Back to Journals » Cancer Management and Research » Volume 12
miR-133a-3p Regulates Hepatocellular Carcinoma Progression Through Targeting CORO1C
Authors Han S, Ding X, Wang S , Xu L, Li W, Sun W
Received 19 March 2020
Accepted for publication 5 August 2020
Published 21 September 2020 Volume 2020:12 Pages 8685—8693
DOI https://doi.org/10.2147/CMAR.S254617
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Beicheng Sun
Shuangxi Han,1,2 Xuemei Ding,1 Shaohong Wang,1 Li Xu,1 Wenxiao Li,1 Wenbing Sun1
1Department of Hepatobiliary Surgery, Beijing Chao-Yang Hospital, Capital Medical University, Beijing 100043, People’s Republic of China; 2Department of Hepatobiliary Surgery, Binzhou Central Hospital, Binzhou 251700, People’s Republic of China
Correspondence: Wenbing Sun
Department of Hepatobiliary Surgery, Beijing Chao-Yang Hospital, Capital Medical University, Beijing 100043, People’s Republic of China
Email [email protected]
Introduction: MicroRNAs (miRNAs) are key modulators for gene expression via inducing translational repression or target gene degradation. miR-133a-3p was reported to stimulate or inhibit cancer progression but its role in hepatocellular carcinoma (HCC) remains to be explored.
Methods: Quantitative real-time PCR (RT-qPCR) was utilized to explore miR-133a-3p expression level in HCC cells. Dual-luciferase activity reporter assay was used to validate the direct interaction between miR-133a-3p and coronin-like actin-binding protein 1C (CORO1C). In addition, we analyzed the expression levels of miR-133a-3p and CORO1C in HCC tissues and normal tissues on the UCALAN website. Functional assays including cell counting kit-8 assay, colony formation assay, flow cytometry analysis and transwell invasion assay were conducted to explore the biological functions of miR-133a-3p in HCC.
Results: miR-133a-3p was found to have downregulated expression in HCC tissues and cells. Meanwhile, we showed that low miR-133a-3p levels were correlated with poorer overall survival of HCC patients. Overexpression of miR-133a-3p suppressed HCC cell growth and invasion but promoted cell apoptosis via targeting CORO1C.
Discussion: Our results revealed a novel mechanism of miR-133a-3p in regulating HCC progression and provided evidence that miR-133a-3p functions as a tumor suppressor in HCC.
Keywords: miR-133a-3p, CORO1C, hepatocellular carcinoma, prognosis
Introduction
Novel treatment measures for cancers are continually being developed and are able to improve the overall survival of cancer patients.1 Hepatocellular carcinoma (HCC) accounts for a large portion of liver cancer cases.2 HCC at advanced stages remains incurable at present as surgical approaches and liver transplantation methods are only feasible for tumors at early stages.3
microRNAs (miRNAs) were firstly identified in 1993 and have attracted more and more attention since then due to their roles in regulating gene expression.4 miRNAs are a type of non-coding RNAs with a length of 18–25 nucleotides and could affect as much as 60% protein coding gene expression.4,5 Moreover, it was found multiple miRNAs can target one mRNA, while various mRNAs can be affected by one miRNA.5
Accumulating studies have indicated that miR-133a-3p serves in a tumor suppressive role in cancers.6–8 For instance, Zhang et al6 found forced miR-133a-3p expression inhibits autophagy to hinder gastric cancer metastasis via blocking GABARAPL1 and ATG13 expression.6 In addition, Yin et al7 showed miR-133a-3p expression level was decreased in esophageal squamous cell carcinoma, and it could negatively regulate COL1A1 expression.7 Functional assays indicated miR-133a-3p was able to inhibit cell growth by inhibiting apoptosis via affecting COL1A1.7 Furthermore, Tang et al revealed miR-133a-3p expression was lower in prostate cancer tissues than in normal tissues.8 Moreover, low miR-133a-3p expression level was revealed as an indicator for worse overall survival.8
Here, we report that miR-133-3p has decreased expression in HCC tissues and cells compared with normal counterparts. Moreover, low miR-133a-3p expression level was revealed as an indicator for poorer overall survival of cancer patients. In addition, overexpression of miR-133a-3p inhibits HCC cell proliferation, colony formation, invasion, and promotes apoptosis.
