Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 17
Molecular and Clinicopathological Characteristics of Lung Cancer Concomitant Chronic Obstructive Pulmonary Disease (COPD)
Authors Ma H, Zhang Q, Zhao Y, Zhang Y, Zhang J, Chen G, Tan Y, Zhang Q, Duan Q, Sun T, Qi C, Li F
Received 22 February 2022
Accepted for publication 25 June 2022
Published 14 July 2022 Volume 2022:17 Pages 1601—1612
DOI https://doi.org/10.2147/COPD.S363482
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Hongxia Ma,1 Qian Zhang,1 Yanwen Zhao,1 Yaohui Zhang,1 Jingjing Zhang,1 Guoqing Chen,1 Yuan Tan,2– 4 Qin Zhang,2– 4 Qianqian Duan,2– 4 Tingting Sun,2– 4 Chuang Qi,2– 4 Fengsen Li1
1Pneumology Department, The Fourth Affiliated Hospital of Xinjiang Medical University, Urumqi, The Xinjiang Uygur Autonomous Region, People’s Republic of China; 2The Medical Department, Jiangsu Simcere Diagnostics Co., Ltd, Nanjing, Jiangsu Province, People’s Republic of China; 3Nanjing Simcere Medical Laboratory Science Co., Ltd, Nanjing, Jiangsu Province, People’s Republic of China; 4The State Key Laboratory of Translational Medicine and Innovative Drug Development, Jiangsu Simcere Diagnostics Co., Ltd, Nanjing, Jiangsu Province, People’s Republic of China
Correspondence: Fengsen Li, Pneumology department, The Fourth Affiliated Hospital of Xinjiang Medical University, Urumqi, The Xinjiang Uygur Autonomous Region, People’s Republic of China, Email [email protected]
Introduction: Chronic obstructive pulmonary disease (COPD) and lung cancer often coexist, but its pathophysiology and genomics features are still unclear.
Methods: In this study, we retrospectively collected lung cancer concomitant COPD (COPD-LC) and non-COPD lung cancer (non-COPD-LC) patients, who performed next generation sequencing (NGS) and had clinicopathological information simultaneously. The COPD-LC data from the TCGA cohort were collected to conduct further analysis.
Results: A total of 51 COPD-LC patients and 88 non-COPD-LC patients were included in the study. Clinicopathological analysis showed that proportion of male gender, older age, and smoking patients were all substantially higher in COPD-LC group than in non-COPD-LC group (all P< 0.01). Comparing the genomic data of the two groups in our cohort, COPD-LC had higher mutation frequency of LRP1B (43% vs 9%, P = 0.001), EPHA5 (24% vs 1%, P = 0.002), PRKDC (14% vs 1%, P = 0.039), PREX2 (14% vs 0%, P = 0.012), and FAT1 (14% vs 0%, P = 0.012), which had a relationship with improved tumor immunity. Immunotherapy biomarker of PD-L1 positive expression (62.5% vs 52.0%, P = 0.397) and tumor mutation burden (TMB, median TMB: 7.09 vs 2.94, P = 0.004) also were higher in COPD-LC. In addition, RNA data from TCGA further indicated tumor immunity increased in COPD-LC. Whereas, COPD-LC had lower frequency of EGFR mutation (19% vs 50%, P = 0.013) and EGFR mutant COPD-LC treated with EGFR-TKI had worse progression-free survival (PFS) (HR = 3.52, 95% CI: 1.27– 9.80, P = 0.01).
Conclusion: In this retrospective study, we first explored molecular features of COPD-LC in a Chinese population. Although COPD-LC had lower EGFR mutant frequency and worse PFS with target treatment, high PD-L1 expression and TMB indicated these patients may benefit from immunotherapy.
Keywords: lung cancer, COPD, mutation landscape, Chinese population, molecular markers
Introduction
Lung cancer (LC) is the leading cause of cancer-related deaths worldwide.1 Despite the development of new diagnostic technology, average 5-year survival rate at early stage is 60%, while average 5-year survival rate at late stage is 22%.2 Chronic obstructive pulmonary disease (COPD) is characterized by persistent respiratory symptoms and airflow limitation. It is a high-profile public health challenge in worldwide and the third leading cause of death in China, with 8.6% prevalence of spirometry-defined COPD.3,4 At present, the spirometry criterion for airflow limitation remains a post-bronchodilator fixed ratio of FEV1/FVC <0.7 which is the gold standard for diagnosis of COPD.5
Lung cancer and COPD are intimately interrelated. Numerous studies have shown that the presence of airflow limitation confers between a 2–6-fold increase in the risk of lung cancer depending on the study design and definition of COPD.6,7 A previous study demonstrated that COPD is a risk factor for lung cancer development, independent of tobacco exposure.8 A meta-analysis of 26 studies revealed that lung cancer concomitant COPD was associated with poorer overall survival (OS), which was independent of tumor stage, diagnostic criteria of COPD or location.9 Although precision medical developments over past decades have sufficiently delineated the genomic landscape of lung cancer and afforded options for targeted and immune therapy for it, our understanding of molecular characteristics and excellent therapeutic options in lung cancer concomitant COPD remains inadequate.
