 4–8g were purchased from Gempharmatech Co., Ltd. (Nanjing, China) and housed under specific pathogen-free environment at the Animal Experimental Center, the First Affiliated Hospital, Sun Yat-sen University. Dams and pups stayed together before pups weaning. All procedures were performed with age-matched mice. Mice received sterile water and WSD (HFHC, Dyets, Inc., Bethlehem, PA), and control diet was given to adult mice (HFHC-C7, Dyets, Inc., Bethlehem, PA) and immature mice (AIN93G, Dyets, Inc., Bethlehem, PA). Detailed information on the three mouse chows is provided in Supplementary Table 1.
We randomly divided the mice into two groups at first: WSD group (n = 16) and AIN93G group (n = 28). Mouse weight was measured daily. After weaning at 3 weeks old, the AIN93G group was fed the AIN93G chow and the WSD group was fed the HFHC chow. The AIN93G chow was replaced with the HFHC-C7 chow in the AIN93G group when the mice grew up (8 weeks old) to meet the energy needs of adulthood; the AIN93G group was thus renamed as AIN93G-HFHCC7 group. The HFHC chow remained unchanged in the WSD group. Blood samples from six random mice in each group were collected for biochemical index testing at 12 weeks old before colitis induction. Sections from the colon, small intestine and liver of those mice were prepared for hematoxylin and eosin (H&E) staining or Masson’s trichrome staining.
The remaining mice in the AIN93G-HFHCC7 group were randomly and equally divided into two groups: AIN93G-HFHCC7-CON (n = 11) and AIN93G-HFHCC7-TNBS (n = 11) after the first round of sampling. The AIN93G-HFHCC7-TNBS and WSD groups received intrarectal administration of TNBS (Sigma, St. Louis, MO, USA), and the AIN93G-HFHCC7-CON group received intrarectal administration of 50% ethanol (Baishi Chemical Co., Ltd., Tianjin, China). The WSD group was also renamed as WSD-TNBS group. Colonic tissue and blood samples were collected on the sixth day after TNBS administration. Colonic tissues were sent for genome-wide microarray and LC-MS/MS analysis. Serum samples were prepared from whole blood samples and used for metabolomics analysis using LC-MS/MS.
Colitis Induction
Prior to TNBS colitis induction, body weight was measured, and the mouse back between the shoulders was presensitized using 1%TNBS solution. Then, 2.5%TNBS solution (equal volume of 5%TNBS was mixed with ethanol) was infused into the lumen of the mouse colon using a catheter and the mouse head was positioned down for 1 minute. PBS instead of 5%TNBS and mixed with equal volume of ethanol was used as a control. The mouse weight was monitored daily.
Histopathological Evaluation of Colon, Small Intestine, and Liver
After being washed with phosphate saline buffer and fixed in 4% paraformaldehyde, the mouse colon, small intestine and liver were embedded with paraffin. Then, paraffin tissue blocks were cut into 5μm-thick sections and subjected to either H&E or Masson’s trichrome staining. Histopathological evaluation of the colonic tissue was performed blindly by two researchers according to a scoring system for inflammation-associated histological changes in TNBS-induced colitis15 (see Supplementary Table 2). Small intestinal tissue histopathological evaluation was based on a modified MacPherson and Pfeiffer histopathological grading system16 (see Supplementary Table 3). Hepatic fibrosis was evaluated using Masson’s trichrome staining and the other parameters in the hepatic histopathological scoring system were evaluated via H&E staining17,18 (see Supplementary Table 4).
Biochemical Index Measurement and Cytokine Detection
Biochemical indexes, including alanine transaminase (ALT), aspartate transaminase (AST), glucose (GLU), high-density lipoprotein cholesterol (HDL-c), low-density lipoprotein cholesterol (LDL-c), total cholesterol (TC), triglyceride (TG), γ-glutamyl transferase (γ-GT), in serum samples from the WSD and AIN93G-HFHCC7 group were measured using an automatic biochemical analyzer (HITACHI 7020, Japan) according to the manufacturer’s instructions. The content of pro-inflammatory cytokines, including interferon-γ (IFN-γ), interleukin-1β (IL-1β), IL-2, IL-5, IL-6, and TNF-α, in serum samples from the WSD-TNBS, AIN93G-HFHCC7-TNBS, and AIN93G-HFHCC7-CON groups were evaluated using a Meso QuickPlex SQ 120 (Meso Scale Discovery, Rockville, MD, USA) according to the manufacturer’s instructions.