Materials and Methods
Cell Culture
HuH-7 and HCCLM3 cells and normal hepatocyte cell L02 were bought from the Cell Bank of the Chinese Academy of Sciences. These cells were incubated with RPMI 1640 in supplement with 10% FBS (both purchased from Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) in a 37°C humified incubator containing 5% CO2.
Plasmid Construction
Sequence of CORO1C was inserted into pcDNA3.1 by GenScript. Upregulated expression of CORO1C was accompanied by transfecting pCORO1C into HCC cells.
Cell Transfection
miR-133a-3p mimic (5ʹ-UUUGGUCCCCUUCAACCAGCUG-3ʹ), miR-133a-3p inhibitor (5ʹ-CAGCUGGUUGAAGGGGACCAAA-3ʹ), and corresponding controls (NC-mimic (5ʹ- UUCUCCGAACGUGUCACGUTT-3ʹ) or NC-inhibitor (5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ)) were synthesized by RiboBio Inc. (Guangzhou, Guangdong, People's Republic of China). Small interfering RNA against CORO1C (si-CORO1C, 5ʹ-GCACAAGACUGGUCGAAUU-3ʹ) and corresponding control (NC-siR, 5ʹ-AUGACGAGUACAAGCUUGC-3ʹ) were also bought from RibiBio Inc. Cells were transfected with miRNAs, siRNAs or constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were collected after 48 hour transfection for functional analyses.
Quantitative Real-Time PCR (RT-qPCR)
Total RNA was extracted from cells using TRIzol (Invitrogen) based on the manufacturer’s instructions and quantified using NanoDrop-1000. An equal amount of RNA sample was reverse transcribed into complementary DNA with PrimeScript Mix (Takara, Dalian, Liaoning, People's Republic of China). RT-qPCR was performed using SYBR Green (Takara) at ABI 7500 system (Applied Biosystems, Foster City, CA, USA) with primers: CORO1C sense, 5′-TCCTCCCTCTGCACAAGACT-3′ and reverse, 5′-GGATCTGCCATACCATGACC-3′; GAPDH sense, 5′-GTCAACGGATTTGGTCTGTATT-3′ and reverse, 5′-AGTCTTCTGGGTGGCAGTGAT-3′; miR-133a-3p sense, 5ʹ-GCCTTTGGTCCCCTTCAAC-3ʹ and reverse, 5ʹ-TATGCTTGTTCTCGTCTCTGTGTC-3ʹ, U6 small nuclear RNA (U6 snRNA) sense, 5ʹ-ATTGGAACGATACAGAGAAGATT-3ʹ and reverse 5ʹ-GGAACGCTTCACGAATTTG-3ʹ. RT-qPCR procedures were: 1 cycle of 95°C for 30 seconds, 40 cycles of 95°C for 5 seconds and 60°C for 30 seconds. Relative gene expression levels were calculated using the 2−ΔΔCt method with GAPDH or U6 snRNA as internal controls.
Western Blot Analysis
Cells were washed with cold PBS and treated with RIPA lysis buffer (Beyotime, Haimen, Jiangsu, People's Republic of China) to extract protein samples. BCA kit (Beyotime) was used to quantify protein concentration. Then 30 μg protein sample was isolated at 10% SDS-PAGE and transferred to PVDF membrane according to the recommended procedures. Membranes were washed with TBST, blocked by fat-free milk, and incubated with primary antibodies (anti-CORO1C: Cat #14749-1-AP, Invitrogen; anti-GAPDH: Cat #MA5-35235, Invitrogen) overnight. Then, membranes were incubated with horseradish peroxidase-linked goat anti-Rabbit secondary antibody (ab6721, Abcam, Cambridge, MA, USA) at room temperature. Finally, protein bands were visualized using BeyoECL kit (Beyotime) and analyzed with ImageJ software.
Cell Counting Kit-8 (CCK-8) Assay
CCK-8 (Beyotime) assay was employed to measure cell proliferation rate. A total of 2,000 cells were seeded into a 96-well plate and then treated with 10 μL CCK-8 solution and further incubated for 2 hours. Optical density at 450 nm was measured using a microplate reader.
Colony Formation Assay
A total of 3,000 cells were seeded onto a 6-well plate and grown for 14 days. Colonies were fixed with methanol, and stained with crystal violet in phosphate-buffered saline (PBS). Colony numbers from five fields were calculated under the microscope.