A retrospective study based on the National Health Insurance Research Database in Taiwan indicated lung cancer patients with EGFR-TKI treatment had a worse survival outcome if patients had pre-existing COPD.10 However, the patients in this study lacked further description of genetic mutations. Another retrospective immunotherapy research investigated the clinical impact of COPD on the treatment response to palliative pembrolizumab in non-small cell lung cancer (NSCLC). Patients with COPD not only had higher response rate (38.2% vs 20.5%, p = 0.028), but also had better OS and progression-free survival (PFS).11 The above results indicate that lung cancer concomitant COPD is different from lung cancer in clinical pathology, efficacy of targeted and immune therapies and even genomic features. Recently, Kenji et al12 demonstrated that the PI3KCA mutation is a distinctive genetic feature of NSCLC and COPD, however, only 72 cancer-related genes were sequenced in this study and without immunotherapy biomarker exploration. Therefore, deep understanding of genomic determinants of lung cancer concomitant COPD is critical for choosing an effective therapy.
In this retrospective study, we first compared the clinical characteristics and serum markers of lung cancer patients with and without COPD. Second, we focused on the potential pathogenic and targeted molecular links and differences between lung cancer concomitant COPD and non-COPD. In addition, TCGA data of lung cancer concomitant COPD was used to explore associations and differences between cancer populations in China and the West.
Materials and Methods
Patients and Study Design
Patients with lung cancer or lung cancer concomitant COPD diagnosed at The Fourth Affiliated Hospital of Xinjiang Medical University (Urumqi, Xinjiang Uygur Autonomous Region, China) between December 2018 and August 2021 and conducted NGS (69/539 cancer related genes panel) to clarify status of driver mutations were enrolled in this study. Patients with other synchronous cancers except for lung cancer were ineligible. COPD was defined as incompletely reversible airflow obstruction associated with post-bronchodilator forced expiratory volume in one second (FEV1) and its ratio to forced vital capacity (FEV1/FVC) <0.7 and persistent respiratory symptoms including cough, dyspnea, and excessive sputum production.13 Clinical and pathological information was obtained from each patient including gender, age, smoking status, pathological type, and details of treatment. Informed written consent was acquired from each patient at the time of sample submission. The demographic and clinical characteristics of all patients are presented in Table 1 and Supplementary Table S1. The study methodologies conformed to the standards set by the Declaration of Helsinki and it was approved by the ethics committee (approved No. 2022XE010).
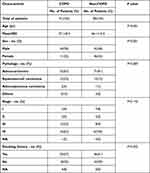 |
Table 1 Clinicopathological Characteristics of the Chinese Cohort |
TCGA Data
The lung cancer samples with explicit lung function data (FEV1/FVC) were chosen in the TCGA database that was downloaded from the cBioPortal (https://www.cbioportal.org). According to the GOLD guidelines,13 (FEV1/FVC) <0.7 in TCGA was defined as COPD-LC. Lung cancer with (FEV1/FVC) ≥0.7 was defined as non-COPD-LC.
Sample Collection, DNA Extraction, and Library Preparation
To be specific, all cancer samples (cfDNA/FFPE/fresh tissue) were tested with 69-genes panel or 539-genes panel. The specific test information of each sample was in the Supplementary Table S1. All samples were sequenced in a College of American Pathologists (CAP)-certified genomic testing facility (Jiangsu Simcere Diagnostics Co., Ltd, Nanjing, China). Genomic profiling of these samples was performed on formalin-fixed paraffin-embedded (FFPE) tumor/plasma biopsy specimens that were obtained from patients who had signed written informed consent. Three commercial kits were used for the DNA extraction. Genomic DNA (gDNA) of formalin-fixed and paraffin-embedded (FFPE) tissues and fresh tissues was extracted using the Tissue sample DNA extraction kit (Kai Shuo). Genomic DNA of leucocytes was extracted using MagMAXTM DNA Multi-Sample Ultra Kit (Thermo). Cell-free DNA (cfDNA) of plasma was extracted using MagMAXTM Cell Free DNA Isolation Kit (Thermo). All of the extraction procedures were performed following the manufacturer’s instructions. DNA was quantified on Qubit Fluorometer with Qubit dsDNA HS Assay kit (Thermo) and its quality was evaluated by Agilent 4200 TapeStation (Agilent). Probe hybridization capture method was used for library construction. Commercial reagents and customized probe were used for library construction and hybridization capture. In brief, 15–200 ng gDNA was sheared into 200~350 bp by fragmentation enzymes. Indexed paired-end adaptors for Illumina platform were self-developed and customized (SimcereDx). End repair, A-tailing and adaptor ligation of sheared DNA and cfDNA was respectively performed by KAPA HyperPlus DNA Library Prep kit (Roche Diagnostics) and VAHTSTM Universal DNA Library Prep Kit for Illumina® (Vazyme). Unligated adaptors were removed by the size selection function of Agencourt AMPure XP beads (Beckman Coulter). The ligation products were PCR amplified to form a pre-library for hybridization. The final library was quantified on Qubit Fluorometer with Qubit dsDNA HS Assay kit (Thermo Fisher) and its quality was evaluated by Agilent 4200 TapeStation (Agilent).