Transcriptomic Profiling
Transcriptomic profiling was performed to detect genes using Agilent microarray (design ID: 026655; platform accession: GPL11202). RNA samples were extracted from colonic tissues of mice in each group. RNA quantification and quality control (QC) were performed by NanoDrop ND-1000. RNA samples were labeled using a Quick Amp Labeling Kit (5190-0442, Agilent Technologies). The labeled RNAs were amplified and transcribed into cRNA using RNeasy Mini Kit (74104, Qiagen). The fluorescent cRNAs were hybridized onto the microarray hybridization chambers (G2530-60029, Agilent Technologies). After the microarray was washed, an Agilent DNA Microarray Scanner (G2565BA, Agilent Technologies) was used to scan the signals. Agilent Feature Extraction software (version 11.0.0.1, Agilent Technologies) was used to collect microarray probe signal values. Agilent GeneSpring GX software (version 12.1, Agilent Technologies) was used for microarray standardization.
The raw data were quantile-normalized and the high-quality probes were selected for further analysis. Differentially expressed genes with statistical significance between each of the two groups were determined using Volcano Plot filtering. Hierarchical clustering was conducted through the R scripts. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) analysis were conducted according to the standard enrichment computation method.
Metabolomic Analysis Using LC-MS/MS
Untargeted metabolomics was performed by Wuhan Metware Biotechnology Co., Ltd., (Wuhan, 430, 070, China). For colonic metabolomics, 500 μL of 70% methanol solution (containing 1ppm of 2-chlorophenylalanine) was added to the colon sample (50 ± 2mg). The homogenized samples were oscillated and iced for 15 min. After centrifugation, 400 μL of the supernatant was collected. Then, 500 μL of ethyl acetate/methanol (volume ratio=1:3) was added to the precipitate of the original centrifuge tube, shaken, and iced for 15 min. After centrifugation, 400 μL of the supernatant was collected. The supernatants were combined and concentrated. Next, 100 μL of 70% methanol solution was added into the dried substance, and 60 μL of supernatant was retained after ultrasound and centrifugation for LC-MS/MS analysis.
For serum metabolomics, 300 μL of pure methanol (including 1ppm 2-chlorophenylalanine) was added to the serum sample (100 μL). After vortexing, the samples were placed in a −20°C refrigerator and incubated for 0.5 h, followed by vortexing and centrifugation. Next, 200 μL of the supernatant was collected, and then iced them for 0.5 h at −20°C. The supernatant was collected again after centrifugation for LC-MS/MS analysis.
The 1290 Infinity LC ultrahigh performance liquid chromatograph was operated with a Waters T3 C18 column (mobile phase A of ultra-pure water and 0.01% formic acid, mobile phase B of acetonitrile) at a column temperature of 35°C, flow rate of 0.3 mL/min, and run time of 14 min. The QTOF/MS-6545 mass spectrometer was operated in ESI positive (with a voltage of 250V) and negative (with a voltage of 1500 V) ion modes.19 The gas flow rate was 8 mL/min, the sheath flow rate was 11 mL/min with gas temperature and sheath temperature of 325°C for both. QC samples were prepared from a mixture of extracted sample to analyze sample repeatability under the same treatment. During instrumental analysis, a QC sample was inserted once for every 11 test samples.
The original data obtained via LC-MS/MS analysis were first processed using Profinder software (Agilent Technologies), and then imported into the Mass Profiler Professional software (Agilent Technologies) in the form of CEF files for peak alignment and statistical analysis. Metabolomics data were analyzed using Student's t-test, principal component analysis (PCA), partial least square discriminant analysis (PLS-DA) and orthogonal PLS-DA (OPLS-DA).
Statistical Analysis
All data except transcriptomics and metabolomics were analyzed using SPSS statistical software (version 25.0, IBM, Chicago, IL, USA) and GraphPad Prism 8.0 (GraphPad Software Inc., CA, USA). Continuous data were compared using the unpaired, two-tailed Student's t-test or analysis of variance. Categorical variables were compared using the Chi-squared test. A two-sided P value less than 0.05 (P < 0.05) suggested statistical significance.
Ethical Considerations
All experimental protocols were approved by the Animal Care and Medical Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University ([2019] No.002). Experimental steps were performed according to the “Guidelines for the Kindly Treatment of Laboratory Animals” issued by the First Affiliated Hospital of Sun Yat-sen University.
