Back to Journals » Infection and Drug Resistance » Volume 16
Microbiological Analysis and Mortality Risk Factors in Patients with Polymicrobial Bloodstream Infections
Authors Xu J, Yuan Y, Wang B, Zhang Q , Wang J, Wang S, Li Y, Yan W
Received 16 March 2023
Accepted for publication 12 June 2023
Published 20 June 2023 Volume 2023:16 Pages 3917—3927
DOI https://doi.org/10.2147/IDR.S412669
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Junhong Xu,1 Youhua Yuan,1 Baoya Wang,1 Qi Zhang,1 Jing Wang,2 Shanmei Wang,1 Yi Li,1 Wenjuan Yan1
1Department of Clinical Laboratory, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, and People’s Hospital of Henan University, Zhengzhou, Henan, 450003, People’s Republic of China; 2Xinyang Third People’s Hospital, Xinyang, Henan, 464000, People’s Republic of China
Correspondence: Yi Li; Wenjuan Yan, Department of Clinical Laboratory, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, and People’s Hospital of Henan University, Zhengzhou, Henan, 450000, People’s Republic of China, Tel +86-371-65580484, Fax +86-371-87160318, Email [email protected]; [email protected]
Purpose: To study the etiological characteristics and risk factors affecting the prognosis of patients with polymicrobial bloodstream infections.
Patients and Methods: Overall, 141 patients with polymicrobial bloodstream infections in Henan Provincial People’s Hospital during 2021 were included. Laboratory test indexes, department of admission, sex, age, intensive care unit (ICU) admission, surgical history, and central venous catheter placement were collected. Patients were divided into surviving and deceased groups based on outcomes at discharge. Mortality risk factors were identified by univariate and multivariable analyses.
Results: Seventy-two of 141 patients survived. Patients were mainly from the ICU and the Departments of Hepatobiliary Surgery and Hematology. Overall, 312 microbial strains were detected: 119 gram-positive, 152 gram-negative, and 13 anaerobic bacteria and 28 fungi. Among the gram-positive bacteria, coagulase-negative staphylococci were most frequent (44/119, 37%), followed by enterococci (35/119, 29.4%). Among coagulase-negative staphylococci, methicillin-resistant coagulase-negative staphylococci incidence was 75% (33/44). Among gram-negative bacteria, Klebsiella pneumoniae was most common (45/152, 29.6%), followed by Escherichia coli (25/152, 16.4%) and Pseudomonas aeruginosa (13/152, 8.6%). Among K. pneumoniae, the incidence of carbapenem-resistant (CR) K. pneumoniae was 45.7% (21/45). On univariate analysis, mortality risk factors included increased white blood cells and C-reactive protein, decreased total protein and albumin, CR strains, ICU admission, central venous catheter, multiple organ failure, sepsis, shock, pulmonary diseases, respiratory failure, central nervous system diseases, cardiovascular diseases, hypoproteinemia, and electrolyte disturbances (P < 0.05). Multivariable analysis showed that ICU admission, shock, electrolyte disorders, and central nervous system diseases were independent mortality risk factors. The survival curve shows that the survival rate of patients with polymicrobial CR bloodstream infections was lower than that of patients with polymicrobial non-CR bloodstream infections (P=0.029).
Conclusion: Patients with polymicrobial bloodstream infections are typically critically ill and harbor multidrug-resistant bacteria. Thus, to minimize mortality rate in critically ill patients, changes in infectious flora should be monitored, antibiotics selected reasonably, and invasive procedures reduced.
Keywords: clinical characteristic, etiology, intensive care unit, polymicrobial bloodstream infections, risk factors
Introduction
Bloodstream infections often occur in patients with primary medical conditions who have high clinical morbidity and mortality rates, as compared with the general population. The risk factors for bloodstream infections include diabetes, chronic liver diseases, cardiovascular diseases, immunosuppressive diseases, hematologic diseases, and tumors.1–3
The pathogens involved in bloodstream infections are typically bacteria and fungi, and patients are mostly infected with a single pathogen. Clarification of pathogen species and drug susceptibility using blood culture can guide the rational use of antibiotics in clinical practice.2 With the advancement of medical technology, the number of critically ill patients has increased, resulting in more complex bloodstream infections. These are no longer limited to a single type of pathogen: polymicrobial bloodstream infections have emerged. Some studies have shown that compared to single bacterial bloodstream infections, polymicrobial bloodstream infections not only have statistical differences in etiology, but also have a more frequent occurrence of septic shock; moreover, the mortality rate is twice that of single bacterial bloodstream infections. Other studies have also shown that pathogens with polymicrobial bloodstream infections have higher multidrug resistance, and the intensive care unit is the main location for patients with polymicrobial bloodstream infections.4,5 However, absence studies have investigated the etiological characteristics of polymicrobial bloodstream infections and mortality risk factors.
