Back to Journals » Infection and Drug Resistance » Volume 16
Microbial Community Characterization and Molecular Resistance Monitoring in Geriatric Intensive Care Units in China Using mNGS
Authors Yang J, Li L, Zhu X, He C, Li T, Qin J, Wang Y
Received 3 June 2023
Accepted for publication 29 July 2023
Published 8 August 2023 Volume 2023:16 Pages 5121—5134
DOI https://doi.org/10.2147/IDR.S421702
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Jilin Yang,1,* Lingyi Li,2,* Xiaolin Zhu,1 Chen He,1 Ting Li,1 Jiahong Qin,1 Yijie Wang1
1Department of Critical Care Medicine, The First Affiliated Hospital of Kunming Medical University, Kunming, People’s Republic of China; 2Department of Medical, Hangzhou Matridx Biotechnology Company, Hangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yijie Wang, Tel +13888126379, Email [email protected]
Background: Surface pathogens in the ICU pose a global public health threat, especially to elderly patients who are immunocompromised. To detect these pathogens, unbiased methods such as metagenomic next-generation sequencing (mNGS) are increasingly utilized for environmental microbiological surveillance.
Methods: In a six-month study from January to July 2022, we investigated microbial communities in Chinese geriatric ICUs by regularly monitoring multiple surfaces at three-month intervals. Using mNGS sequencing, we analyzed microorganisms present at eight specific locations within the ICU. Additionally, we compared pathogen profiles and drug resistance genes between patient cultures and environmental samples collected during the same period.
Results: The microbial composition remained relatively stable over time, but significant differences in alpha diversities were observed among various surfaces such as floors, hands, pumps, trolleys, and ventilator inlets/outlets. Surfaces with high contact frequency for healthcare workers, including workstations, ventilator panels, trolleys, pumps, and beds, harbored pathogenic microorganisms such as Acinetobacter baumannii, Cutibacterium acnes, Staphylococcus haemolyticus, Pseudomonas aeruginosa, and Enterococcus faecium. Acinetobacter baumannii, particularly the carbapenem-resistant strain (CRAB), was the most frequently identified pathogen in geriatric ICU patients regardless of testing method used. The mNGS approach enabled detection of viruses, fungi, and parasites that are challenging to culture. Additionally, an abundance of drug resistance genes was found in almost all environmental samples.
Conclusion: The microbial composition and abundance in the ICU remained relatively constant over time. The floor exhibited the highest microbial diversity and abundance in the ICU environment. Drug-resistant genes in the ICU environment may migrate between patients. Overall, mNGS is an emerging and powerful tool for microbiological monitoring of the hospital environment.
Keywords: geriatric intensive care unit, environmental microorganisms, mNGS, infection and prevention and control, hospital-acquired infections
Introduction
The intensive care units (ICU) environmental microbial communities are highly diverse, consisting primarily of various bacterial and archaeal phyla, as shown by the 16S study.1 Microorganisms found in hospital environments primarily originate from human skin or outdoor air, and can include potential human pathogens as well as beneficial microorganisms with positive effects on the host.2
Although the ICU is typically subject to more stringent decontamination procedures than other parts of the hospital, patients with severe illnesses still contract hospital-acquired infections in the ICU.3 Improving disease infection management in hospitals, particularly in geriatric ICUs, is crucial. The risk of nosocomial secondary infections in geriatric ICU patients is progressively severe as they age due to co-morbidities, thymic atrophy insufficiency, and weakened T-lymphocyte proliferative response. In fact, infection is the leading cause of death for one third of individuals aged 65 and over.4 Current nosocomial infections are caused by several factors, including inadequate disinfection and sterilization of instruments, partial aerosol transmission of pathogenic bacteria, insufficient disinfection of pathogens in the environment, neglect of hand hygiene and medication-related factors, while hospital environmental surfaces can be neglected due to their non-living hosts.5