Results
WSD Fueled More Severe Inflammation and Increased Susceptibility to TNBS-Induced Colitis
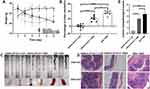
The body weight of mice in the WSD-TNBS group started decreasing from the second day of experimental colitis induction (Figure 1A). From day 1 to day 4 after colitis induction, the body weight of mice in the WSD-TNBS group was significantly higher than that of mice in the AIN93G-HFHCC7-TNBS group. At day 5, there was no significant difference in body weight between the two groups, and the weight in the WSD-TNBS group was significantly lower than that in the AIN93G-HFHCC7-TNBS group on day 6. The body weight of mice in the AIN93G-HFHCC7-CON group remained significantly higher than that of mice in the AIN93G-HFHCC7-TNBS group from day 2 of experimental colitis induction (Figure 1A).
The colon shortening percentage in the WSD-TNBS group and AIN93G-HFHCC7-TNBS group was significantly higher than that in the AIN93G-HFHCC7-CON group (Figure 1B). Furthermore, the colon shortening percentage in the WSD-TNBS group was significantly higher than that in the AIN93G-HFHCC7-TNBS group (Figure 1B). As shown in Figure 1C, colonic tissue was obviously congested, with severe inflammation and ulcers in the WSD-TNBS group. H&E staining of colonic tissue in the AIN93G-HFHCC7-CON group showed that the crypts were normal and goblet cells were abundant (Figure 1D). In the AIN93G-HFHCC7-TNBS group, the mucus layer in the colonic tissue was thin, and goblet cell count was lower than that in the AIN93G-HFHCC7-CON group (Figure 1D). There were serious gland disorders, abnormal crypts, less goblet cells, and high inflammatory cell infiltration in the colonic tissue in the WSD-TNBS group (Figure 1D). Colonic histopathological scores were significantly higher in the AIN93G-HFHCC7-TNBS and WSD-TNBS group than in the AIN93G-HFHCC7-CON group (Figure 1E). These scores of the WSD-TNBS group were significantly higher than those of the AIN93G-HFHCC7-TNBS group (Figure 1E).
The concentration of pro-inflammatory cytokines (IFN-γ, IL-1β, IL-2, IL-5, IL-6, and TNF-α) was significantly higher in the WSD-TNBS and AIN93G-HFHCC7-TNBS groups than in the AIN93G-HFHCC7-CON group (see Supplementary Figure 1). The contents of IL-1β, IL-2, IL-5 and IL-6 were significantly higher in the WSD-TNBS group than in the AIN93G-HFHCC7-TNBS group (see Supplementary Figure 1). In summary, we analyzed the TNBS-induced colitis model and found that pre-illness WSD induced a more severe clinical and pathological damage and enhanced susceptibility to experimental IBD.
WSD Had Little Effect on the Intestine of Mice Before Colitis Induction
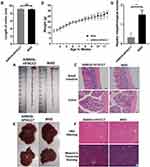
WSD is associated with intestinal inflammation, atherosclerosis, obesity, insulin resistance, and fatty liver.20–22 Samples from the serum, colon, small intestine, and liver of mice in the WSD and AIN93G-HFHCC7 group were collected at 12 weeks old before colitis induction. There was no clear distinction in colon length between the WSD and AIN93G-HFHCC7 groups (Figure 2A and B), as well as in small intestinal pathology (Figure 2C). The glands in the colonic tissue in the WSD group were slightly shorter, but no inflammatory cells and ulcers were found, with abundant goblet cells and intact intestinal epithelium in the colonic tissues of both groups (Figure 2C).
There was no significant difference in body weight between the WSD group and AIN93G-HFHCC7 group from 3 to 7.5 weeks of age, while the weight of mice in the WSD group was significantly higher than that of mice in the AIN93G-HFHCC7 group from 8 weeks of age (Figure 2D). There was no significant difference in liver appearance between the WSD group and AIN93G-HFHCC7 group (Figure 2E). However, H&E staining showed that hepatocytes were orderly arranged, but a few of them were ballooning, and hepatic sinusoids were clearly visible in the WSD group (Figure 2F). Masson’s trichrome staining showed no evidence of hepatic tissue fibrosis in the WSD and the AIN93G-HFHCC7 groups (Figure 2F). Thus, the hepatic histopathological scores were significantly higher in the WSD group than in the AIN93G-HFHCC7 group (Figure 2G). Higher levels of AST in serum were seen in the WSD group than in the AIN93G-HFHCC7 group, while the levels of ALT, GLU, HDL-c, LDL-c, TC, TG, and γ-GT between the WSD group and AIN93G-HFHCC7 group were not significantly different (see Supplementary Figure 2).