Therefore, this study retrospectively collected the laboratory test results and clinical data of patients with polymicrobial bloodstream infections in Henan Provincial People’s Hospital within a 1-year period, to analyze the etiological characteristics of patients with polymicrobial bloodstream infections and mortality risk factors to provide a basis for reducing the mortality rate of hospitalized patients with polymicrobial bloodstream infections.
Materials and Methods
Patients
We analyzed 1318 patients with positive blood culture and found that 141 patients had polymicrobial bloodstream infections in Henan Provincial People’s Hospital from 1 January 2021 to 31 December 2021; these patients were selected as study participants. The inclusion criterion was that at least two types of bacteria were detected in the same blood culture. The exclusion criteria were inadequate clinical or laboratory data, including transfer or discharge without relevant clinical diagnosis and treatment process, resulting in insufficient clinical data and unclear patient outcomes. For example, when we collected the clinical and laboratory results of patients with bloodstream infection, if the analyzed variables (such as underlying diseases history or total protein result) were missing, that particular patient was excluded from the analysis.
Methods
Collection of Clinical Data
The clinical data of patients with polymicrobial bloodstream infections, including sex, age, department, length of hospitalization, surgical history, central venous catheter placement, and underlying diseases (tumors, malignant hematologic diseases, multiple organ failure, hypertension, diabetes, sepsis, shock, pulmonary diseases, respiratory failure, hepatobiliary diseases, central nervous system diseases, cardiovascular diseases, renal diseases, bone and joint diseases, anemia, hypoproteinemia, and electrolyte disorders), were collected using the hospital’s system based on Donghua software. The definition of admission diagnosis was based on the patient’s diagnosis on admission, classified according to the International Classification of Diseases-11 (ICD-11).6 Sepsis was defined as life-threatening organ dysfunction caused by a dysregulated host response to infection.7 The death or survival of a patient at discharge defined the treatment outcome of a patient.
Collection of Laboratory Test Results
Information on the pathogen species and drug susceptibility were collected from the blood culture results of patients with polymicrobial bloodstream infections, using the laboratory information system, based on Ruimei software. A BACTEC FX automatic blood culture instrument was used for blood culture, a Germany Bruker mass spectrometer (MALTI-TOF) was used for strain identification, and a PHOENIX100 automatic drug sensitivity identification system (American BD) was used for bacterial drug sensitivity. The antifungal susceptibility test was performed with ATB FUNGUS 3 strips (Vitek, bioMérieux, Marcy-l’Étoile, France); fastidious bacteria and anaerobic bacteria were not used for drug sensitivity. The antibiotic sensitivity breakpoint was referenced according to the criteria of the Clinical and Laboratory Standards Institute (CLSI). The quality control strains were the standard strains purchased by the Ministry of Health, including Escherichia coli ATCC25922, Streptococcus pneumoniae ATCC49619, Staphylococcus aureus ATCC29213, Pseudomonas aeruginosa ATCC27853, and Enterococcus faecalis ATCC29212. Laboratory findings were obtained within 24 h of blood culture collection, including white blood cell count (WBC), neutrophil percentage, platelet count, C-reactive protein (CRP), procalcitonin, total protein (TP), and albumin (ALB). The SYSMEX-CS5100 (Siemens Healthineers, Erlangen, Germany), a fully automated blood coagulation analyzer, and VITROS-5600 (Ortho Clinical Diagnostics; Raritan, NJ, USA), a fully automated biochemical and immunoassay analyzer, were used.