Cell Apoptosis Assay
Flow cytometry assay was used to detect cell apoptosis rate using an Annexin V-FITC/PI kit purchased from Beyotime. Briefly, cells were collected, suspended in binding buffer, and stained with Annexin V-FITC and PI solution in the dark for 30 minutes. Cell apoptosis rate was measured using flow cytometry equipped with FlowJo software.
Cell Invasion Assay
Cell invasion ability was measured using a Matrigel (BD Biosciences, San Jose, CA, USA) coated 24-well transwell chamber (Corning, Inc., Corning, NY, USA). A total of 50,000 cells suspended in serum-free medium were seeded in the upper chamber of the transwell chamber. FBS-containing medium was added to the lower chamber. After 48 hours of incubation, non-invasive cells were removed, while invaded cells were washed with PBS, fixed by paraformaldehyde, stained with crystal violet, and counted under a microscope from five independent fields.
Luciferase Activity Reporter Assay
Targets for miR-133a-3p were analyzed using TargetScan software. Wild-type and mutant sequence of CORO1C was inserted into pMIR-REPORT to obtained CORO1C-wt/mt luciferase vectors. Cells were seeded in 24-well plates and transfected with miRNAs and luciferase constructs using Lipofectamine 2000 for 48 hours. Relative luciferase activity was measured using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) and normalized to Renilla luciferase.
Bioinformatical Analysis
Kaplan-Meier website (http://kmplot.com/analysis/index.php?p=background) was used to explore the clinical significance of miR-133a-3p in HCC. UALCAN (http://ualcan.path.uab.edu/analysis.html) was used to explore miR-133a-3p and CORO1C expression levels in HCC tissues and normal tissues.
Animal Experiments
Male BABL/c mice (5–6 weeks, HFK Biotechnology, People's Republic of China) were incubated in a specific pathogen-free room. Animal study protocol was approved by animal care and use committee of Beijing Chao-Yang Hospital, Capital Medical University. Each mouse was injected with miR-133a-3p mimic or NC-mimic transfected Huh-7 cells in the back. A slide caliper was used to measure tumor length and width every 7 days. Tumor volume was calculated with the formula of length× width2/2. After 21 days, the mice were killed to measure tumor weight.
Statistical Analysis
Student’s t-test, one-way ANOVA and Tukey post hoc test were used to analyze differences in groups using SPSS 16.0 (SPSS Inc., Chicago, IL, USA). A P-value less than 0.05 was identified as statistically significant.
Results
Low miR-133a-3p Expression in HCC
miR-133a-3p expression level in HCC tissues and normal tissues was firstly analyzed on the UALCAN website and the results showed miR-133a-3p had decreased expression in HCC tissues compared with normal tissues (Figure 1A). Next, we analyzed miR-133a-3p expression level in HCC cells and normal cells using RT-qPCR. Our results showed the miR-133a-3p expression level was lower in HCC cells compared with normal cells (Figure 1B). Moreover, Kaplan-Meier curve showed that low miR-133a-3p expression levels were a predictor for poorer overall survival of HCC patients (Figure 1C).
 |
Figure 1 Low expression of miR-133a-3p in HCC. |
miR-133a-3p Overexpression Inhibits HCC Cell Growth and Invasion
Gain-of-function experiments were conducted to explore the effects of miR-133a-3p on HCC cell growth and invasion. We found miR-133a-3p mimic transfection increased miR-133a-3p levels in HCC cells (Figure 2A). CCK-8 assay showed miR-133a-3p overexpression inhibits HCC cell proliferation (Figure 2B). Colony formation assay showed that, compared with the NC-mimic transfected group, colony formation ability was significantly suppressed by miR-133a-3p mimic (Figure 2C). Flow cytometry assay indicated introduction of miR-133a-3p mimic promoted cell apoptosis (Figure 2D). Transwell invasion assay showed cell invasion ability was significantly inhibited by miR-133a-3p mimic (Figure 2E). Besides that, we found HCC cells transfected with NC-mimic did not alter cell proliferation and invasion compared with those without transfection (Supplementary Figure 1A-D).
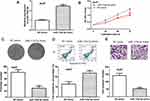 |
Figure 2 miR-133a-3p overexpression inhibits HCC cell growth and invasion. |
miR-133a-3p Knockdown Promotes HCC Cell Growth and Invasion
Then, we performed loss-of-function experiments and found transfection of miR-133a-3p inhibitor reduced expression levels of miR-133a-3p in HCC cells (Figure 3A). CCK-8 assay, colony formation assay, and flow cytometry assay indicated that knockdown of miR-133a-3p stimulates HCC cell growth (Figure 3B–D). Moreover, transwell invasion assay demonstrated knockdown of miR-133a-3p promotes cell invasion (Figure 3E).