The above results indicated that WSD significantly promoted weight gain, increased the risk of insulin resistance and cardiovascular adverse events, and resulted in partial ballooning of hepatic cells, but had little effect on the intestinal tract before colitis induction. It showed that the effects of WSD on different metabolism-related organs and systems are quite different.
Transcriptomic Profiling of the Colonic Tissue of Colitis
Transcriptomic profiling of 8 colon samples was performed. The RNA quantity and quality of each sample were assessed. The WSD-TNBS group, AIN93G-HFHCC7-TNBS group, and AIN93G-HFHCC7-CON group each contained 4 samples. The heat maps show the genes expressed at different levels among the three groups (Figure 3A). The statistically significant genes with a P value <0.05, and |fold change| ≥ 2 were selected. There was a total of 2540 up-regulated and 2737 down-regulated genes in colonic tissue in the WSD-TNBS group compared with those in the AIN93G-HFHCC7-TNBS group. KEGG pathway analysis (Figure 3B and C) and GO analysis (Figure 3D and E) were performed using differentially expressed genes to infer their pathway involvement. For example, several classical cellular signaling pathways, such as Hippo, Wnt, Hedgehog, TGF-β, cAMP, and MAPK signaling pathway, were activated in experimental colitis with WSD compared with those in the absence of WSD (see Supplementary Table 5). Insulin resistance, insulin secretion, cholesterol metabolism, bile secretion, glutathione metabolism, linoleic acid metabolism, cell adhesion molecules, adherens junction, sensory perception of chemical stimulus, G-protein coupled receptor signaling pathway, and cell communication were also involved in experimental colitis with WSD compared with those for other diet (see Supplementary Table 5). Collectively, pre-illness WSD induced changes in transcription products in colonic tissue of colitis.
Metabolomic Profiling in Mouse Colonic Tissue
Metabolite profiling of 18 colon samples was performed using LC-MS/MS under ESI negative and positive ion modes. The WSD-TNBS group, AIN93G-HFHCC7-TNBS group, and AIN93G-HFHCC7-CON group each contained 6 samples. All QC samples were gathered in a similar location (Figure 4A and B). This showed that the detection instrument had high stability and repeatability. The PCA plots revealed separation between the WSD-TNBS group and AIN93G-HFHCC7-TNBS group under ESI negative and positive ion modes (Figure 4C and D). The differential compound was defined as metabolite with variable importance in projection (VIP) ≥1, P value <0.05, and fold change ≥2 or ≤0.5. The heatmaps (Figure 4E and F) directly unveiled the differences in colonic metabolites between the WSD-TNBS group and AIN93G-HFHCC7-TNBS group using the Masshunter software. We found 17 eligible metabolites in ESI negative ion mode, and 24 eligible metabolites in the positive ion mode in the WSD-TNBS group compared with those in the AIN93G-HFHCC7-TNBS group (see Supplementary Table 6).
KEGG pathway enrichment analysis of differential metabolites in the WSD-TNBS group and AIN93G-HFHCC7-TNBS group under negative ion mode mainly involved in steroid hormone biosynthesis (P = 0.015), while under positive ion mode mainly involved in sphingolipid metabolism (P = 0.020), lysine degradation (P = 0.023), and lysine biosynthesis (P = 0.042).
Metabolomic Profiling in Mouse Serum
Metabolite profiling of 18 serum samples were performed. The WSD-TNBS group, AIN93G-HFHCC7-TNBS group, and AIN93G-HFHCC7-CON group each contained 6 samples. All QC samples were gathered in a similar location (Figure 5A and B). The PCA plots revealed obvious separation between the WSD-TNBS group and AIN93G-HFHCC7-TNBS group under ESI negative and positive ion modes (Figure 5C and D). We used the same model and selection criteria as for colonic metabolomics to process serum metabolomic data. The heatmaps (Figure 5E and F) directly unveiled the differences in serum metabolites between these two groups. We found 39 eligible metabolites in ESI negative ion mode, and 17 eligible metabolites in the positive ion mode in the WSD-TNBS group compared with those in the AIN93G-HFHCC7-TNBS group (see Supplementary Table 7).
KEGG pathway enrichment analysis of differential metabolites comparing the WSD-TNBS group and AIN93G-HFHCC7-TNBS group under negative ion mode mainly involved in butanoate metabolism (P = 0.005), retinol metabolism (P = 0.029), and ubiquinone and other terpenoid-quinone biosynthesis pathway (P = 0.050). Positive ion mode results were mainly involved in nitrogen metabolism (P = 0.001), porphyrin and chlorophyll metabolism (P = 0.007), glycine, serine and threonine metabolism (P = 0.009), cyano-amino acid metabolism (P = 0.029), and methane metabolism (P = 0.044).