Statistical Analysis
SPSS v20.0 software (IBM Inc., Armonk, NY, USA) was used for data analysis in this study. The enumeration data were described by frequency and percentage, whereas the measurement data were described by the mean and standard deviation or median and quartiles. The t-test and rank-sum test were used for univariate analysis of measurement data, depending on whether the data conformed to a normal distribution, whereas the chi-squared test was used for qualitative data. Variables with P values <0.05 in the univariate analyses were selected for possible inclusion in in the multivariate logistic regression. We used backward stepwise bivariate logistic regression to select variables for inclusion in the final multivariate logistic regression model. P < 0.05 indicated that a difference was statistically significant. GraphPad Prism (GraphPad Inc., La Jolla, CA, USA) was used to plot survival curves.
Results
Characteristics of Strains in Patients with Polymicrobial Bloodstream Infections
Among 1318 patients of blood culture positive, 141 had polymicrobial bloodstream infections (incidence, 10.1% [141/1318]). Among the 141 patients with polymicrobial bloodstream infections, two strains were detected in 117 patients, three strains were detected in 18 patients, and four strains were detected in six patients. Of the 312 total strains, 152 were gram-negative bacteria, 117 were gram-positive bacteria, 28 were fungi (11 Candida albicans, 8, C. tropicals, 6 C. prapsilosis, and 3 C. glabrata),13 were anaerobic bacteria, and 2 were Actinomycetes (Figure 1).
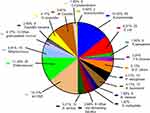 |
Figure 1 Distribution of strains in patients with polymicrobial bloodstream infection. |
Drug Susceptibility of Strains in Polymicrobial Bloodstream Infections
The isolation rate of carbapenem-resistant (CR) Enterobacteriaceae was 22.2% (22/99) among 99 Enterobacteriaceae strains, including one isolate of Enterobacter cloacae, one isolate of Serratia marcescens and 20 isolates of Klebsiella pneumoniae (CRKPN). The incidence rate (44.4%, 20/45) of CRKPN was significantly higher than that of other strains. There were 25 strains of carbapenem-sensitive K. pneumoniae, and the incidence rate of extended spectrum beta-lactamases (ESBLs) was 28% (7/25). Of the 25 strains of E. coli without detectable CR, the incidence rate of ESBLs was 68% (17/25). The isolation rate of CR P. aeruginosa (CRPA) was 38.5% (5/13). Nine of the 13 Acinetobacter baumannii strains were pan-resistant and only susceptible to polymyxin B and tigecycline.
Among the 54 strains of staphylococci, the incidence rates of methicillin-resistant S. aureus (MRSA) and methicillin-resistant coagulase-negative staphylococci (MRSCN) were 40% (4/10) and 75% (33/44), respectively. There were 35 strains of enterococci, with a penicillin susceptibility rate of 37.1% (13/35), and the positivity rate for high-concentration gentamicin screening was 40% (14/35).
The drug susceptibility test for the 28 fungal strains revealed a susceptibility rate of 100% for caspofungin, amphotericin B, and 5-fluorocytosine; 93.1% for voriconazole; 79.3% for itraconazole; and 65.5% for fluconazole.
Distribution of the Departments of Origin of Patients with Polymicrobial Bloodstream Infections
The 141 patients with polymicrobial bloodstream infections were mainly distributed in ICU and Departments of Hepatobiliary Surgery, Hematology, Orthopedics, Infection, and Gastrointestinal Surgery (Figure 2).
 |
Figure 2 Distribution of departments of patients with polymicrobial bloodstream infections. |
Analysis of Mortality Risk Factors in Patients with Polymicrobial Bloodstream Infections
Univariate analysis showed that increased WBC and CRP and decreased TP and ALB had significant effects on the prognosis of patients with polymicrobial bloodstream infections (P < 0.05) (Table 1). Admission to the ICU, central venous catheter placement, multiple organ failure, sepsis, shock, pulmonary diseases, respiratory failure, central nervous system diseases, cardiovascular diseases, hypoproteinemia, electrolyte disturbances, and detection of CR strains were mortality risk factors (P < 0.05) (Table 2).
 |
Table 1 Univariate Analysis of Mortality Risk Factors in Patients with Polymicrobial Bloodstream Infections |
 |
Table 2 Univariable Analysis of Mortality Risk Factors in Patients with Polymicrobial Bloodstream Infections |
The results of the multivariable analysis showed that a history of central nervous system diseases, admission to the ICU, shock, and electrolyte disturbances were independent mortality risk factors in patients with polymicrobial bloodstream infections (Figure 3).
