Back to Journals » Cancer Management and Research » Volume 12
MiR-802 Suppresses Colorectal Cancer Cell Viability, Migration and Invasion by Targeting RAN
Authors Feng H, Liu L, Xu L, Wang H, Hua Q, He P
Received 19 September 2019
Accepted for publication 21 February 2020
Published 26 March 2020 Volume 2020:12 Pages 2291—2300
DOI https://doi.org/10.2147/CMAR.S231709
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Hailong Feng,1 Lingling Liu,1 Laijing Xu,1 Haili Wang,1 Qiuju Hua,2 Peng He1
1Department of General Surgery, The First Affiliated Hospital of Xinxiang Medical University, Weihui City, Henan Province 453100, People’s Republic of China; 2Hospital of Nephrology, The First Affiliated Hospital of Xinxiang Medical University, Weihui City, Henan Province 453100, People’s Republic of China
Correspondence: Peng He
Department of General Surgery, The First Affiliated Hospital of Xinxiang Medical University, Weihui City, Henan Province 453100, People’s Republic of China
Tel +86-373-4404713
Email [email protected]
Purpose: Colorectal cancer is one of the most malignant tumors in the world, and the incidence is increasing every year. MicroRNAs (miRNA) are small non-coding RNAs that are involved in a variety of physiological or pathological processes. Abnormal expression of microRNA-802 (miR-802) has been demonstrated in various types of cancer. However, the expression and biological role of miR-802 in human colorectal cancer remain largely unknown.
Methods: Here, we used quantitative real-time PCR (qRT-PCR) to measure miR-802 expression levels in colorectal cancer tissues and cell lines. Cell Counting Kit-8 (CCK-8) was used to assess the effect of miR-802 on colorectal cancer cell viability. Migration and invasion assays were performed to determine the effect of miR-802 on metastasis of colon tumor cells by transwell analysis. Luciferase activity assays were used to confirm the target of miR-802.
Results: The results show that miR-802 is significantly downregulated in colorectal cancer tissues and cell lines. Overexpression of miR-802 profoundly inhibited viability, migration and invasion of colorectal cancer cells. In addition, we have newly discovered that the Ras-associated nucleus (RAN) is a direct target of miR-802 which could reverse the effects induced by miR-802 overexpression in colorectal cancer cells.
Conclusion: In conclusion, our study shows that miR-802 is downregulated in colorectal cancer, and overexpression of miR-802 inhibits colorectal cancer cell viability, migration and invasion by directly targeting RAN.
Keywords: miR-802, colorectal cancer, RAN, viability, migration, invasion
Introduction
Colorectal cancer, also known as colon cancer or bowel cancer which is developed from colon or rectum is one of the most malignant tumors worldwide and represents the second leading cause for cancer-related death in the world.1 In China, the colorectal cancer is listed as the sixth commonly diagnosed cancer in women while in United States it is one of the top 5 cancers that leading to cancer-related death.2,3 Although huge improvements in preventing strategies, screening programmes, and combination of different therapeutic approaches that include surgery, radiation and chemotherapy may improve the survival, the median survival of colorectal cancer patient is only 20 months with advanced colorectal cancer.4 In recent years, personalized and targeted therapies are developed rapidly and achieved dramatic improvement for the treatment of colorectal cancer. However, these therapies rely on status of the cancer and also the response to the treatment. Thus, exploring more efficient therapeutic targets and prognostic biomarkers are critical and important to improve the overall treatment.