Library Sequencing and Bioinformatics Analysis
The qualified DNA libraries were sequenced on Illumina NovaSeq6000 platform (Illumina, San Diego, CA) and generated 150 bp paired end reads. Base calls from Illumina NovaSeq6000 were conducted to FASTQ files. The software fastp (v.2.20.0) was used for adapter trimming and filtering of low-quality bases.14 The BWA-MEM (v.0.7.17) algorithm was performed to align to the reference genome (UCSC’s hg19 GRCh37).15 Duplicate reads from PCR were excluded using Dedup with Error Correct. SNVs/InDels were called and annotated via VarDict (v.1.5.7)16 and InterVar,17 then the variants were filtered against the common SNPs in public database including 1000 Genome Project (Aug 2015) and Exome Aggregation Consortium (ExAC) Browser28 (v.0.3). CNVs and fusions were analyzed by CNVkit (dx1.1)18 and factera (v1.4.4),19 respectively.
Immunohistochemistry (IHC) Staining of PD-L1
Tissue biopsies of this study were performed with immunohistochemical (IHC) staining of PD-L1 expression with the VENTANA PD-L1 (SP263) Assay (Ventana Medical Systems, Inc., Tucson,
Arizona) according to the protocol reported previously.20 PD-L1 IHC was evaluated by a pathologist based on tumor cell score (TC score) with membranous and/or cytoplasmic staining, and divided into three groups: no-expression (<1% of tumor cells), low expression (1–49%) and high expression (≥50%).
TMB Analysis
Tumor mutation burden (TMB) was defined as the total number of non-synonymous somatic, coding, base substitution, and indel mutations per megabase (Mb) of genome in both our clinical cohort and The Cancer Genome Atlas (TCGA) cohort. For whole-exome sequencing (WES) data in TCGA, 38 Mb was adopted as the estimated exome size and calculated using maftools.21 In our Chinese cohort, 539 panel data were used to calculate TMB and 2 Mb was adopted as the estimated exome size.
Tumor Immunogenicity and Immune Characteristics Analysis
The CIBERSORT web portal (https://cibersort.stanford.edu/) was used to assess 22 types of infiltrating immune cells that were based on normalized gene expression data. The CIBERSORT immune infiltration proportions and immune-associated gene list were acquired from the immune landscape implemented by Thorsson et al.22
Gene Set Enrichment Analysis
The gene set enrichment analysis (GSEA) was conducted using the Java GSEA 4.1.0 Desktop Application (http://www.gsea-msigdb.org/gsea/index.jsp) to identify immune-related gene signatures between the COPD-LC and non-COPD-LC.23 R package DESeq2 was conducted to produce properly normalized RNA-seq Counts data which are compatible with GSEA.24 The gene sets examined in GSEA of Hallmark gene sets were obtained from Molecular Signatures Database (MSigDB database v7.4) (http://www.gsea-msigdb.org/gsea/downloads.jsp). This analysis involved 1000 random permutations for gene set and weighted enrichment statistic. The normalized enrichment score (NES) is the primary statistic for examining gene set enrichment results. The proportion of false-positives was controlled by calculating the false discovery rate (FDR). FDR estimates the probability that a gene set with a given NES represents a false-positive finding. A significantly enriched gene set was expected at FDR < 0.05.
Statistical Analysis
Results are reported as percentage, median (interquartile range (IQR)) or mean with standard deviation. Fisher’s exact or Chi-square test was performed to compare categorical variables. The Student’s t-test was used for analyzing quantitative data between two groups, and one-way ANOVA was used for comparisons of more than two groups. PFS of EGFR-TKIs was defined as the time from EGFR-TKIs therapy to documented progression or death from any cause or until the time of the last follow-up. The Kaplan-Meier (K-M) curve analysis of PFS was compared using the Log rank test. The Cox proportional hazards model was applied for univariable and multivariate survival analysis, and available confounding factors were adjusted. All the data were analyzed via R statistics package (R version 4.0.3). All statistical tests were two-sided, and P-value of < 0.05 was considered significant.