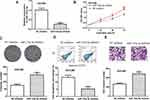 |
Figure 3 miR-133a-3p knockdown promotes HCC cell growth and invasion. |
CORO1C was a Direct Target for miR-133a-3p
Furthermore, we predicted the potential target of miR-133a-3p in HCC using TargetScan, and we selected CORO1C for analyses as it ranks top among all predicted targets and serves as an oncogene in cancers (Figure 4A). UALCAN analysis indicated CORO1C had increased expression in HCC tissues compared with normal tissues (Figure 4B). RT-qPCR analysis showed CORO1C expression level was increased in HCC cells compared with normal cells (Figure 4C). Luciferase activity reporter assay revealed miR-133a-3p mimic transfection could reduce relative luciferase activity of cells with CORO1C-wt transfection (Figure 4D). In addition, RT-qPCR assay showed miR-133a-3p mimic transfection reduced mRNA levels of CORO1C in HCC cells (Figure 4E). Western blot assay confirmed CORO1C protein level could also be decreased by miR-133a-3p mimic (Figure 4F).
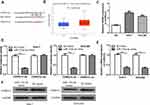 |
Figure 4 CORO1C was a direct target for miR-133a-3p. |
miR-133a-3p Regulates HCC Cell Behaviors by Regulating CORO1C
Finally, rescue experiments were conducted as to whether CORO1C was the functional target of miR-133a-3p in HCC. It was found CORO1C overexpression could stimulate HCC cell growth and invasion (Figure 5A–D). Importantly, we showed miR-133a-3p overexpression could partially reverse the stimulatory effects of CORO1C on HCC cell growth and invasion (Figure 5A–D).
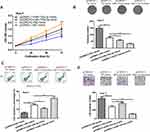 |
Figure 5 CORO1C overexpression reversed the effects of miR-133a-3p overexpression on HCC cell behaviors. |
Moreover, we found that silencing the expression of CORO1C could inhibit HCC cell growth and invasion (Figure 6A–D). Interestingly, it was found that silencing the expression of miR-133a-3p attenuated the inhibitory effects of si-CORO1C on HCC cell behaviors (Figure 6A–D).
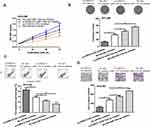 |
Figure 6 CORO1C knockdown reversed the effects of miR-133a-3p knockdown on HCC cell behaviors. |
miR-133a-3p Suppresses Tumor Growth in vivo
We further explored the roles of miR-133a-3p on tumor growth in vivo using an animal model. The results showed miR-133a-3p treatment significantly suppressed tumor volume and weight (Figure 7A and B).
 |
Figure 7 miR-133a-3p overexpression inhibits HCC tumor growth. |
Discussion
miRNAs can function either in a tumor suppressive role or an oncogenic role in cancerprogression.9–11 The appearance of the first comprehensive analysis of miRNA expression profile in HCC was reported in 2006.12 Since then, more and more studies have been performed to investigate the role of miRNAs in HCC. A recent work explored abnormal expression status of miRNAs in drug resistance of HCC cells linked to miRNAs and overall survival of cancer patients.13
miR-133a-3p was previously reported to serve as tumor suppressive miRNA in several cancers but its role in HCC was not reported. Here, we showed miR-133a-3p had reduced expression in HCC, and correlated with the overall survival of HCC patients. Gain-of and loss-of function experiments showed miR-133a-3p inhibits HCC cell proliferation, colony formation, invasion, and promotes cell apoptosis. The in vivo experiment results suggested that forcing the expression of miR-133a-3p suppresses HCC tumor growth in vivo. These results suggested a tumor suppressive role of miR-133a-3p in HCC, which is consistent with its role in other cancer types.6–8
CORO1C contains a conserved region that is enclosed by Gly-His and Trp-Asp and could promote cancer progression.14 For example, high CORO1C expression was found in gastric cancer patients with advanced tumor stages, poorer relapse-free survival, and worse overall survival.15 In addition, knockdown of CORO1C suppresses gastric cancer cell growth, mitosis, and metastasis.15 Besides that, CORO1C was found to be regulated by YBX1 to promote breast cancer metastasis.16 Moreover, CORO1C was found to be negatively regulated by miR-206 in triple‐negative breast cancer and thus to affect cancer cell migration and proliferation.17 In addition, this work reported targeting of CORO1C results in a remodeling of the actin filaments and thereby altered cancer cell morphology and migratory behavior, which may be a mechanism for the roles of CORO1C in cancer.17 Here, we showed CORO1C overexpression could promote HCC cell growth and invasion, while the knockdown of CORO1C caused the opposite effects on HCC cell behaviors. These results suggested the oncogenic role of CORO1C. Moreover, we found CORO1C overexpression could reverse the inhibitory effects of miR-133a-3p overexpression on HCC cell behaviors, indicating CORO1C was a functional target of miR-133a-3p. We have to admit one limitation of this work which is we did not further explore the downstream signaling pathways or molecules responsible for the roles of CORO1C in regulating cancer progression. In future, the clinical significance of miR-133a-3p and CORO1C and their correlations with the clinicopathological parameters of HCC patients should be explored to validate the findings of our study.