 |
Figure 3 Multivariable analysis of mortality in patients with polymicrobial bloodstream infections. |
Among the 141 patients with polymicrobial bloodstream infections, 21 of 32 patients with polymicrobial CR bloodstream infections died (mortality rate: 65.6%), which was higher than that of patients with polymicrobial non-CR bloodstream infections (48/109, mortality rate: 44.0%). Survival curve analysis with 30 hospitalization days after positive blood culture showed that the survival rate of patients with polymicrobial CR bloodstream infections was lower than that of patients with polymicrobial non-CR bloodstream infections (P = 0.029) (Figure 4).
Discussion
It is noteworthy that previous literature only analyzed patients with a single bacterial bloodstream infection. No previous study in China has categorized patients with polymicrobial bloodstream infections and analyzed their clinical characteristics and mortality risk factors. Among the gram-positive bacteria, coagulase-negative staphylococci were most frequent, and of these, three-quarters were MRCNS. Among gram-negative bacteria, K. pneumoniae was most common, and of these, almost half were CRKPN. On multivariable analysis, ICU admission, shock, electrolyte disorders, and central nervous system diseases were independent risk factors for mortality. Survival rates with 30 hospitalization days after positive blood culture were lower in patients with polymicrobial CR bloodstream infections than in those with polymicrobial non-CR bloodstream infections (P = 0.029).
The incidence of polymicrobial bloodstream infections in our hospital was similar to that reported in the literature (average level, 10.9% to 15.5%).4 It has been noted in the literature that the 1-year mortality rate of patients with bloodstream infections ranges from 8% to 48%.1 In the present study, the mortality rate of 141 patients with polymicrobial bloodstream infections was 49% (69/141). It is thus inferred that the mortality rate for polymicrobial bloodstream infections in our hospital was higher than the average mortality rate for bloodstream infections in other hospitals. This may be because our hospital is one of the large tertiary hospitals in Henan Province, and thus is the referral hospital for more critically ill patients from various cities. In addition, these data are also a reminder that patients with polymicrobial bloodstream infections may have more complex clinical manifestations and treatment processes due to the diversity of infection strains; this may be the reason for the high mortality rate. However, additional data is needed to confirm this.
Patients were mainly distributed in the ICU, Department of Hepatobiliary Surgery, and Department of Hematology. This may be because ICU patients are critically ill, with systemic involvement and have mostly experienced traumas and major surgeries that typically involve invasive procedures.2,3 The Department of Hepatobiliary Surgery is the main ward for patients with digestive diseases, who are mostly at the postoperative stage, have mucosal damage caused by serious primary diseases and surgeries, and have translocation of normal flora, which causes endogenous infections. The digestive system contains the largest number of species and the largest quantity of flora in the human body, which is the main factor leading to polymicrobial infections. Patients in the Department of Hematology are also at high risk for infections due to their extremely low immunity and placement of a peripherally inserted central catheter.
The research results show that the main pathogens isolated from patients with polymicrobial blood infection include E. coli, K. pneumoniae and S. aureus and that these were associated with a high incidence rate and mortality.8 It has been reported that Bacteroides is the most isolated anaerobe in bloodstream infection, while Clostridium has the highest mortality rate (36.5%).9,10 The anaerobes we detected were mainly these two kinds of bacteria; Clostridium was more frequently detected than bacillus. There were two patients with mixed anaerobe blood infection, respectively from hepatobiliary surgery and gastrointestinal surgery. In addition, we also detected relatively rare strains of bloodstream infection, such as Aggregatibacter, Haemophilus, Eiken, and Actinomycetes,11–13 and we also detected a strain of Neisseria meningitidis14–16 from a 7-year-old patient with hematological disease, which was also complicated by Candida tropicalis infection. Moreover, four pathogens were detected among six patients with bloodstream infection, which suggests the severity of infection among these patients and emphasizes the need for physicians to prioritize blood culture and rapidly implement antibiotic treatment. For these patients with polymicrobial bloodstream infection, we speculate that the source of bacteria was mostly from damaged organs such as the digestive or respiratory tract.17–20
According to the Blood Bacterial Resistant Investigation Collaborative System report of 2014–2019, gram-negative bacteria were found to be significantly more common than gram-positive bacteria among the pathogens involved in bloodstream infections.21 However, gram-negative bacteria were only slightly more common than gram-positive bacteria among the 312 strains detected in this study, which thus differs from the literature. The isolation rate of coagulase-negative staphylococci was the highest among that of gram-positive bacteria, which was also inconsistent with the literature regarding most isolation rates of pathogens. This may be because most patients with polymicrobial bloodstream infections were from the ICU and had low immunity and because invasive operations had led to the invasion by environmental bacteria as well as normal flora from the human body.22,23 For example, the coagulase-negative staphylococci and enterococci, which demonstrated higher isolation rates, were mostly isolated from the ICU patient population.