Results
Clinicopathological Characteristics of COPD-LC Patients in Our Chinese Cohort and TCGA Cohort
Between December 2018 and August 2021, 51 patients diagnosed with lung cancer concomitant COPD (defined as COPD-LC) and 88 lung cancer patients without COPD (defined as non-COPD-LC) at The Fourth Affiliated Hospital of Xinjiang Medical University were enrolled in this study (Supplementary Table S1 and Supplementary Figure S1). The clinicopathologic characteristics are summarized in Table 1. Comparison of the clinical features between COPD-LC and non-COPD-LC revealed that COPD-LC had a significantly higher ratio of male patients (78% vs 48%, P <0.001) and elderly patients (mean ± SD: 73.1±8.0 vs 66.1±10.6, P< 0.001) than non-COPD-LC. Meanwhile, the patients with COPD-LC had a higher proportion of smoking history (57% vs 41%, P = 0.002), which is not surprising because COPD is a major independent risk factor for lung carcinoma in smokers and increases the risk of lung cancer up to 4.5-fold.25 Moreover, there were also some differences between COPD-LC and non-COPD-LC in some serum markers and tumor markers, such as C-reaction protein and albumin levels (details in Supplementary Table S2). In the TCGA cohort, 69 COPD-LC and 126 non-COPD-LC cases were included in the analysis. The clinicopathological features of the TCGA cohort are shown in Supplementary Table S3. In the TCGA cohort, pathologic types and smoking history differed between the two groups.
The Genomic Landscape of COPD-LC Patients in Our Chinese Cohort
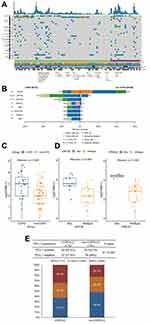
In order to do a comprehensive analysis, 21 COPD-LC and 68 non-COPD-LC tissue samples that conducted 539 cancer-related genes were eventually collected in the landscape analysis (Supplementary Figure S1). The mutational landscape of tissue samples in the two groups is shown in Figure 1A. In general, the high frequency mutations of all patients happened on TP53 (54%), EGFR (43%), MUC16 (27%), LRP1B (17%), PIK3CA (10%), and RBM10 (10%). While in COPD-LC, recurrent mutant genes were TP53 (71%), LRP1B (43%), MUC16 (33%), EPHA5 (24%), EGFR (19%), and RBM10 (19%). The mutation frequency and Fisher’s exact test results of 539 genes within the two groups are shown in Supplementary Table S4. Notably, LRP1B (43% vs 9%, P = 0.001), EPHA5 (24% vs 1%, P = 0.002), PRKDC (14% vs 1%, P = 0.039), PREX2 (14% vs 0%, P = 0.012), and FAT1 (14% vs 0%, P = 0.012) mutated more frequently in COPD-LC than non-COPD-LC (Figure 1B), which were reported to be associated with better immunotherapy efficacy in NSCLC or pan-cancer.26–31 Interestingly, EGFR, which was a targetable driver gene and had relationship with worse response to immunotherapy, had lower mutation frequency in COPD-LC group than non-COPD-LC group (19% vs 50%, P = 0.013). Therefore, we investigated TMB and PD-L1 expression status in the two groups. COPD-LC had higher TMB than non-COPD-LC (median [IQR]: 7.09 [2.84–11.03] vs 2.94 [1.46–4.55], P = 0.004, Figure 1C). And it was observed that patients with PREX2 mutation (median [IQR]: 20.59 [18.75–22.35] vs 6.25 [2.37–9.04], P = 0.008) and LRP1B (median [IQR]: 11.03 [9.22–16.91] vs 3.91 [2.01–7.27], P = 0.005) had significantly higher TMB (Figure 1D). A gastric cancer study found mutation status of PREX2 in baseline ctDNA influenced the PFS of immunotherapy.27 But there have been no reports that this gene is associated with lung cancer immunotherapy and TMB. Therefore, the association of this gene with high TMB and immunotherapy warrants further investigation. Patients with mutations in other mutant genes except for EGFR had a trend toward a higher TMB value (Supplementary Figure S2). In addition, COPD-LC had a higher proportion of PD-L1 positive expression (TC≥1%) than non-COPD-LC (62.5% vs 52.0%, P = 0.397, Figure 1E). However, significant difference of PD-L1 positive expression was not observed, probably due to the limited sample size. These results indicated that COPD-LC may benefit from immunotherapy.