Previous studies typically monitored bacterial community characteristics in ICUs using 16S rRNA sequencing or culture methods.6 While 16S rRNA sequencing cannot accurately predict pathogenic strains, it has confirmed the presence of hospital-acquired bacteria and common opportunistic pathogens such as Acinetobacter, Burkholderia, Propionibacterium, and Escherichia, many of which were previously isolated from human clinical samples.7 Culture-based studies have identified the most common intestinal flora, including the Proteobacteria, in the ICU, indicating the direct presence of microorganisms in patients and the environment through the flow of healthcare workers or visitors.8 Escherichia coli, Staphylococcus aureus, and Klebsiella are frequently reported pathogens in long-term hospitalized patients.9 Skin-associated genera, such as Propionibacterium, Corynebacterium, Streptococcus, and Staphylococcus, are frequently deposited through skin contact due to their presence on healthcare workers’ hands and high abundance on hospital equipment surfaces.6 However, these genera are also commensal species for many healthy populations. These findings emphasize the importance of environmental monitoring in the ICU. Additionally, the proportion of nosocomial infections caused by multidrug-resistant organisms in the ICU is increasing, which limits treatment options and results in longer hospital stays, higher mortality rates, and increased costs.10 A study demonstrated that 40% of hospital wards had environmental contamination with multidrug-resistant bacteria, with Vancomycin resistant Enterococcus (VRE) being the most common.11 Furthermore, some nasal pathogens of drug-resistant microorganisms can persist on dry surfaces, such as Methicillin-resistant Staphylococcus aureus (MRSA). BlaKPC-carrying Klebsiella pneumoniae was cultivable on plastic and steel for up to 5 to 6 days.12 The prolonged survival of drug-resistant bacteria increases the likelihood of contaminated sites acting as sources of transmission.
Our study utilized Metagenomic next-generation sequencing (mNGS) methodology to monitor microbial communities on different surfaces within the geriatric ICU for six months. The primary objective was to describe the microbiota dynamics within various environments. Next-generation sequencing technology provides a greater understanding of the entire geriatric ICU microbial community and a new perspective on the hospital environment. It can vastly improve monitoring and guide medical processes for rational disinfection, providing a solid foundation for effective prevention, control, and management of hospital infections.
Materials and Methods
Study Design and Sample Collection
- From January 2022 to July 2022, geriatric ICU patients were selected every three months from a tertiary hospital in Yunnan Province, China. Sampling sites included medical workstation (healthcare professionals perform various tasks and activities related to patient care, reviewing medical records and monitoring vital signs.), healthcare workers’ hands (the hands of healthcare personnel who come into contact with patients and various surfaces within the ICU), beds (ICU beds include attachments for medical equipment and monitoring devices), trolley (a mobile cart used for transporting medical supplies, medications, and equipment within the ICU.), infusion pumps (devices used for delivering fluids, medications, or nutrients directly into a patient’s bloodstream in a controlled manner, which the nurses have the most daily contact with), respiratory machine panels (parameters like oxygen concentration and ventilation rate can be managed through the panels, one patient disposable sticker to prevent cross contamination), respiratory machine inlets and outlets (inlets connect to oxygen or air sources, while outlets expel exhaled gases from the patient’s respiratory system, it is a single-use plastic product), and floors (the horizontal surface of the ICU beneath patients, healthcare workers, and equipment, floors can accumulate microorganisms and require regular cleaning and disinfection). The images were labeled as workstation, hands, bed, trolley, pump, ventilator panel, ventilator inlets and outlets and floor. The number 1 indicates that the sample was collected in January; the number 2 indicates that the sample was collected in April; the number 3 indicates that the sample was collected in July. The sampler wore sterile gloves and wiped the selected area surface. A clean, double-ended sterile cotton swab was rotated onto the surface for 10 times, then immediately placed in a sterile, water-soaked centrifuge tube. All samples were stored at −80°C. The samples were transported to the laboratory and stored on dry ice until DNA extraction. (2) A retrospective statistical analysis was performed comparing the pathogenic microbial spectrum detected by mNGS in clinically diagnosed patients between January 2022 and July 2022 with the results of various specimens culture during the same period to determine the pathogenic microbial spectrum and drug resistance phenotype. This study was approved by the Ethics Committee of First Affiliated Hospital of Kunming Medical University (2022-L-50). The guidelines outlined in the Declaration of Helsinki were followed and legal representatives of all patients signed the informed consent before participation.