Conclusion
In conclusion, we showed miR-133a-3p could inhibit HCC tumor growth in vitro and in vivo through regulating CORO1C, suggesting a tumor suppressive role of miR-133a-3p.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hu Z, Ott PA, Wu CJ. Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat Rev Immunol. 2018;18(3):168–182. doi:10.1038/nri.2017.131
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
3. Imai N, Ishigami M, Ishizu Y, et al. Transarterial chemoembolization for hepatocellular carcinoma: A review of techniques. World J Hepatol. 2014;6(12):844–850. doi:10.4254/wjh.v6.i12.844
4. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi:10.1016/0092-8674(93)90529-Y
5. Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31(13):1609–1622. doi:10.1038/onc.2011.354
6. Zhang X, Li Z, Xuan Z, et al. Novel role of miR-133a-3p in repressing gastric cancer growth and metastasis via blocking autophagy-mediated glutaminolysis. J Exp Clin Cancer Res. 2018;37(1):320. doi:10.1186/s13046-018-0993-y
7. Yin Y, Du L, Li X, Zhang X, Gao Y. miR-133a-3p suppresses cell proliferation, migration, and invasion and promotes apoptosis in esophageal squamous cell carcinoma. J Cell Physiol. 2019;234(8):12757–12770. doi:10.1002/jcp.27896
8. Tang Y, Pan J, Huang S, et al. Downregulation of miR-133a-3p promotes prostate cancer bone metastasis via activating PI3K/AKT signaling. J Exp Clin Cancer Res. 2018;37(1):160. doi:10.1186/s13046-018-0813-4
9. Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi:10.1073/pnas.242606799
10. Bajan S, Hutvagner G. Regulation of miRNA processing and miRNA mediated gene repression in cancer. Microrna. 2014;3(1):10–17. doi:10.2174/2211536602666140110234046
11. He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi:10.1038/nature03552
12. Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25(17):2537–2545. doi:10.1038/sj.onc.1209283
13. Pratama MY, Pascut D, Massi MN, Tiribelli C. The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann Transl Med. 2019;7(20):577. doi:10.21037/atm.2019.09.142
14. Rosentreter A, Hofmann A, Xavier CP, Stumpf M, Noegel AA, Clemen CS. Coronin 3 involvement in F‐actin‐dependent processes at the cell cortex. Exp Cell Res. 2007;313(5):878–895. doi:10.1016/j.yexcr.2006.12.015
15. Cheng X, Wang X, Wu Z, Tan S, Zhu T, Ding K. CORO1C expression is associated with poor survival rates in gastric cancer and promotes metastasis in vitro. FEBS Open Bio. 2019;9(6):1097–1108. doi:10.1002/2211-5463.12639
16. Lim JP, Shyamasundar S, Gunaratne J, Scully OJ, Matsumoto K, Bay BH. YBX1 gene silencing inhibits migratory and invasive potential via CORO1C in breast cancer in vitro. BMC Cancer. 2017;17(1):201. doi:10.1186/s12885-017-3187-7
17. Wang J, Tsouko E, Jonsson P, et al. miR-206 inhibits cell migration through direct targeting of the actin-binding protein coronin 1C in triple-negative breast cancer. Mol Oncol. 2014;8(8):1690–1702. doi:10.1016/j.molonc.2014.07.006
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