Integration of Transcriptome and Metabolome
As shown in Supplementary Tables 6 and 7, some metabolites do not have KEGG pathway annotation and therefore are not suitable for transcriptome and metabolome integration analysis based on KEGG annotation and enrichment results. Therefore, Pearson correlation analysis was conducted to calculate the possible correlation between all differential transcripts and differential metabolites in the comparison of the WSD-TNBS and AIN93G-HFHCC7-TNBS group. We displayed the relationships that met the Pearson Correlation Coefficient (PCC) > 0.8 or < −0.8, and the Pearson Correlation Coefficient P value (PCCP) < 0.05 (see Supplementary Table 8).
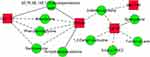
When combining transcriptome and metabolome data of colonic tissue, we found that co-expression networks between certain steroid metabolizing enzymes [hydroxysteroid (17-beta) dehydrogenase 6 (Hsd17b6); steroid 5 alpha-reductase 3 (Srd5a3)], cytochrome P450 enzymes [cytochrome P450, family 1, subfamily a, polypeptide 1 (Cyp1a1); cytochrome P450, family 11, subfamily b, polypeptide 1 (Cyp11b1)], glycan synthesis and metabolism (Saccharopine; 1,2-Dehydroreticuline; Biliverdin; Sphinganine), and steroid hormone biosynthesis (19-Hydroxytestosterone; Aldosterone) were strongly related to WSD-induced increased susceptibility to experimental IBD (Figure 6).
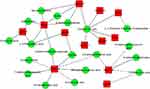
Integrating analysis of colonic transcriptomic data and serum metabolomic data, we found that co-expression networks of retinol metabolizing enzyme [retinol dehydrogenase 5 (Rdh5)], CYP enzymes (cytochrome P450, family 2, subfamily c, polypeptide 70 (Cyp2c70); cytochrome P450, family 4, subfamily a, polypeptide 14 (Cyp4a14); cytochrome P450, family 2, subfamily c, polypeptide 29 (Cyp2c29); cytochrome P450, family 3, subfamily a, polypeptide 11 (Cyp3a11); Cyp1a1), UGTs [UDP glucuronosyltransferase 2 family, polypeptide B5 (Ugt2b5); UDP glucuronosyltransferase 2 family, polypeptide B35 (Ugt2b35); UDP glucuronosyltransferase 2 family, polypeptide B37 (Ugt2b37)], some dehydrogenases [alcohol dehydrogenase 4 (class II), pi polypeptide (Adh4); alcohol dehydrogenase 1 (class I) (Adh1); Hsd17b6], glycan synthesis and metabolism (Normetanephrine; Uridine; L-1-Pyrroline-3-hydroxy-5-carboxylate; 4-Hydroxyphenylpyruvic acid; 4-acetamidobutanoate; 3R-hydroxy-butanoic acid; p-Coumaric acid), and lysine degradation (5-Acetamidovalerate, 2-Keto-6-acetamidocaproate) mediated the high experimental IBD susceptibility under WSD (Figure 7).
Discussion
This study showed that pre-illness WSD may be related to susceptibility to experimental colitis, at least in part due to changes in transcriptomic and metabolomic characteristics. The comparison between Figures 4 and 5 showed that the difference in serum metabolites was more obvious than that in colonic tissue metabolites. Furthermore, we integrated transcriptomics and metabolomics and conducted interaction network analysis of differential transcripts and metabolites to investigate the underlying mechanism of WSD in enhancing susceptibility to experimental colitis. Besides, compared with the control diet, WSD induced weight gain and hepatic cell edema in mice, but did not affect the intestine before colitis induction. Therefore, the general population accustomed to WSD perhaps tend to ignore its effects on the gut and put more emphasis on cardiovascular, liver, and endocrine care. The differences in the influences of WSD on different metabolism-related organs and systems deserve further study. To the best of our knowledge, this is the first study to explore the effect of pre-illness WSD on the susceptibility to experimental colitis via in-depth multi-omics analysis.