COPD-LC Had Lower EGFR Mutation Frequency and Worse Clinical Outcomes of EGFR-TKI Treatment
Notably, in our Chinese cohort, the COPD group had a much lower mutation frequency of the driver gene EGFR (19% vs 50%, P = 0.013) than the non-COPD group (Figure 1B). Then we followed treatment status of all EGFR mutant patients who conducted NGS tests. Finally, EGFR-TKI first-line therapy information of 27 advanced patients (7 COPD-LC, 20 non-COPD-LC) was achieved. Their clinical information is shown in Supplementary Tables S5 and S6. Survival analysis revealed that EGFR mutant COPD-LC was significantly associated with worse PFS (mPFS: 21.7 vs 48.0 weeks, HR = 3.52, 95% CI: 1.27–9.80, long-rank P = 0.01, Figure 2A). In the multivariable Cox proportion hazards regression model, taking account of age, sex, smoking history, PD-L1 expression and TP53 co-mutation, COPD was also an adverse factor of PFS (HR = 6.86; 95% CI: 1.37–34.36; P = 0.02, Figure 2B).
The Genomic Landscape of COPD-LC Patients in TCGA Cohort
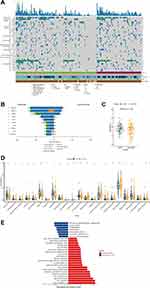
In the TCGA cohort, 69 COPD-LC and 126 non-COPD-LC were included in the analysis. The clinicopathological features of TCGA cohort are shown in Supplementary Table S3. In the TCGA cohort, COPD-LC had a higher proportion of smoking patients than non-COPD-LC (100% vs 93%, P = 0.028), while COPD-LC had a lower proportion of lung adenocarcinoma than non-COPD-LC group (42% vs 67%, P = 0.001). Although the proportion of adenocarcinoma in our Chinese cohort was lower in the COPD-LC group, it did not reach a statistical difference (63% vs 81%, P = 0.087). Interestingly, in the TCGA cohort, we found no differences in age and sex between the COPD-LC and non-COPD-LC groups. For somatic mutation analysis, 539 genes, same as genes in our clinical panel, were used, and the mutational landscape of these patients is shown in Figure 3A. In general, the high frequency mutations of all patients occurred on TP53 (62%), MUC16 (46%), LRP1B (39%), SPTA1 (23%), and KRAS (21%). In COPD-LC, top mutant genes were TP53 (68%), MUC16 (52%), LRP1B (38%), SPTA1 (22%), KEAP1 (19%), and KRAS (17%). EGFR is not shown in the heatmap because of its low mutation frequency (6%, 11/195). The mutation frequency and Fisher’s exact test of 539 genes in the two groups are shown in Supplementary Table S7. Differentially mutant genes between COPD-LC and non-COPD-LC are shown in Figure 3B. Notably, there was no significant difference between the two groups on tumor immunity related genes, such as LRP1B, EPHA5, PRKDC, and PREX2. Further, comparing TMB between the two groups, although COPD-LC had higher TMB (median [IQR]: 10.89 [7.00–16.94] vs 10.08 [4.67–15.76]), the difference was not statistically significant (P = 0.097, Figure 3C). These differences in molecular characteristics between the TCGA cohort and Chinese cohort were probably due to clinicopathological differences of the two groups.
Herein, we used transcriptome data from the TCGA cohort to explore the underlying mechanisms associated with COPD-LC and immunotherapy. We used CIBERSORT to explore 22 types of infiltration of immune cells and the results are shown in Figure 3D. As expected, improved anti-tumor immunity was observed in COPD-LC. Eosinophils, plasma cells, and activated mast cells were more abundant in the COPD-LC group. Resting mast cells, resting CD4 memory T cells and Tregs cells were lower in COPD-LC. Since COPD is a consequence of complex host-environment interactions that occur over time, COPD-LC had more active mast cells (P = 0.0087). In addition, the inflammatory response in COPD involves both neutrophilic inflammation and eosinophilic inflammation in COPD.32 The eosinophils in COPD-LC have higher expression (P = 0.0101). Since essentially all smokers develop macrophage and neutrophil infiltration in their lungs,33 no difference in neutrophil and macrophage cells expression was observed between the two groups. In addition, the results of enrichment analysis showed that many pathways varied significantly between the two groups (Figure 3E). Some signal pathways were upregulated in COPD-LC, such as DNA repair and G2/M checkpoint, which further indicated these patients may benefit from immunotherapy. Perturbation of DNA repair machinery could increase genomic instability, generate neoantigens, upregulate the expression of programmed death ligand 1 (PD-L1) which may promote the efficacy of immunotherapy. The G2/M phase, involving energy dependent reactions for structural rearrangements of cell division, is associated with T cell proliferation.34 Therefore, an upregulated G2/M checkpoint pathway may promote T cell proliferation.