Sample Processing and Sequencing
1.2 mL of swab soaking solution was taken and placed into a 2 mL shaking tube for cell disruption. The NGS Automatic Library Preparation System was used for both DNA extraction and library preparation. Different reagents were used based on the type of sample, including those from the Nucleic Acid Extraction Kit (Cat#MD013, Hangzhou Matridx Biotechnology Co., Ltd.) and Total DNA Library Preparation Kit (body fluid samples) (Cat# MD001T, Hangzhou Matridx Biotechnology Co., Ltd.). The pooled library was sequenced using the 75 bp single-end sequencing kit (KAPA Library Quantification Kits) on the Illumina NextSeq 550.13 The qualified result for each sample is no less than 15M reads and Q30≥90%. Negative control samples were processed and sequenced in parallel during each sequencing run for quality control.14
Bioinformatics Analysis
Bowtie215 was used for mapping reads, followed by removal of adapter sequences and low-quality reads. Reads with a final length less than 50bp were removed as well as human host sequences [mapped to human reference genome hg19 (GRCh38.p13 https://www.ncbi.nlm.nih.gov/assembly/2334371), only unique mapped reads were selected). Non-human reads were classified using Kraken216 with comparison to the NCBI nt.17 Any inconsistencies between Kraken2 and Bowtie2 were validated using BLAST. Species annotation was performed using Kraken2 and a self-built microbial nucleic acid database with filtering of bacterial, fungal, archaeal, and viral sequences from the NCBI nt and RefSeq18 whole genome database. The Bracken19 was used for estimation of species abundance in the sample. Bray-Curtis Alpha and beta diversity analyses were performed using Vegan20 and Ggplot221 packages in R (version 3.6.1) based on species abundance table for sample clustering analysis. LEfSe biomarker analysis was performed using the online website https://github.com/biobakery/lefse for mining differences in species composition and functional composition between the samples based on grouping information for different time points and sampling positions. The microbial taxa that demonstrated significant differences (using the Kruskal–Wallis Rank-Sum test) among the groups of samples were visualized. Spearman correlation analysis was performed using the Corrplot (https://cran.r-project.org/web/packages/corrplot/index.html) package in R. CARD (http://arpcard.mcmaster.ca) database was used for analysis of antibiotic resistant gene fragments in the samples. Heatmap was plotted by https://www.bioinformatics.com.cn (last accessed on 20 Feb 2023), an online platform for data analysis and visualization. All statistical tests were two-sided, and *P < 0.05 was considered statistically significant.
Data Availability
Metagenomic sequencing data (FASTQ files) have been deposited in the National Center for Biotechnology Information Sequence Read Archive as BioProject PRJNA 994670.
Results
Samples and Sequencing Results
Twenty-four surface samples were collected from eight designated areas within the ICU, medical workstation, healthcare workers’ hands, beds, trolley, infusion pumps, respiratory machine panels, respiratory machine inlets and outlets, and floors (Figure 1). Each location was sampled in January, April and July 2022, and a total of 24 samples were sequenced. The sequencing of the samples generated a total of 334,996,101 reads, with an average of 13,958,171±6,714,317 reads per sample. To objectively evaluate the issues of daily cleaning and the microbial ecosystem of the ICU environment, no targeted cleaning was performed prior to or after sampling. We consequently analyzed each sample’s microbial composition and compared the microbial communities at different times within each region and between different regions.