MicroRNAs (miRNA) are small non-coding RNAs of 18–24 nucleotides in length. MiRNAs bind to the 3ʹ-UTR of its specific target mRNAs which causes binding RNA degradation or inhibition of its translation, thus silencing its mRNA targets.5,6 More and more evidence showed that miRNA plays important role in many physiological and pathological processes which include cell growth, proliferation, differentiation and apoptosis.7 Aberrant expression of miRNAs was reported in different types of cancers and these miRNAs act as either tumor suppressor or oncogenes, dependent on the cellular function of their targets.8 In colorectal cancer, numerous studies investigated the expression and the biologic role of miRNA and they proved that the expression of these miRNAs is consistently altered in colorectal cancer.9 The elevated expression was found in almost 2/3 of these miRNAs and the reduced expression was reported in other 1/3 of miRNAs.9 The expression of miR-143 and miR-145 proved to be decreased in colorectal cancer and proposed as tumor suppressors, while miR-21 was reported to be elevated in colorectal cancer by several groups and served as oncogenic miRNA.9–12 Among these miRNAs, miR-802 located on chromosome 21, was reported involved in several cancer development and progression, which include breast cancer, prostate cancer, gastric cancer, lung carcinoma, etc.13–16 The biologic role of miR-802 in cancer development and progression is dependent on cancer type. MiR-802 promoted lung carcinoma proliferation by targeting menin16 while suppressing breast tumor cell proliferation by downregulation of FoxM1.13 It was also reported miR-802 inhibits human prostate cancer growth by targeting flotillin-2,14 as well as suppressing gastric cancer cell growth via targeting RAB23.15 However, the role of miR-802 in colorectal cancer development and progression is still largely unknown.
In this study, we aimed to investigate the expression profile and the biological function of miR-802 in the development and progression of human colorectal cancer. Our results showed that miR-802 expression is markedly downregulated in colon tumor tissue compared with adjacent normal tissue, and its expression is also downregulated in the colon tumor cell lines compared with normal mucosa in colon. More interestingly, overexpression of miR-802 significantly inhibited colon tumor cell viability, migration and invasion. In addition, we also proved that the Ras-associated nucleus (RAN) is a direct target of miR-802 which could reverse the effects caused by miR-802 overexpression in colorectal cancer cells. These results suggested that miR-802 is a potential target for the treatment of colorectal cancer.
Materials and Methods
Patients and Clinical Specimens
Paired colorectal cancer tumors and adjacent normal mucosa tissues were obtained postoperatively from 38 patients at the Department of General Surgery, The First Affiliated Hospital of Xinxiang Medical University between September 2010 and September/April 2013. The tumor stage was classified according to the 7th edition of the AJCC Cancer Staging Manual (2010). All experimental protocols were approved by the Ethics Committee at The First Affiliated Hospital of Xinxiang Medical University, and that was conducted in accordance with the Declaration of Helsinki. The written informed consent was obtained from all patients. All samples were kept in liquid nitrogen immediately after resection and stored at −80°C for further molecular detection. None of the patients received neoadjuvant chemo-or radiotherapy prior to surgery. The clinical-pathological features of 38 patients are summarized in Table 1.
 |
Table 1 Correlation Between miR-802 Expression and Clinical Features of the Colon Cancer Patients |
Cell Culture and Transfection
Three human colorectal cancer cell lines (HCT-116, LS174T and SW480), and a normal human colon mucosal epithelial cell line (NCM460) were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences and cultured in DMEM medium containing 10% FBS (Gibco, USA), 100 U/mL penicillin, 100 μg/mL streptomycin and incubated in a humidified incubator (37°C, 5% CO2).
For transfection, miR-802 mimic and corresponding negative control (miR-NC) were obtained from GenePharma (Shanghai, China). The sequences used were as follows: miR-802 mimic, 5ʹ-CAGUAACAAAGAUUCAUCCUUGU-3ʹ; miR-NC, 5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ. The pcDNA-3.1-Ras-related nuclear (RAN) plasmid (RAN vector) and blank vector (pcDNA-3.1) (RAN NC) were designed by RiboBio (Guangzhou, China). HCT-116 cells were plated into 6-well plates at the density of 5×105 cells per well. After overnight culture, cells density reached at 75–80% of the confluence and then cells were transfected with related plasmids and miRNAs. Cell transfections were performed using Lipofectamine 2000 kit obtained from Invitrogen (Carlsbad, CA, USA) following the manufacturer’s instructions. The transfection efficiency for miR-802 mimic was determined by qRT-PCR. The cells were divided into two groups for the following function studies, that included miR-NC group and miR-802 mimic group.