Discussion
For the past few years, extensive studies have focused on mutation landscape, innovational target and immunotherapies of lung cancer.35–37 However, our understanding of lung cancer with various complications including COPD is inadequate. In this retrospective study, we first found COPD-LC patients had unique features of clinicopathology. In addition, some serum and tumor markers also differed between COPD-LC and non-COPD-LC groups. Then we explored the genomic differences between two groups in our Chinese cohort. In terms of molecular characteristics, compared with non-COPD-LC, COPD-LC had higher mutation frequency of some immune-related genes (LRP1B, EPHA5, PRKDC, PREX2, and FAT1).26,29,38 Notably, in these differential mutant genes, COPD-LC patients with mutations in LRP1B (11.03 vs 3.91, P = 0.005) and PREX2 (20.59 vs 6.25, P = 0.007) had significantly higher TMB value than non-COPD-LC patients. LRP1B (low-density lipoprotein receptor-related protein 1B) is one of the most frequently mutated genes in tumor samples and have been proved to associate with TMB and survival prognosis in NSCLC patients treated by ICIs.29 PREX2 (Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 2) is capable of inhibiting the activity of PTEN and thus regulates the downstream PI3K signaling pathway. As a result, PREX2 is considered to be an oncoprotein.39 Previous study of gastric cancer hinted that PREX2 mutation had a relationship with better PFS in immunotherapy but no further mechanism was researched or discussed.27 Considering the association of PREX2 mutation and TMB, and high mutation frequency of PREX2 in COPD-LC group, the function of PREX2 in COPD-LC patients needs more research. Further, we compared the results to the matching molecular atlas in the TCGA database to explore the differences. However, there was no significant difference between two groups on tumor immunity related genes and TMB. There were differences in clinicopathological characteristics between our Chinese cohort and the TCGA cohort, which may be a possible factor leading to the differences in gene mutations between the two cohorts. In addition, racial difference may be another reason. A French study40 performed WES sequencing on 22 advanced NSCLC patients (11 with COPD vs 11 without COPD) who received immunotherapy and found no significant difference in TMB between the two groups. However, mutations in these COPD-LC patients were not disclosed in the study. Therefore, to the best of our knowledge, this study is the first to elucidate the comprehensive molecular profile including driver mutation and common immunotherapy related biomarkers of lung cancer concomitant COPD in the Chinese population and TCGA cohort.
In addition to the mutation differences between COPD-LC and non-COPD-LC, we also investigated the differences between the two groups in treatment outcomes among patients with EGFR targetable mutations. The COPD-LC group had a much lower mutation frequency of EGFR (19% vs 50%, P = 0.013) than the non-COPD. And results of EGFR mutant patients with EGFR-TKI therapy revealed that COPD-LC had worse PFS than non-COPD-LC (HR = 3.52, 95% CI: 1.27–9.80, P = 0.01). This result is consistent with the results of the prior large-scale clinical study.10 However, that study did not describe more genetic information. Previous literatures have reported that TP53 is an unfavorable factor for the prognosis and targeted therapy of lung cancer.41–43 In addition, Inomata et al44 found patients with negative tumor PD-L1 expression showed better PFS of first-line treatment with an EGFR-TKI. After taking account of these two molecular factors and some clinical factors, only COPD was the worse factor to EGFR treatment (HR = 6.86; 95% CI: 1.37–34.36; P = 0.02). Because of the limited sample size, influence of mutant differential genes (such as LRP1B, EPHA5, and PRKDC) and TMB on PFS needs more research.
This retrospective analysis has several limitations. First, although COPD-LC was characterized by infiltration of some T-cell immune cells and some immune-related pathways, population and molecular gaps exist between the TCGA cohort and our Chinese cohort. More underpinnings with further clinical study and experimental work are needed. Second, this study was conducted by a single center with a limited number of COPD-LC patients, thus further investigation to validate our findings in a prospective study and multicenter collaboration are necessary.
Conclusion
In conclusion, our retrospective study is the first to reveal the molecular differences between COPD-LC and non-COPD-LC in both a Chinese population and TCGA cohort. In the Chinese cohort, COPD-LC patients had features including low mutation frequency of EGFR, but high mutation frequency of immune-related genes, high PD-L1 expression, and high TMB which showed these patients may benefit from immunotherapy. Notably, there was no significant difference between the two groups on EGFR gene, tumor immunity related genes and TMB in the TCGA cohort. These findings could pave the way to improving treatment planning for lung cancer coexisting COPD patients, especially in Chinese COPD-LC patients.