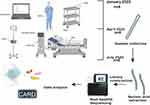 |
Figure 1 Surface samples were collected from eight designated areas within the ICU and Complete mNGS assay workflow. |
Microbial Composition in Geriatric ICU
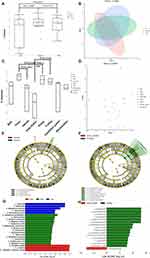
The species classification results of 24 samples indicate that microbial communities’ composition varies among different samples at the species level, as illustrated in Figure 2A. Mainly bacteria and fungi, the top 10 microbial species identified from surface swab samples of the geriatric ICU environment are Cutibacterium_acnes, with the highest average relative abundance. Subsequently, Acinetobacter baumannii, Moraxella osloensis, Pseudomonas aeruginosa, Staphylococcus epidermidis, Malassezia restricta, Ralstonia_mannitolilytica, Chryseobacterium indologenes, Staphylococcus hominis, Acinetobacter johnsonii, and Klebsiella pneumoniae were identified. Cutibacterium acnes accounted for an average of 27.04%, 13.7%, and 14.2% in the first, second, and third samplings, accounting for each percentage, respectively. We calculated the correlation between samples using the Spearman correlation coefficient, which was based on the relative abundance of each species. The results showed a positive correlation between samples in the ICU environment (Figure 2B). These correlations reflected the associations between sampling locations, as evidenced by a significant positive correlation between bed1 and floor1 (r = 0.601, P<0.05). Additionally, we observed a strong correlation between hand samples hand2 and hands3 (r=0.689, P<0.05), possibly due to frequent contact with medical workstations. Furthermore, hands3 showed a significant positive correlation with Workstation3 (r=0.64, P<0.05), indicating potential transmission between them. Trolley2 and Ventilator panel2 also showed a significant positive correlation (r=0.606, P<0.05). Hierarchical clustering analysis of the top 30 taxa in all samples revealed no clustering based on sampling location over time (Figure 2C). Samples of hands, ventilator panel, and trolley clustered together in the first sampling in January 2022, while workstation and floor samples from three different time points clustered together based on their species composition. There were variations in species abundance at different locations in terms of taxonomic diversity as illustrated in Figure 2D. On the bed surface, Acinetobacter baumannii (23.72%) was the most abundant species; on the floor, Moraxella osloensis (31.27%) was the most abundant; on respiratory machine inlets and outlets, Luteibacter anthropi (26.30%) was the most abundant; on the hands, Cutibacterium acnes (43.18%) was among the three most abundant species; on the infusion pump, Cutibacterium acnes (16.38%) was the most abundant species; on the trolley, Acinetobacter baumannii (14.54%) was the most abundant species; on the respiratory machine panel, Cutibacterium acnes (25.86%) was the most abundant species, and at the workstation, Acinetobacter baumannii (38.76%) was the most abundant species. In general, highly abundant species in the geriatric ICU environment were mostly common opportunistic pathogens like Acinetobacter baumannii, Enterococcus faecium, and Klebsiella pneumoniae. The proportion of human skin commensal bacteria was relatively high, such as Cutibacterium acnes, Malassezia furfur, Moraxella osloensis, Candida albicans, and Staphylococcus epidermidis. Bacteria and fungi dominated the environment, but we also found a small number of viruses enriched in the ICU environment, like Betapapillomavirus, Human papillomavirus, Human alphaherpesvirus, Human gammaherpesvirus 4, and Human polyomavirus. Most of the other viruses in the environmental samples were phages from the intestinal flora, such as the uncultured Caudovirales phage, which may be related to the progression of liver cancer and cirrhosis.22 Probably due to the presence of abundant opportunistic pathogenic bacteria in the ICU environment, the viral species are also mostly corresponding phages, such as Staphylococcus phage derived from coagulase-negative staphylococci,23 Pseudomonas phage, Klebsiella phage, Acinetobacter phage, Faecalibacterium virus, Streptococcus phage, Lactococcus phage. There is also Torque teno virus, which marks the patient as immunocompromised.24
We assessed whether there were temporal changes in the microbial community of the geriatric ICU using alpha diversity analysis at the species level based on the Shannon index. Sampling took place every three months, and there was no appreciable impact of time on the environmental microbial diversity across the three samplings, as the difference was not statistically significant (p>0.05). Furthermore, we observed no significant differences in alpha diversity, determined by abundance, between January and April (p=0.1469), January and July (p=0.2371), and April and July (p=0.7751) (Figure 3A). Our beta diversity investigation showed no significant changes in microbial composition among different months and sampling locations (p > 0.01) based on comparisons performed using nonmetric multidimensional scaling (NMDS) analysis (Figure 3B). However, alpha diversity analysis suggested significant variations in Shannon’s index between different sampling points, with notably higher indices for the floor, infusion pump, and small trolley locations compared to other sites (Figure 3C). The two-dimensional stress value was 0.0648, indicating reliable results, since stress values less than 0.2 are considered dependable (Figure 3D).