Among the 99 Enterobacteriaceae in this study, K. pneumoniae was detected most frequently. Forty-five strains were detected, including 20 strains of CRKPN, with an isolation rate of 44.4%, which was higher than the national level of 16.1%. Eighteen of the CRKPN strains were obtained from ICU patients. In recent years, the detection rate of CRKPN has been increasing annually. It causes severe infections that are difficult to treat and poses a challenge to healthcare workers.24 Carbapenem antibiotics are the first-line drugs for clinical treatment of patients with severe infections. The CR mechanism mainly involves the production of different types of carbapenem enzymes, combined with mutation of porins, leading to the emergence of K. pneumoniae that are highly resistant to carbapenem. This study included the minimum inhibitory concentration (MIC) of imipenem and meropenem. Their MIC values were > 8 ug/mL or > 32 ug/mL, respectively, indicating highly resistant strains.25 The resistance mechanism should be further investigated. The most commonly isolated non-fermenters were P. aeruginosa and A. baumannii, with severe drug resistance.
Among the gram-positive bacteria, 54 strains of staphylococci were isolated, including 44 strains of coagulase-negative staphylococci. The incidence rate of MRSCN was 75%, which was higher than that of MRSA (40%), which was consistent with the drug resistance rates reported in the literature. Coagulase-negative staphylococci are known to be normal flora of human skin, mucous membranes, and environment. These strains that were previously used for calculating the rate of blood culture contamination have evolved into one of the pathogens of bloodstream infections in recent years.26–29 The high isolation rate in this data analysis may be because most patients with polymicrobial bloodstream infections are critically ill and immunocompromised, and coagulase-negative staphylococci then become pathogens by invading the bloodstream or these patients become infected by other pathogens that invade the bloodstream. Another reason is that the blood cultures of patients with polymicrobial infections had elevated inflammatory indexes and corresponding clinical signs, and thus it is not possible to determine whether coagulase-negative staphylococci were contaminants or pathogens in the cases selected for this trial.
With advances in medical technology and the diversity of medical treatment, although critically ill patients are saved, the number of patients with polymicrobial bloodstream infections is increasing, when compared with those with bloodstream infections caused by a single strain that were commonly seen in the past, leading to clinical treatment complexity.
In this study, patients with polymicrobial bloodstream infections were divided into surviving and deceased groups for analysis of indicators of prognosis. Univariate analysis showed that deceased patients had significant (P < 0.05) increases in WBC and CRP and decreases in TP and ALB. A significant increase in WBCs, which are important components of the immune system, suggests that a patient has a severe infection. CRP, which is a sensitive inflammatory indicator, tends to remain at high levels in the early and progressive stages of a disease and is an important laboratory reference indicator. The decrease in TP in patients with polymicrobial bloodstream infections is mostly associated with more excessive protein catabolism and less protein synthesis. Albumin maintains the normal oncotic pressure, serves as a carrier for transporting different substances to detoxification organs, and is an important component of the immunological barrier. A decrease in ALB causes an increase in the permeability of the vascular wall, which makes it easy for exogenous substances, such as bacteria, to invade the bloodstream, leading to infections. A decrease in ALB also causes a decrease in transport capacity, leading to accumulation of toxins in the blood and causing a further decrease in immunity and damage to organ functions. It has been demonstrated that increasing the serum ALB is an effective means of reducing the mortality rate of patients.30,31