Quantitative Real Time-PCR (qRT-PCR)
Total RNA from tumor tissue and tumor cell lines was extracted by using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocols. RNA was reverse transcribed into cDNA with the PrimeScript RT reagent Kit 128 (Promega, Madison, WI, USA) under the condition of: 42°C for 50 min and 70°C for 15 min and the mRNA levels of related genes were detected by qRT-PCR. All qRT-PCRs were performed in triplicates on an ABI 7500 Real-time PCR system (Applied Biosystems, Foster City, CA, USA). Primers of miR-802, RAN, U6 and GADPH were purchased from Ambion (Applied Biosystems, Foster City, CA, USA). Three-step cycling protocols were used for qRT-PCR: 50°C for 2 mins, 94°C for 10 mins, 40 cycles of 94°C for 15 s and 60°C for 30 s; hold at 4°C. The relative expression of each gene was calculated using the 2−ΔΔCt method and normalized to U6 or GAPDH.17 U6 snRNA was used as a normalization control for the detection of miR-802. Control gene GAPDH was used as a normalization control for the detection of RAN. The following primers were used for reverse transcription and qRT-PCR:
- miR-802-forward 5ʹ-CGTTGTGTAGCTTATCAGACTG-3ʹ;
- miR-802-reverse 5ʹ-AATGGTTGTTCTCCACACTCTC-3ʹ;
- U6-forward 5′-CTCGCTTCGGCAGCACA-3′;
- U6-reverse 5′-AACGCTTCAGGAATTTGCGT-3′;
- RAN-forward 5′-TGAGAAGAAGTATGTAGCCACC-3′;
- RAN-reverse 5′-GATTCTTCTTCCGGTGGAAGAC-3′;
- GAPDH-forward 5′-CGTCTTCACCACCATGGAGAAGGC-3′;
- GAPDH-reverse 5′-AAGGCCATGCCAGTGAGCTTCCC-3′;
Cell Viability Assay
24 hrs after HCT-116 cells transfected with 25 pmol of miR-NC or miR-802 mimic in 6-well plates, cells were seeded into 96-well plates at a density of 2 × 103 cells/well and cultured for 0, 24, 48 and 72 h. Then, 10 μL of Cell Counting Kit-8 (CCK-8, Beyotime, China) reagent was added to each well. The 96-well plate was placed in a 37°C, 5% CO2 incubator, and the absorbance was measured at 450 nm using a microplate reader after 2 h incubation.
Cell Migration and Invasion Assay
24 hrs after HCT-116 cells transfected with 25 pmol of miR-NC or miR-802 mimic in 6-well plates, cells were cultured in serum-free medium for 12 h. Then, cells were collected and the density was adjusted to 4–5 × l05/mL. A transwell chamber with 8-μm pores (Corning, Corning, NY, USA) was used for migration assay. DMEM medium (500 μL) containing 10% FBS was placed in the lower layer, and 200 μL of the cell suspension in serum-free media was placed in the upper chamber. Then, the cells were incubated for 10 h. The cells on the bottom surface of the chamber were fixed with glacial acetic acid for 15–30 min and stained with crystal violet for 30 min, and 10 fields were selected randomly to count by using light microscope with a magnification of 200 ×. The same method was used to perform cell invasion assay, except the chamber insert has Matrigel on it (BD, Franklin, NJ, USA).
Dual-Luciferase Reporter Assay
The potential targets of miR-802 were analyzed by TargetScan7.0 (http://www.targetscan.org) and miRanda (http://www.microrna.org) softwares. The wild type (WT) RAN and the target site mutant (MT) 3ʹ-UTR sequence were amplified by PCR, cloned into the dual-luciferase reporter plasmid (Promega, Madison, WI, USA) and named as pGL3-RAN-3ʹ-UTR-WT (WT vector) (5ʹ-CTAGAAATTAGGATTAATAAAGATGGCACTTTCCCGTTTTATTCCAGTT-3ʹ) and pGL3-RAN-3ʹ-UTR-MT (MT vector) (5ʹ-CTAGAAATTAGGATTAATAAAGATGTTTGCACCCCGTTTTATTCCAGTT-3ʹ). The mutated putative miR-802 binding site in the RAN 3ʹUTR was generated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, Cedar Creek, USA) according to the manufacturer’s protocols. HCT-116 cells in logarithmic growth phase were plated into 96-well plates at 1.5 × 104 cells/well. And then HCT-116 cells were co-transfected with WT or MT vectors and miR-802 mimics or miR-NC using Attractene Transfection Reagent (Qiagen). After 48 hrs of transfection, the ratio of firefly to Renilla luciferase activity was measured using a dual-luciferase reporter system (Promega, Madison, WI, USA).