Linear Discriminant Analysis Effect Size (LEfSe) was used to compare microbial compositions between two groups in order to determine specific major microbiota at different sampling sites. Ultimately, 22 taxa were identified as significantly discriminative for the floor vs hands vs pump (Figure 3E), with Bacteroidia, Bacteroidales, Sphingomonas echinoides, Ophiocordyceps, Ophiocordyceps sinensis, and Sphingomonas guangdongensis being the major enriched taxa in the pump, Gleimia europaea, Patulibacter minatonensis, Megasphaera elsdeni, Pseudomonas dryadis, Methyloversatilis thermotolerans, Citrobacter koseri, Solirubrobacter, Solirubrobacteraceae, Stappia indica, Microbacterium mangrove, Thermomicrobia, Exophiala lecanii corni, and Sphingobium cloacae being the major enriched taxa in the hands, and Burkholderia stabilis, Luteibacter, and Luteibacter anthropi being more abundant in the floor samples (Figure 3G). For the inlet outlet vs trolley, 16 taxa were identified as significantly discriminative (Figure 3F), with Actinobacteria, Propionibacteriales, Cutibacterium, Acinetobacter johnsonii, Micrococcales, Micrococcaceae, Lactobacillus crispatus, Cutibacterium namnetense, Leuconostocaceae, Leuconostoc, and Leuconostoc lactis being the major enriched taxa in the inlet outlet, and Mycobacteriaceae being more abundant in the trolley (Figure 3H).
Analysis of the Pathogenic Profile of Patients in the Same Period
During the first seven months of 2022, mNGS testing was performed on samples collected from 17 geriatric ICU patients (Figure 4A), the mean age of the 17 samples was 58.47, with a median age of 64 (Supplementary 1). The samples included bronchoalveolar lavage fluid (8 cases), peripheral blood (5 cases), cerebrospinal fluid (3 cases), and ascites (1 case). Acinetobacter baumannii was detected in three cases, making it the bacteria with the highest detection frequency. Escherichia coli was found in two cases. Each of the fungi, Candida albicans and Pneumocystis jirovecii, were detected twice. In terms of viruses, Human betaherpesvirus 6B, Human gammaherpesvirus 4 (Epstein-Barr virus), Human betaherpesvirus 5 (cytomegalovirus), Human alphaherpesvirus, and Human alphaherpesvirus 1 were detected. Interestingly, Balamuthia mandrillaris, a clinically rare parasitic organism, was detected in the cerebrospinal fluid.
 |
Figure 4 Results of mNGS (A) and culture-detected (B) pathogenic microorganisms in geriatric ICU patients, January-July 2022. |
In the meantime, ninety suspected infection cases were sent to our hospital’s laboratory for culture examination, resulting in the isolation of pathogenic bacteria (Figure 4B), the mean age of the 90 samples was 64.91, with a median age of 65. Among them, Acinetobacter baumannii had the highest detection rate in geriatric ICU patients in both culture and mNGS testing. The commonly found and easily cultivated pathogenic bacteria, such as Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli were identified from the cultures. As the respiratory system is a semi-open environment with numerous commensal microorganisms or opportunistic pathogens, bronchoalveolar lavage fluid and sputum samples were selected for analysis due to their higher positivity rates in both culture and mNGS testing. Compared to culture, mNGS identified more herpes viruses, non-culturable fungus Pneumocystis jirovecii, Ureaplasma urealyticum and parasitic Balamuthia mandrillaris. Blood culture is typically informative in identifying the causative agent but peripheral blood mNGS can also detect viruses such as cytomegalovirus and Epstein-Barr virus, which may be in a latent state or activate during periods of immunosuppression, without causing viral sepsis.
Antimicrobial Resistance Gene Profiles in Geriatric ICU Microbial Community Samples
Environmental samples taken from the geriatric ICU were tested for antimicrobial resistance genes every three months. All 24 samples (100%) tested positive for at least one gene, with the intersection of antimicrobial resistance genes detected in three samples across eight positions shown in Figure 5A. These genes may persist on environmental surfaces for a prolonged period. Among the samples, bed contained ErmC, ErmB, ErmX, mecR1, mecA, and mecI, while hand samples had ErmX alone. Trolley contained ErmC, mecR1, mecA, CfxA2, tet(W/N/W), mecI, sul1, tetQ, ErmX, tetM, and sul2. Workstation had tetW, ErmC, mecR1, mecA, CfxA2, OXA-195, ErmF, tetQ, ErmX, and tetM. Floor had the largest number of antimicrobial resistance genes, including vanHM, mecR1, tet(W/N/W), sul1, Erm(36), sul4, sul2, ErmC, ErmB, mecA, aadA17, NDM-6, mecI, IMP-9, IMP-26, tetQ, ErmX, and tet. Pump contained ErmC, mecA, and ErmX, while sul1, ErmC, mecR1, mecA, and ErmX were found on ventilator panel samples, and ventilator inlet-outlet had the ErmX. The ErmX was found on the surfaces of all eight locations.Moreover, mecA and mecR1 from the Mec family were detected in multiple positions, signifying the prevalence of Mec in the ICU environment (Figure 5B).