The results of the univariate analysis also showed that admission to the ICU and central venous catheter placement were the risk factors for the prognosis of patients and that multiple organ failure, sepsis, shock, respiratory failure, hypoproteinemia, electrolyte disturbances, detection of CR strains, pulmonary diseases, central nervous system diseases, and cardiovascular diseases that present during the course of disease, were risk factors of mortality. These results were similar to those of previous studies of risk factors for bloodstream infections.32 These patients tend to have a rapid disease progression and are prone to complications that prolong the disease course and further reduce the immunity of patients, allowing multiple bacteria to invade into the bloodstream and causing polymicrobial bloodstream infections. The results of the multivariable analysis showed that a history of underlying central nervous system diseases, admission to ICU, shock, and electrolyte disturbances were the main mortality risk factors in these patients.33,34 Consistent with the literature, a history of central nervous system disease is a risk factor for patients with polymicrobial blood flow infections, and we also confirmed that it is a risk factor for mortality, thus this should garner clinical attention.19 It serve as a risk indicator to help clinical doctors identify patients with polymicrobial bloodstream infections. Our data analysis also showed that risk factors associated with electrolyte disorders in 56 patients with polymicrobial bloodstream infections included shock (30/39, 76.9%), respiratory failure (19/27, 70.4%), sepsis (23/36, 63.9%), and kidney disease (21/35, 60.0%), which may suggest the effect of disease progression of patients in clinical practice. The results of the survival curve analyses also showed that patients infected with CR strains had significantly lower survival rates than did those with non-CR infections, which was similar to the results of previous studies.35
This study also had some limitations. Firstly, due to it was a retrospective observatory study, information bias would occur. Secondly, the sample size was not sufficiently large. In the future, we will collect more samples of inpatients with polymicrobial bloodstream infection to further analyze mortality risk factors, with a final aim of decreasing mortality in these patients.
Conclusion
In conclusion, the results of this study showed that polymicrobial bloodstream infections in our hospital occurred mostly in critically ill patients and that coagulase-negative staphylococci were significantly predominant among the isolated pathogens, unlike the overall reports in the literature. In the future, the number of observed patients can be increased to observe any changes in pathogen trends. Analysis of mortality risk factors showed that admission to the ICU, central nervous system diseases, shock, and electrolyte disturbances were the main risk factors for mortality in these patients. This suggests that clinicians should pay marked attention to patients with such factors and that symptomatic treatment and improvement of these factors have the potential to reduce the mortality rate of critically ill patients.
Abbreviations
ALB, albumin; CR, carbapenem-resistant; CRP, C-reactive protein; ESBLs, extended spectrum beta-lactamases; ICU, intensive care unit; MRCNS, methicillin-resistant coagulase-negative staphylococci; MRSA, methicillin-resistant Staphylococcus aureus; TP, total protein; WBC, white blood cell count.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval and Informed Consent
This study was approved by the Ethics Committee of Henan Provincial People’s Hospital, Henan, China (2022-1-233). No personally identifiable information was collected in this study. The requirement for informed consent from patients was also waived. We ensured this study complied with the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by Henan Provincial Key Programs in Science and Technology [grant number: 202102310355].
Disclosure
The authors declare no conflicts of interest in this work.