Ethics Approval
This study was approved by The Fourth Affiliated Hospital of Xinjiang Medical University, Urumqi, Xinjiang Uygur Autonomous Region, China. (No. 2022XE0106).
Consent to Participate
Written informed consent was obtained from each patient upon sample collection.
Acknowledgment
The authors thank Wanglong Deng, Guanghua Lu, Ran Ding, Qiong Wang, Yidan Jing and Liang Liu for their contributions to the data collection of this research.
Funding
This work was supported by Xinjiang key laboratory of respiratory disease research (No. 2020D04030).
Disclosure
Y.T, Q.Z, Q.D, T.S, and C.Q are the employees of Jiangsu Simcere Diagnostics Co., Ltd and Nanjing Simcere Medical Laboratory Science Co., Ltd. The authors report no other potential conflicts of interest in relation to this work.
References
1. Mouronte-Roibás C, Leiro-Fernández V, Ruano-Raviña A, et al. Predictive value of a series of inflammatory markers in COPD for lung cancer diagnosis: a case-control study. Respir Res. 2019;20(1):198. doi:10.1186/s12931-019-1155-2
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi:10.3322/caac.21708
3. Labaki WW, Rosenberg SR, Chronic obstructive pulmonary disease. Ann Intern Med. 2020;173(3):Itc17–itc32. doi:10.7326/AITC202008040
4. Wang C, Xu J, Yang L, et al. Prevalence and risk factors of chronic obstructive pulmonary disease in China (the China Pulmonary Health [CPH] study): a national cross-sectional study. Lancet. 2018;391(10131):1706–1717. doi:10.1016/s0140-6736(18)30841-9
5. Halpin DMG, Criner GJ, Papi A, et al. Global Initiative for the diagnosis, management, and prevention of chronic obstructive lung disease. The 2020 GOLD science committee report on COVID-19 and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2021;203(1):24–36. doi:10.1164/rccm.202009-3533SO
6. Rabe KF, Watz H. Chronic obstructive pulmonary disease. Lancet. 2017;389(10082):1931–1940. doi:10.1016/s0140-6736(17)31222-9
7. Young RP, Hopkins RJ. Chronic obstructive pulmonary disease (COPD) and lung cancer screening. Transl Lung Cancer Res. 2018;7(3):347–360. doi:10.21037/tlcr.2018.05.04
8. Sandri BJ, Kaplan A, Hodgson SW, et al. Multi-omic molecular profiling of lung cancer in COPD. Eur Respir J. 2018;52(1):1702665. doi:10.1183/13993003.02665-2017
9. Gao YH, Guan WJ, Liu Q, et al. Impact of COPD and emphysema on survival of patients with lung cancer: a meta-analysis of observational studies. Respirology. 2016;21(2):269–279. doi:10.1111/resp.12661
10. Wu CC, Rau KM, Lee WC, et al. Presence of Chronic Obstructive Pulmonary Disease (COPD) impair survival in lung cancer patients receiving Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitor (EGFR-TKI): a nationwide, population-based cohort study. J Clin Med. 2019;8(7):1024. doi:10.3390/jcm8071024
11. Shin SH, Park HY, Im Y, et al. Improved treatment outcome of pembrolizumab in patients with nonsmall cell lung cancer and chronic obstructive pulmonary disease. Int J Cancer. 2019;145(9):2433–2439. doi:10.1002/ijc.32235
12. Sawa K, Koh Y, Kawaguchi T, et al. PIK3CA mutation as a distinctive genetic feature of non-small cell lung cancer with chronic obstructive pulmonary disease: a comprehensive mutational analysis from a multi-institutional cohort. Lung Cancer. 2017;112:96–101. doi:10.1016/j.lungcan.2017.07.039
13. Mirza S, Clay RD, Koslow MA, Scanlon PD. COPD guidelines: a review of the 2018 GOLD report. Mayo Clin Proc. 2018;93(10):1488–1502. doi:10.1016/j.mayocp.2018.05.026
14. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi:10.1093/bioinformatics/bty560
15. Hwang KB, Lee IH, Li H, et al. Comparative analysis of whole-genome sequencing pipelines to minimize false negative findings. Sci Rep. 2019;9(1):3219. doi:10.1038/s41598-019-39108-2
16. Lai Z, Markovets A, Ahdesmaki M, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44(11):e108. doi:10.1093/nar/gkw227
17. Li Q, Wang K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100(2):267–280. doi:10.1016/j.ajhg.2017.01.004
18. Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12(4):e1004873. doi:10.1371/journal.pcbi.1004873
19. Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30(23):3390–3393. doi:10.1093/bioinformatics/btu549