A major health issue of public concern is the effect of diet on disease. To provide reliable experimental data for dietary health recommendations to the public, we studied the effect and potential mechanism of pre-illness WSD. Compared with the diet of the WSD-TNBS group, the feed of the AIN93G-HFHCC7-TNBS group did not contain sucrose, cream and cholesterol. The diet of the WSD-TNBS group was composed of 40% kcal from fat, 43% carbohydrate, and 17% protein, which reflect the typical high-fat, high-sugar dietary pattern of Western societies. WSD may increase the susceptibility to experimental IBD, which may be relatable for people with a family history of CD. A positive family history of CD is associated with a higher risk of disease,23 a more aggressive clinical course,24,25 and is an important determinant of early immunosuppressive therapy.26 People with a family history of CD probably should avoid WSD as much as possible, which may improve the course of disease if they get sick. However, the individual diet is very complex, and dietary management should not be generalized, but customized.
The enzymatic reactions involved in biotransformation are mainly divided into two-phase reactions. The Phase I reaction includes oxidation, reduction, and hydrolysis reaction. Conjugation occurs in the Phase II reaction. The CYP enzyme system is the most important in oxidation reaction and participates in the hydroxylation of many substances, including steroid hormone synthesis. Alcohol dehydrogenase is also an active oxidase, which can catalyze the oxidation of alcohols to aldehydes. Glucuronic acid binding catalyzed by UGT is the most important and common binding reaction. Most UGT substances are the products of the CYP enzyme system. The water solubility of products catalyzed by UGT is not enough to make them excreted smoothly. In this study, WSD may have changed the nutritional status, led to disorders in biotransformation enzyme system of the body, and then increased the susceptibility of the intestine to harmful stimuli and more serious colitis.
Glycans are oligosaccharides or polysaccharides that are covalently linked to proteins or lipids to form glycoprotein, proteoglycans, or glycolipids. As one of the main components of intestinal mucosa, epithelial glycans participate in the interaction between microbiota and epithelial cells, which is also regulated by host genome and environmental factors.27,28 Our results lead us to speculate that the mechanism of WSD aggravation of experimental colitis involves abnormal glycan synthesis and metabolism. Many studies have reported glycosylation disturbance in the intestinal epithelium of IBD patients, resulting in changes in the intestinal immune environment, abnormal glycan-lectin and host-microbial interactions, and affecting the synthesis and stability of Mucin 2.29 This study enriched the understanding of the impact of glycans on IBD: WSD may have a negative effect on intestinal glycosylation, thereby increasing the risk for intestinal health.
Aldosterone and 19-hydroxytestosterone are steroid hormone metabolites. Steroid hormone biosynthesis in the colonic tissue was one of the pathways of WSD-induced severe experimental colitis in this study. Townsend et al30 reported that steroid hormones are important in disease prevention and treatment for humans and animals and are involved in inflammatory response. Sex steroids can regulate the immune system differently by controlling the expression level of pro- and anti-inflammatory cytokines.31 The steroid hormones in colonic tissues in the WSD-TNBS group observed in this study seem to be associated with inflammation. Further studies on steroid hormones and sex steroids may be valuable for patients with IBD.
This study has several limitations. First, it is unclear whether mice with colitis fed WSD could be rescued by dietary modification. Second, this study is based entirely on a mouse model, and the actual effects of WSD on humans are unknown. Third, colitis induction in mice generally occurs at 6–16 weeks old.15 Thus, WSD duration can be extended to 12 or 16 weeks, and the effect of the different WSD durations on the intestine could also be studied. The above limitations need to be addressed by well-designed trials in the future. However, at the same time, our results lay a foundation and provide a clear direction for further research on the onset and development mechanism of IBD in the future. Collectively, deciphering the implications of WSD based on multi-omics analysis will facilitate a deeper understanding of the onset pathogenesis and preventive strategies for IBD.
Conclusion
In this study, pre-illness WSD had little effect on the intestine before colitis induction but induced serious inflammation in experimental colitis. Integrating the changes in gene expression and metabolite characteristics, we provide several molecular insights into the potential alteration of experimental colitis susceptibility. WSD may damage the body’s ability to metabolize harmful stimuli, destroy the integrity of the intestinal mucosal epithelium, or induce a high inflammatory environment in the intestine by regulating gene expression and metabolic pathways (Box 1).
 |
Box 1 Hypotheses of WSD Increased Experimental Colitis Susceptibility |
Summary
Pre-illness Western-style diet increased experimental colitis susceptibility. Transcriptomic and metabolomic analysis revealed that a vast number of genes and metabolites are involved. Co-expression networks can be obtained by integrating omics information.