Western Blot Analysis
Lysates from the tissues and cells were prepared by using RIPA buffer (50 mM Tris-HCl, 150 mM sodium chloride, 1mM EDTA, 1% NP-40, 1% sodium deoxycholic acid and 0.1% SDS). The protein concentration of each sample was measured by using the BCA kit (Pierce, USA) and adjusted to a uniform concentration. 30 µg of total proteins from each sample were separated on 10% SDS polyacrylamide gels (SDS-PAGE) and transferred to polyvinylidene fluoride membranes at 100 V for 2.5 h. The membrane was blocked with 5% fat-free milk in TBST and incubated with rabbit anti-RAN primary antibody (ab53775, Abcam, Cambridge, UK) (1:500) at 4°C for overnight. Then, horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG secondary antibody (ZB-2301, ZSGBBIO, Beijing, China) (1:5000) were added and incubated for 2 h at room temperature. The results were visualized using chemiluminescence (Millipore, MA, USA). Image J software (National Institutes of Health, Bethesda, USA) was used to quantify the protein expression levels. GAPDH (TA-08, ZSGBBIO, Beijing, China) (1:1000) was used as the control.
Statistical Analysis
Statistical analysis was performed using the SPSS 17.0 software. Data were expressed as mean ± SD. Student’s t-test was used for comparisons between the two groups. A one-way ANOVA test was performed, followed by Bonferroni’s posthoc test, to analyze difference among more than two groups. The correlation between miR-802 and clinical features was evaluated by using the Chi-Square test. The Kaplan–Meier method was used to calculate the survival curve, and log-rank test to determine statistical significance. All the experiments were repeated for at least three times with similar results. P < 0.05 was considered significant.
Results
MiR-802 Is Downregulated in Colorectal Cancer Tissues and Cell Lines
To investigate the expression profile of miR-802 in colorectal cancer tissue and colorectal cancer cell lines, we first checked its expression in 38 paired colorectal cancer specimens and corresponding adjacent normal tissue specimens from colorectal cancer patients by qRT-PCR. The results showed that miR-802 was significantly downregulated in colorectal cancer tissues compared to matched adjacent normal tissues (P<0.01) (Figure 1A). In addition, we also detected the expression of miR-802 in tumor tissue from patients with different colorectal cancer stages by using qRT-PCR. The results showed that patients with advanced colorectal cancer stages (III and IV) had lower expression of miR-802 compared with patient in lower stages (I and II) (Figure 1B). Besides, the correlation between the expression of miR-802 and patients’ survival rate was analyzed. The information and disease stages of the enrolled patients were listed in Table 1. There were 17 patients with high miR-802 expression and 21 patients with low miR-802 expression according to the miR-802 median expression (Table 1). The correlation of miR-802 expression and the OS of the patients were analyzed and the results showed that the low expression of miR-802 was apparently associated with shorter OS of patients with colorectal cancer (P<0.01) (Figure 1C). Moreover, we also measured miR-802 expression in three colorectal cancer cell lines (HCT-116, LS174T and SW480) as well as a normal human colon mucosal epithelial cell line (NCM460). Compared with NCM460 cell, miR-802 expression was significantly downregulated in three colorectal cancer lines (P<0.01) (Figure 1D). These data indicated that miR-802 expression was significantly downregulated in both colorectal cancer tissue and colorectal cancer cell lines and associated with the survival of patients. MiR802 might serve as a tumor suppressor in human colorectal cancer. HCT-116 cell line had the lowest expression of miR-802 among three colorectal cancer cell lines, and thus was selected for the following studies.