During the same period, bacterial cultures from geriatric ICU patients indicated that 43 cases (47.78%) were antibiotic-resistant strains, with resistance genes mainly identified in blood, sputum, and broncho alveolar lavage fluid (BALF) samples that were evenly distributed (Figure 5C). The antimicrobial-resistant genes included Carbapenem-resistant Acinetobacter baumannii (CRAB), Class A carbapenemases, carbapenem-resistant Enterobacterales (CRE), Extended Spectrum Beta-Lactamase (ESBL), and beta-lactamases. Both Acinetobacter baumannii and Acinetobacter nosocomialis were CRAB. Four strains of Klebsiella pneumoniae carried both CRE and Class A carbapenemases. The main beta-lactamase-positive strains were Staphylococcus epidermidis, Staphylococcus aureus, and one strain of Haemophilus influenzae. Klebsiella pneumoniae and one strain of Enterobacter cloacae were CRE, while ESBL were mainly found in Klebsiella pneumoniae and Escherichia coli. Pseudomonas aeruginosa were resistant to all carbapenems (CRPA).
Discussion
Elderly patients in the ICU are susceptible to nosocomial infections due to comorbidities and immunosuppression.25,26 Analyzing the microbial community on non-living surfaces within hospital ICU is of significant importance in understanding, preventing, and reducing infections among elders. In this study, we monitored the microbial community at a specific geriatric ICU for six months using mNGS. The findings indicate that there were negligible differences in microbial composition over time, implying that the microbiota present in the ICU environment was generally stable under the prevailing routine disinfection regime. Results revealed high abundances of Cutibacterium acnes, Acinetobacter baumannii, Moraxella osloensis, Pseudomonas aeruginosa, Staphylococcus epidermidis, Malassezia restricta, and Klebsiella pneumoniae, which are bacteria and fungi colonizing human skin surfaces. This finding is consistent with previous research employing 16S and ITS analyses, suggesting that microbial communities on environmental ICU surfaces primarily comprise microorganisms affecting healthcare-associated infections and skin and intestinal biota.27,28 Previous studies using 16S rRNA could only identify the genus level. Unlike fungi and bacteria, viruses do not have specific conservative regions, making broad-spectrum virus detection difficult using amplicon-based techniques.29 However, mNGS can significantly enhance taxonomic resolution, accuracy, and diversity of species detection.