References
1. McNamara JF, Righi E, Wright H, Hartel GF, Harris PNA, Paterson DL. Long-term morbidity and mortality following bloodstream infection: a systematic literature review. J Infect. 2018;77(1):1–8. doi:10.1016/j.jinf.2018.03.005
2. Lin H, Yang L, Fang J, et al. Clinical characteristics of bloodstream infection in immunosuppressed patients: a 5-year retrospective cohort study. Front Cell Infect Microbiol. 2022;12:796656. doi:10.3389/fcimb.2022.796656
3. Gerver SM, Mihalkova M, Bion JF, et al. Surveillance of bloodstream infections in intensive care units in England, May 2016–April 2017: epidemiology and ecology. J Hosp Infect. 2020;106(1):1–9. doi:10.1016/j.jhin.2020.05.010
4. Pavlaki M, Poulakou G, Drimousis P, et al. Polymicrobial bloodstream infections: epidemiology and impact on mortality. J Glob Antimicrob Resist. 2013;1(4):207–212. doi:10.1016/j.jgar.2013.06.005
5. Chen Q, Zheng Z, Shi Q, Wu H, Li Y, Zheng C. Multidrug-resistant Acinetobacter baumannii may cause patients to develop polymicrobial bloodstream infection. Can J Infect Dis Med Microbiol. 2022;2022:8368578. doi:10.1155/2022/8368578
6. Kazlauskas E, Gegieckaite G, Eimontas J, Zelviene P, Maercker A. A brief measure of the international classification of diseases-11 adjustment disorder: investigation of psychometric properties in an adult help-seeking sample. Psychopathology. 2018;51(1):10–15. doi:10.1159/000484415
7. Rhodes A, Evans LE, Alhazzani W, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Crit Care Med. 2017;45(3):486–552. doi:10.1097/CCM.0000000000002255
8. Diekema DJ, Hsueh PR, Mendes RE, et al. The microbiology of bloodstream infection: 20-year trends from the SENTRY antimicrobial surveillance program. Antimicrob Agents Chemother. 2019;63(7). doi:10.1128/AAC.00355-19
9. Watanabe T, Hara Y, Yoshimi Y, et al. Application of MALDI-TOF MS to assess clinical characteristics, risk factors, and outcomes associated with anaerobic bloodstream infection: a retrospective observational study. Ann Clin Microbiol Antimicrob. 2021;20(1):42. doi:10.1186/s12941-021-00449-4
10. Di Bella S, Antonello RM, Sanson G, et al. Anaerobic bloodstream infections in Italy (ITANAEROBY): a 5-year retrospective nationwide survey. Anaerobe. 2022;75:102583. doi:10.1016/j.anaerobe.2022.102583
11. Rubin LG, Moxon ER. Pathogenesis of bloodstream invasion with Haemophilus influenzae type b. Infect Immun. 1983;41(1):280–284. doi:10.1128/iai.41.1.280-284.1983
12. Chien YC, Huang YT, Liao CH, Chien JY, Hsueh PR. Clinical characteristics of bacteremia caused by Haemophilus and Aggregatibacter species and antimicrobial susceptibilities of the isolates. J Microbiol Immunol Infect. 2021;54(6):1130–1138. doi:10.1016/j.jmii.2020.12.002
13. Hsiao YC, Lee YH, Ho CM, Tseng CH, Wang JH. Clinical characteristics of actinomyces viscosus bacteremia. Medicina. 2021;57(10):1064. doi:10.3390/medicina57101064
14. Chiou CS, Liao YS, Chen BH, et al. Demographic features of invasive meningococcal disease in Taiwan, 1993 to 2020, and genetic characteristics of Neisseria meningitidis isolates, 2003 to 2020. Microbiol Spectr. 2022;10(4):e0088222. doi:10.1128/spectrum.00882-22
15. Melican K, Dumenil G. Vascular colonization by Neisseria meningitidis. Curr Opin Microbiol. 2012;15(1):50–56. doi:10.1016/j.mib.2011.10.008
16. Staudacher JJ, Wittenberg M, Schneider T. Bloodstream infection by Neisseria meningitidis. Dtsch Arztebl Int. 2022;119(33–34):557. doi:10.3238/arztebl.m2022.0156
17. Facchin G, Candoni A, Lazzarotto D, et al. Clinical characteristics and outcome of 125 polymicrobial bloodstream infections in hematological patients: an 11-year epidemiologic survey. Support Care Cancer. 2022;30(3):2359–2366. doi:10.1007/s00520-021-06640-9
18. Karakonstantis S, Kritsotakis EI. Systematic review and meta-analysis of the proportion and associated mortality of polymicrobial (vs monomicrobial) pulmonary and bloodstream infections by Acinetobacter baumannii complex. Infection. 2021;49(6):1149–1161. doi:10.1007/s15010-021-01663-0
19. Yo CH, Hsein YC, Wu YL, et al. Clinical predictors and outcome impact of community-onset polymicrobial bloodstream infection. Int J Antimicrob Agents. 2019;54(6):716–722. doi:10.1016/j.ijantimicag.2019.09.015