MiR-802 Overexpression Inhibits Colorectal Cancer Cell Viability, Migration and Invasion
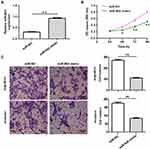
To determine the role and function of miR-802 on colorectal cancer cell viability and growth, we established miR-802 overexpressed cells by transfection of miR-802 mimic into HCT-116 cells. The qRT-PCR result confirmed that transfection of miR-802 mimic could significantly increase the expression of miR-802 in HCT-116 cells (P<0.01) (Figure 2A). To check the effects of miR-802 on viability of colorectal cancer cells, CCK-8 assay was performed. As shown in Figure 2B, overexpression of miR-802 significantly inhibited viability of HCT-116 cells (P<0.05, P<0.01) (Figure 2B). To further investigate the role of miR-802 involved in colorectal cancer metastasis, migration and invasion assays were performed in HCT-116 cells transfected with miR-802 mimic or miR-NC by transwell migration and invasion analysis. The data showed that overexpression of miR-802 remarkably suppressed migration and invasion of HCT-116 cells (P<0.01) (Figure 2C). Taken together, these results indicated that overexpression of miR-802 suppressed colorectal cancer cell viability, migration, and invasion.
RAN Is a Direct Target of miR-802 in Colorectal Cancer Cells
To explore the molecular mechanisms of how miR-802 affects the colon tumor cell viability, growth and metastasis, we searched for the target genes of miR-802 using TargetScan7.0 and miRanda. Ras-related nuclear protein (RAN) was selected as one of the candidate targets of miR-802, based on putative target sequences at 293–299 bp of RAN (Figure 3A). RAN, a small G protein that is essential for the transport of RNA and proteins through the nuclear pore complex, has been reported to belong to the Ras superfamily which is used to control DNA synthesis and cell cycle progression, and it has been found that RNA mutations disrupt DNA synthesis.18,19 To further confirm the relationship between RAN and miR-802, luciferase activity assay was performed. Luciferase activity assay results showed that overexpression of miR-802 mimic in HCT-116 cells significantly inhibited WT RAN 3ʹUTR reporter activity (P<0.01), while had no inhibitory effect on the MT RAN 3ʹUTR reporter activity (P>0.05) (Figure 3B). In addition, the mRNA and protein levels of RAN were detected in colorectal cancer cells transfected miR-802 mimic or miR-NC. Our results showed that overexpression of miR-802 significantly downregulated the mRNA and protein levels of RAN in HCT-116 cells (P<0.01) (Figure 3C and D). These results suggested that miR-802 suppressed RAN expression by binding to 3ʹUTR of RAN mRNA.
Overexpression of RAN Reverses the Effects Induced by miR-802 Overexpression in HCT-116 Cells
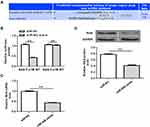
To evaluate the effect of RAN on miR-802 overexpression induced colorectal cancer cells viability and metastasis, HCT-116 cells were co-transfected with miR-802 mimic or miR-NC and RAN NC or RAN vector. Then, cell viability, migration and invasion assays were performed. Western blot assay showed that HCT-116 cells co-transfection of miR-802 mimic plus RAN NC significantly decreased RAN protein expression, while cells co-transduced with miR-802 mimic plus RAN vector presented higher RAN protein expression (P<0.01) (Figure 4A). In addition, we also found that overexpression of RAN attenuated the inhibitory effect of miR-802 mimic on cell viability, migration and invasion in HCT-116 cells (P<0.05, P<0.01) (Figure 4B and C). Thus, these results indicated that miR-802 exerts its biological roles by targeting RAN in colorectal cancer.