Abbreviations
WSD, Western-style diet; IBD, inflammatory bowel disease; CD, Crohn’s disease; UC, ulcerative colitis; TNF, tumor necrosis factor; TNBS, trinitro-benzene sulfonic acid; LC-MS/MS, liquid chromatography-tandem mass spectrometry; H&E, hematoxylin-eosin; ALT, alanine transaminase; AST, aspartate transaminase; GLU, glucose; HDL-c, high-density lipoprotein cholesterol; LDL-c, low-density lipoprotein cholesterol; TC, total cholesterol; TG, triglyceride; γ-GT, γ-glutamyl transferase; IFN-γ, interferon-γ; IL, interleukin; QC, quality control; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; min, minute; PCA, principal component analysis; PLS-DA, partial least square discriminant analysis; OPLS-DA, orthogonal PLS-DA analysis; VIP, variable importance in projection; PCC, Pearson Correlation Coefficient; PCCP, Pearson Correlation Coefficient significance P value; Srd5a3, steroid 5 alpha-reductase 3; Hsd17b6, hydroxysteroid (17-beta) dehydrogenase 6; Cyp1a1, cytochrome P450, family 1, subfamily a, polypeptide 1; Cyp11b1, cytochrome P450, family 11, subfamily b, polypeptide 1; Rdh5, retinol dehydrogenase 5; Ugt2b37, UDP glucuronosyltransferase 2 family, polypeptide B37; Cyp2c70, cytochrome P450, family 2, subfamily c, polypeptide 70; Cyp4a14, cytochrome P450, family 4, subfamily a, polypeptide 14; Ugt2b5, UDP glucuronosyltransferase 2 family, polypeptide B5; Cyp2c29, cytochrome P450, family 2, subfamily c, polypeptide 29; Cyp3a11, cytochrome P450, family 3, subfamily a, polypeptide 11; Ugt2b35, UDP glucuronosyltransferase 2 family, polypeptide B35; Adh4, alcohol dehydrogenase 4 (class II), pi polypeptide; Adh1, alcohol dehydrogenase 1 (class I).
Acknowledgments
The authors thank Wuhan Metware Biotechnology Co., Ltd. for metabolomics services and Editage for English language editing.
Funding
This study was funded by a grant from the National Natural Science Foundation of China (#82070538, #81870374, #81670607), Guangdong Science and Technology (#2017A030306021, #2021A1515220107), Guangdong basic and Applied Basic Research Fund (2021A1515220107).
Disclosure
The authors have no conflicts of interest to disclose.
References
1. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474(7351):307–317. doi:10.1038/nature10209
2. Levine A, Boneh RS, Wine E. Evolving role of diet in the pathogenesis and treatment of inflammatory bowel diseases. Gut. 2018;67(9):1726–1738. doi:10.1136/gutjnl-2017-315866
3. Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140(6):1785–1794. doi:10.1053/j.gastro.2011.01.055
4. Wilson J, Hair C, Knight R, et al. High incidence of inflammatory bowel disease in Australia: a prospective population-based Australian incidence study. Inflamm Bowel Dis. 2010;16(9):1550–1556. doi:10.1002/ibd.21209
5. Rizzello F, Spisni E, Giovanardi E, et al. Implications of the Westernized diet in the onset and progression of IBD. Nutrients. 2019;11(5):1033. doi:10.3390/nu11051033
6. Amre DK, D’Souza S, Morgan K, et al. Imbalances in dietary consumption of fatty acids, vegetables, and fruits are associated with risk for Crohn’s disease in children. Am J Gastroenterol. 2007;102(9):2016–2025. doi:10.1111/j.1572-0241.2007.01411.x
7. Li T, Qiu Y, Yang HS, et al. Systematic review and meta-analysis: association of a pre-illness Western dietary pattern with the risk of developing inflammatory bowel disease. J Dig Dis. 2020;21(7):362–371. doi:10.1111/1751-2980.12910
8. Mayr L, Grabherr F, Schwrzler J, et al. Dietary lipids fuel GPX4-restricted enteritis resembling Crohn’s disease. Nat Commun. 2020;11(1):1775. doi:10.1038/s41467-020-15646-6
9. Andersen V, Hansen AK, Heitmann BL. Potential impact of diet on treatment effect from anti-TNF drugs in inflammatory bowel disease. Nutrients. 2017;9(3):286. doi:10.3390/nu9030286
10. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi:10.1038/s41586-019-1237-9