20. Williams GH, Nicholson AG, Snead DRJ, et al. Interobserver reliability of programmed cell death ligand-1 scoring using the VENTANA PD-L1 (SP263) assay in NSCLC. J Thorac Oncol. 2020;15(4):550–555. doi:10.1016/j.jtho.2019.11.010
21. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi:10.1101/gr.239244.118
22. Huang TX, Fu L. The immune landscape of esophageal cancer. Cancer Commun. 2019;39(1):79. doi:10.1186/s40880-019-0427-z
23. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
24. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8
25. Caramori G, Ruggeri P, Mumby S, et al. Molecular links between COPD and lung cancer: new targets for drug discovery? Expert Opin Ther Targets. 2019;23(6):539–553. doi:10.1080/14728222.2019.1615884
26. Chen Y, Li Y, Guan Y, et al. Prevalence of PRKDC mutations and association with response to immune checkpoint inhibitors in solid tumors. Mol Oncol. 2020;14(9):2096–2110. doi:10.1002/1878-0261.12739
27. Jin Y, Chen DL, Wang F, et al. The predicting role of circulating tumor DNA landscape in gastric cancer patients treated with immune checkpoint inhibitors. Mol Cancer. 2020;19(1):154. doi:10.1186/s12943-020-01274-7
28. Chen Z, Chen J, Ren D, et al. EPHA5 mutations predict survival after immunotherapy in lung adenocarcinoma. Aging. 2020;13(1):598–618. doi:10.18632/aging.202169
29. Chen H, Chong W, Wu Q, et al. Association of LRP1B Mutation with tumor mutation burden and outcomes in melanoma and non-small cell lung cancer patients treated with immune check-point blockades. Front Immunol. 2019;10:1113. doi:10.3389/fimmu.2019.01113
30. Yang Y, Zhang J, Chen Y, et al. MUC4, MUC16, and TTN genes mutation correlated with prognosis, and predicted tumor mutation burden and immunotherapy efficacy in gastric cancer and pan-cancer. Clin Transl Med. 2020;10(4):e155. doi:10.1002/ctm2.155
31. Fang W, Ma Y, Yin JC, et al. Comprehensive genomic profiling identifies novel genetic predictors of response to Anti-PD-(L)1 therapies in non-small cell lung cancer. Clin Cancer Res. 2019;25(16):5015–5026. doi:10.1158/1078-0432.Ccr-19-0585
32. Brightling C, Greening N. Airway inflammation in COPD: progress to precision medicine. Eur Respir J. 2019;54(2):1900651. doi:10.1183/13993003.00651-2019
33. Houghton AM. Mechanistic links between COPD and lung cancer. Nat Rev Cancer. 2013;13(4):233–245. doi:10.1038/nrc3477
34. MacPherson S, Kilgour M, Lum JJ. Understanding lymphocyte metabolism for use in cancer immunotherapy. Febs j. 2018;285(14):2567–2578. doi:10.1111/febs.14454
35. Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol. 2019;16(6):341–355. doi:10.1038/s41571-019-0173-9
36. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi:10.1016/s0140-6736(16)30958-8
37. Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: current status and perspectives. Crit Rev Oncol Hematol. 2021;157:103194. doi:10.1016/j.critrevonc.2020.103194
38. Bai H, Duan J, Li C, et al. EPHA mutation as a predictor of immunotherapeutic efficacy in lung adenocarcinoma. J Immunother Cancer. 2020;8(2):e001315. doi:10.1136/jitc-2020-001315
39. Yang MH, Yen CH, Chen YF, et al. Somatic mutations of PREX2 gene in patients with hepatocellular carcinoma. Sci Rep. 2019;9(1):2552. doi:10.1038/s41598-018-36810-5
40. Biton J, Ouakrim H, Dechartres A, et al. Impaired tumor-infiltrating T cells in patients with chronic obstructive pulmonary disease impact lung cancer response to PD-1 blockade. Am J Respir Crit Care Med. 2018;198(7):928–940. doi:10.1164/rccm.201706-1110OC
41. Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer. 2019;19(9):495–509. doi:10.1038/s41568-019-0179-8
42. Xu F, Lin H, He P, et al. A TP53-associated gene signature for prediction of prognosis and therapeutic responses in lung squamous cell carcinoma. Oncoimmunology. 2020;9(1):1731943. doi:10.1080/2162402x.2020.1731943
43. VanderLaan PA, Rangachari D, Mockus SM, et al. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: correlation with clinical outcomes. Lung Cancer. 2017;106:17–21. doi:10.1016/j.lungcan.2017.01.011
44. Inomata M, Azechi K, Takata N, et al. Association of tumor PD-L1 expression with the T790M mutation and progression-free survival in patients with EGFR-mutant non-small cell lung cancer receiving EGFR-TKI therapy. Diagnostics. 2020;10(12). doi:10.3390/diagnostics10121006
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.