Discussion
MicroRNAs belong to short noncoding RNAs that have been reported to play important role in regulating gene expression by binding to the 3ʹ-UTR of their targeting mRNAs.6 Human miRNAs are frequently located at the fragile sites and genomic regions which are related to cancer.20 MiRNAs play pivotal role in the development and progression of different types of cancer by targeting tumor suppressor gene or oncogenes.5,6 In colorectal cancer, a series of miRNAs have been investigated to serve either as a tumor suppressor role or oncogenic driver.21 Among these miRNAs, miR-143 and miR-145 are initially explored to play a tumor suppressor role in colorectal cancer.10,22 In addition, miR-101, miR-133b, miR-126 and miR-142-3p are all served as a tumor suppressor in colorectal cancer, while miR-21 and miR-106a expression are significantly higher in the stool of the colorectal cancer patients.23–26 MiR-802 has been reported to play either a tumor suppressor or a congenic driver in cancers dependent on the cancer type.13–16 However, its role and function in colorectal cancer are still under investigation.
In this study, we investigated miR-802 expression and biological function in colorectal cancer. We screened 38 pairs of colorectal cancer patient sample and adjacent normal tissue and our data showed that miR-802 expression significantly downregulated in tumor tissue compared with normal adjacent tissue. The expression of miR-802 in three colorectal tumor cell lines was also downregulated. These results for the first time suggested that miR-802 might be a potential tumor suppressor in colorectal cancer. To investigate the biological role of miR-802 in colorectal cancer, CCK-8 assay was performed to check the effect of overexpression of miR-802 on the viability of colorectal cancer cell. The results proved that overexpression of miR-802 profoundly inhibited the tumor cell viability. Further migration and invasion studies indicated that overexpression of miR-802 inhibited the colon cell metastasis. All together these results first timely confirmed the tumor suppressor role of miR-802 in colorectal cancer.
More interestingly, our data indicated that Ras-associated nucleus (RAN) was directly targeted by miR-802. RAN also known as GTP-binding nuclear protein RAN, is a small G protein which is essential for the translocation of RNA and proteins through the nuclear pore complex. RAN which belong to the Ras superfamily, was reported to be applicated in the control of DNA synthesis and cell cycle progression, and mutations of RNA have been found to disrupt DNA synthesis.18,19 In our study, we found that miR-802 directly targets RAN and overexpression of miR-802 significantly downregulated the expression of RAN in colon tumor cells. Co-expression of RNA with miR-802 attenuate miR-802 induced suppression of colon tumor cell viability and metastasis. Thus, miR-802 might directly target RNA to regulate colorectal cancer cell viability and growth. Since each miRNA might have multiple target genes for the exertion of its function, other target/targets may also be investigated to verify its biological function in colorectal cancer. This is the first report to uncover the expression and the biologic role of miR-802 in colorectal cancer. Moreover, we also identified a new target gene RAN as a direct target of miR-802, indicating the molecular mechanism of miR-802 involvement in the regulation of colorectal cancer progression in vitro, which supplies the fundamental understanding for the future in vivo study and clinical-related studies in the far future.
Our studies uncovered that miR-802 serves as a tumor suppressor in both colon tumor and colorectal cancer cells. Moreover, miR-802 directly targets RNA to regulate the tumor cell viability, growth and metastasis. Thus, these findings suggested that miR-802 may be a potential biomarker for the diagnosis of colorectal cancer because of its downregulation in colorectal cancer. Moreover, it also has the potential to be explored as therapeutic target for treatment of colorectal cancer since its inhibition on colorectal cancer cell growth. However, there is still limitation for our study. Since CCK-8 assay is limited to assess the viability of colorectal cancer cells, in the future studies, BrdU incorporation will be conducted to further verify the effect of miR-802 on the proliferation of colorectal cancer cells. The effect of miR-802 on colorectal cancer cell cycle will also be conducted in the future studies. In addition, although HCT-116 cell line has been widely used for exploration of the different miRNAs role in colorectal cancer,27,28 more colorectal cancer cell lines such as LOVO, SW620, etc., and associated normal colon cell lines will be used for the future studies to verify and confirm the expression and biological function of miR-802 in the development and progression of colorectal cancer.