20. Claeys KC, Heil EL, Pogue JM, et al. The Verigene dilemma: gram-negative polymicrobial bloodstream infections and clinical decision making. Diagn Microbiol Infect Dis. 2018;91(2):144–146. doi:10.1016/j.diagmicrobio.2018.01.012
21. Chen Y, Ji J, Ying C, et al. Blood bacterial resistant investigation collaborative system (BRICS) report: a national surveillance in China from 2014 to 2019. Antimicrob Resist Infect Control. 2022;11(1):17. doi:10.1186/s13756-022-01055-5
22. Timsit JF, Ruppé E, Barbier F, Tabah A, Bassetti M. Bloodstream infections in critically ill patients: an expert statement. Intensive Care Med. 2020;46(2):266–284. doi:10.1007/s00134-020-05950-6
23. Abe T, Ogura H, Shiraishi A, et al. Characteristics, management, and in-hospital mortality among patients with severe sepsis in intensive care units in Japan: the FORECAST study. Crit Care. 2018;22(1):322. doi:10.1186/s13054-018-2186-7
24. Yuan Y, Wang J, Yao Z, et al. Risk factors for carbapenem-resistant Klebsiella pneumoniae bloodstream infections and outcomes. Infect Drug Resist. 2020;13:207–215. doi:10.2147/IDR.S223243
25. Bulman ZP, Krapp F, Pincus NB, et al. Genomic features associated with the degree of phenotypic resistance to carbapenems in carbapenem-resistant Klebsiella pneumoniae. mSystems. 2021;6(5):e0019421. doi:10.1128/mSystems.00194-21
26. El Houssaini Z, Harrar N, Zerouali K, Belabbes H, Elmdaghri N. Prévalence des staphylocoques à coagulase négative dans les hémocultures au Centre Hospitalier Universitaire Ibn Rochd de Casablanca [Prevalence of coagulase-negative staphylococci in blood cultures at the Ibn-Rochd University Hospital in Casablanca]. Pan Afr Med J. 2019;33. French. doi:10.11604/pamj.2019.33.193.18552
27. Asai N, Sakanashi D, Suematsu H, et al. Clinical characteristics and relevance of coagulase-negative Staphylococci other than S epidermidis by positive blood culture. J Microbiol Immunol Infect. 2021;54(4):632–638. doi:10.1016/j.jmii.2020.03.001
28. Almeida Junior ER, Braga IA, Filho PPG, Ribas RM. Multicentre surveillance of epidemiologically important pathogens causing nosocomial bloodstream infections and pneumonia trials in Brazilian adult intensive care units. J Med Microbiol. 2023;72(2). doi:10.1099/jmm.0.001654
29. Pouget C, Chatre C, Lavigne JP, Pantel A, Reynes J, Dunyach-Remy C. Effect of antibiotic exposure on Staphylococcus epidermidis responsible for catheter-related bacteremia. Int J Mol Sci. 2023;24(2):1547. doi:10.3390/ijms24021547
30. Bekhit OE, Yousef RM, Abdelrasol HA, Mohammed MA. Serum albumin level as a predictor of outcome in patients admitted to pediatric intensive care units. Pediatr Emerg Care. 2021;37(12):e855–e860. doi:10.1097/PEC.0000000000002567
31. Shannon CM, Ballew SH, Daya N, et al. Serum albumin and risks of hospitalization and death: findings from the atherosclerosis risk in communities study. J Am Geriatr Soc. 2021;69(10):2865–2876. doi:10.1111/jgs.17313
32. Hu F, Yuan L, Yang Y, et al. A multicenter investigation of 2773 cases of bloodstream infections based on China antimicrobial surveillance network (CHINET). Front Cell Infect Microbiol. 2022;12:1075185. doi:10.3389/fcimb.2022.1075185
33. Giannella M, Pascale R, Ferraro G, et al. Risk factors for treatment failure in patients receiving β-lactam/β-lactamase inhibitor combinations for Enterobacteriaceae bloodstream infection: a retrospective, single-centre, cohort study. Int J Antimicrob Agents. 2019;53(5):574–581. doi:10.1016/j.ijantimicag.2019.01.005
34. Ju M, Huang Y, Xu X, et al. Predictors of mortality in adult patients with methicillin-resistant Staphylococcus aureus bloodstream infection: a meta-analysis and systematic review. Ann Palliat Med. 2021;10(8):8617–8627. doi:10.21037/apm-21-932
35. Song F, Zhang K, Huang J, et al. Clinical characteristics, risk factors, and outcomes of patients with polymicrobial Klebsiella pneumoniae bloodstream infections. Biomed Res Int. 2021;2021:6619911. doi:10.1155/2021/6619911
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