11. Girgenti MJ, Duman RS. Transcriptome alterations in posttraumatic stress disorder. Biol Psychiatry. 2018;83(10):840–848. doi:10.1016/j.biopsych.2017.09.023
12. Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–269. doi:10.1038/nrm3314
13. Perse M, Cerar A. Dextran sodium sulphate colitis mouse model: traps and tricks. J Biomed Biotechnol. 2012;2012:718617. doi:10.1155/2012/718617
14. Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu Rev Immunol. 2002;20:495–549. doi:10.1146/annurev.immunol.20.100301.064816
15. Wirtz S, Popp V, Kindermann M, et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. 2017;12(7):1295–1309. doi:10.1038/nprot.2017.044
16. Soares PMG, Mota JM, Gomes AS, et al. Gastrointestinal dysmotility in 5-fluorouracil-induced intestinal mucositis outlasts inflammatory process resolution. Cancer Chemoth Pharm. 2008;63(1):91–98. doi:10.1007/s00280-008-0715-9
17. Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi:10.1002/hep.20701
18. Matsumoto M, Hada N, Sakamaki Y, et al. An improved mouse model that rapidly develops fibrosis in non-alcoholic steatohepatitis. Int J Exp Pathol. 2013;94(2):93–103. doi:10.1111/iep.12008
19. Zou W, She J, Tolstikov VV. A comprehensive workflow of mass spectrometry-based untargeted metabolomics in cancer metabolic biomarker discovery using human plasma and urine. Metabolites. 2013;3(3):787–819. doi:10.3390/metabo3030787
20. Xu XY, Yuan XX, Li NJ, Dewey WL, Li PL, Zhang F. Lysosomal cholesterol accumulation in macrophages leading to coronary atherosclerosis in CD38(-/-) mice. J Cell Mol Med. 2016;20(6):1001–1013. doi:10.1111/jcmm.12788
21. Wu HZ, Ghosh S, Perrard XD, et al. T-cell accumulation and regulated on activation, normal T cell expressed and secreted upregulation in adipose tissue in obesity. Circulation. 2007;115(8):1029–1038. doi:10.1161/CIRCULATIONAHA.106.638379
22. Kassel KM, Sullivan BP, Cui W, Copple BL, Luyendyk JP. Therapeutic administration of the direct thrombin inhibitor argatroban reduces hepatic inflammation in mice with established fatty liver disease. Am J Pathol. 2012;181(4):1287–1295. doi:10.1016/j.ajpath.2012.06.011
23. Borren NZ, Conway G, Garber JJ, et al. Differences in clinical course, genetics, and the microbiome between familial and sporadic inflammatory bowel diseases. J Crohns Colitis. 2018;12(5):525–531. doi:10.1093/ecco-jcc/jjx154
24. Hwang SW, Kwak MS, Kim WS, et al. Influence of a positive family history on the clinical course of inflammatory bowel disease. J Crohns Colitis. 2016;10(9):1024–1032. doi:10.1093/ecco-jcc/jjw063
25. Du P, Sun C, Ashburn J, et al. Risk factors for Crohn’s disease of the neo-small intestine in ulcerative colitis patients with total proctocolectomy and primary or secondary ileostomies. J Crohns Colitis. 2015;9(2):170–176. doi:10.1093/ecco-jcc/jju014
26. De Greef E, Mahachie John JM, Hoffman I, et al. Profile of pediatric Crohn’s disease in Belgium. J Crohns Colitis. 2013;7(11):e588–e598. doi:10.1016/j.crohns.2013.04.016
27. Arike L, Holmen-Larsson J, Hansson GC. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiol. 2017;27(4):318–328. doi:10.1093/glycob/cww134
28. Kudelka MR, Hinrichs BH, Darby T, et al. Cosmc is an X-linked inflammatory bowel disease risk gene that spatially regulates gut microbiota and contributes to sex-specific risk. Proc Natl Acad Sci U S A. 2016;113(51):14787–14792. doi:10.1073/pnas.1612158114
29. Kudelka MR, Stowell SR, Cummings RD, Neish AS. Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD. Nat Rev Gastroenterol Hepatol. 2020;17(10):597–617. doi:10.1038/s41575-020-0331-7
30. Townsend EA, Miller VM, Prakash YS. Sex differences and sex steroids in lung health and disease. Endocr Rev. 2012;33(1):1–47. doi:10.1210/er.2010-0031
31. Garcia-Gomez E, Gonzalez-Pedrajo B, Camacho-Arroyo I. Role of sex steroid hormones in bacterial-host interactions. Biomed Res Int. 2013;2013:928290. doi:10.1155/2013/928290
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.