Conclusion
In conclusion, our study shows that miR-802 is downregulated in colorectal cancer, and overexpression of miR-802 inhibits colorectal cancer cell viability, migration and invasion by directly targeting RAN.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
2. Rozic G, Paukov L, Cohen Z, et al. STK405759 as a combination therapy with bortezomib or dexamethasone, in in vitro and in vivo multiple myeloma models. Oncotarget. 2018;9(59):31367–31379. doi:10.18632/oncotarget.25825
3. Chen W. Cancer statistics: updated cancer burden in China. Chin J Cancer Res. 2015;27(1):1.
4. Dattatreya S. Metastatic colorectal cancer-prolonging overall survival with targeted therapies. South Asian J Cancer. 2013;2(3):179–185. doi:10.4103/2278-330X.114152
5. Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3(3):83–92.
6. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
7. Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23(1):175–205. doi:10.1146/annurev.cellbio.23.090506.123406
8. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi:10.1038/nrc1997
9. Luo X, Burwinkel B, Tao S, Brenner H. MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011;20(7):1272–1286. doi:10.1158/1055-9965.EPI-11-0035
10. Michael MZ, O’Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1(12):882–891.
11. Akao Y, Nakagawa Y, Hirata I, et al. Role of anti-oncomirs miR-143 and −145 in human colorectal tumors. Cancer Gene Ther. 2010;17(6):398. doi:10.1038/cgt.2009.88
12. Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi:10.1073/pnas.0510565103
13. Yuan F, Wang W. MicroRNA-802 suppresses breast cancer proliferation through downregulation of FoxM1. Mol Med Rep. 2015;12(3):4647–4651. doi:10.3892/mmr.2015.3921
14. Wang D, Lu G, Shao Y, Xu D. microRNA-802 inhibits epithelial-mesenchymal transition through targeting flotillin-2 in human prostate cancer. Biosci Rep. 2017;37(2). doi:10.1042/BSR20160521
15. Zhang XY, Mu JH, Liu LY, Zhang HZ. Upregulation of miR-802 suppresses gastric cancer oncogenicity via targeting RAB23 expression. Eur Rev Med Pharmacol Sci. 2017;21(18):4071–4078.
16. Wang LQ, Chen G, Liu XY, Liu FY, Jiang SY, Wang Z. microRNA802 promotes lung carcinoma proliferation by targeting the tumor suppressor menin. Mol Med Rep. 2014;10(3):1537–1542. doi:10.3892/mmr.2014.2361
17. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
18. Moore MS, Blobel G. A G protein involved in nucleocytoplasmic transport: the role of Ran. Trends Biochem Sci. 1994;19(5):211–216. doi:10.1016/0968-0004(94)90024-8
19. Sazer S, Dasso M. The ran decathlon: multiple roles of Ran. J Cell Sci. 2000;113(Pt 7):1111–1118.
20. Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi:10.1073/pnas.0307323101
21. Azizian A, Epping I, Kramer F, et al. Prognostic value of MicroRNAs in preoperative treated rectal cancer. Int J Mol Sci. 2016;17(4):568. doi:10.3390/ijms17040568
22. Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16(4):845–850.
23. Link A, Balaguer F, Shen Y, et al. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19(7):1766–1774. doi:10.1158/1055-9965.EPI-10-0027
24. Schee K, Boye K, Abrahamsen TW, Fodstad O, Flatmark K. Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC Cancer. 2012;12(1):505. doi:10.1186/1471-2407-12-505
25. Xuan Y, Yang H, Zhao L, et al. MicroRNAs in colorectal cancer: small molecules with big functions. Cancer Lett. 2015;360(2):89–105. doi:10.1016/j.canlet.2014.11.051
26. Shen WW, Zeng Z, Zhu WX, Fu GH. MiR-142-3p functions as a tumor suppressor by targeting CD133, ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl). 2013;91(8):989–1000. doi:10.1007/s00109-013-1037-x
27. Gomes SE, Pereira DM, Roma-Rodrigues C, Fernandes AR, Borralho PM, Rodrigues CMP. Convergence of miR-143 overexpression, oxidative stress and cell death in HCT116 human colon cancer cells. PLoS One. 2018;13(1):e0191607. doi:10.1371/journal.pone.0191607
28. Wang XH, Yu XM, Jiang H, Luo C. Differential microRNA expression profiles in HCT116 colorectal cancer cell lines located in the lung and colon. Mol Med Rep. 2015;11(4):2903. doi:10.3892/mmr.2014.3010
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.