Although the sampling of all locations was non-uniform, consistent trends were observed in many sites. Notably, the floor had the highest Shannon index, possibly due to its frequent use by a diverse range of individuals walking in the hospital. During the primary sample analysis, hand surfaces, respiratory machine panels, and trolley clustered together, displaying similar microbial compositions. These three sites are areas that medical staffs frequently touch, as hands, respiratory machine panels, and trolley. Workstations and floors also exhibited similar microbiome compositions, indicating their relative stability in fixed locations. Conversely, other sites experienced more significant microbial composition changes due to medical decisions and healthcare personnel activities. Microbial communities in bed, hands, and trolley displayed significant positive correlations, suggesting that microbes can migrate between high-hand-contact areas. The Shannon index was low for respiratory machine panels, and their correlation with other samples was weak, indicating that the present disinfection and replacement strategy for respiratory machine parts in the ICU is appropriate. Although many common species were found on hand surfaces, the Shannon index for hands remained low because of regular disinfection and hand washing by medical staff. Moreover, Lefse analysis identified specific microbiota found on hands that are widespread in aquatic environments, such as Sphingomonas Guangdongensis,30 Sphingobium cloacae.31 There were also Gram-positive bacteria Gleimia europaea, which is also widely present in the human skin microbiota and has been reported to cause brain abscesses.32 Citrobacter koseri is a Gram-negative bacterium of the family Enterobacteriaceae isolated by hand and commonly found in water, soil, and human intestine,33 which causes brain abscesses,34 skeletal muscle,35 and skin infections36 in immunocompromised populations such as neonates as a conditional pathogen. The fungi were Exophiala lecanii corni which infects the skin37 and cornea38 and Megasphaera elsdenii which causes endocarditis.39 Despite strict adherence to hand hygiene protocols by healthcare personnel in the ICU, there remains a possibility of transmission of potentially weakly pathogenic microorganisms to bed areas and infusion pumps during medical procedures, resulting in infections among immunocompromised elderly patients. The detection of Ophiocordyceps sinensis specifically in infusion pumps can be attributed to the inclusion of certain traditional Chinese medicine preparations in medications.40 Floor specific isolated Luteibacter anthropi may cause blood stream infection.41 Specific isolation of multi-drug resistant Acinetobacter lwoffii in trolley,42 Acinetobacter are a major pathogen,43 so there is a need for improved control measures and to establish a more appropriate cleaning systems. Leuconostoc lactis in Trolley is commonly isolated from foods such as dairy products and is widely used in the fermentation industry, but is also rare in bloodstream infections,44 and is not part of the normal human flora. The trolley was found to contain beneficial microorganisms, including Lactobacillus crispatus, a commonly isolated beneficial microbe in the vaginal microbiota of healthy individuals. This particular microorganism is known for its ability to produce significant amounts of lactic acid, making it an effective broad-spectrum antimicrobial and immunomodulatory agent.45 The high abundance of Cutibacterium acnes that was detected on the trolley was likely due to frequent contact with hands. Although this bacterium is commonly associated with ordinary acne, it can also cause joint prosthesis infections,46 infections after implantation of devices in various parts of the body,47 post-brain surgery infections caused by instrument invasion,48 and even pneumonia in immunocompromised patients.49 Consequently, while most microorganisms commonly found in hospital environments are skin and mucosal commensal bacteria such as Staphylococcus epidermidis and Candida albicans, some microorganisms come from water sources. Even though these microorganisms are relatively weakly pathogenic, they still pose a threat to hospitalized patients, especially elderly patients in the ICU. Microorganisms such as bacteria, fungi, and viruses are omnipresent in the environment and can easily adapt to hospital environments, increasing their ability to invade the human body. The Human papillomavirus (HPV) was detected at low levels in almost every sampling site, with variations in pathogenicity and tissue specificity observed among different subtypes.50 Notably, members of the Herpesviridae family, including Human alphaherpesvirus types 1, 2, and 3, and 4, were frequently found in the environment. While Human alphaherpesvirus type 1, 2, and 3 primarily infect the skin and may remain asymptomatic, rare cases have reported complications such as encephalitis in immunocompromised individuals.51 Detecting HSV1, HSV2, and EBV through nucleic acid is typically inconsequential, as their activation may be due to other diseases or a viral latent state.52 In contrast, certain types of Human polyomavirus are continuously excreted in the urine or feces of infected or healthy individuals, showing moderate resistance to high temperatures, chlorine, ultraviolet light, and low pH values, with the DNA persisting in water for several months.53 Although human polyomaviruses are commonly detected in clinical mNGS, they often do not have clinical implications and may indicate environmental contamination or latent infections, only leading to pathogenic diseases, such as progressive multifocal leukoencephalopathy (PML), under severe immunosuppressive conditions.54 Viruses exist in various forms outside their hosts, ranging from naked particles to those shielded in shedding tissues or corpses and over time. All of these factors impact the potential for viral spread from the environment. Since the emergence of COVID-19, studies regarding the virus’s half-life on the surfaces of inanimate objects have also escalated.55,56 Thus, understanding the viral types present in the environment and their stability in that environment can help refine requisite conditions to guide virus control measures. Continuing the use of newer antibiotics fosters the rise and spread of antibiotic resistance. It is imperative to improve control measures and raise public awareness to curtail the dissemination of these microorganisms in a hospital setting. For the detection of pathogenic microorganisms in patients, mNGS exhibits high concurrence with bacteria detected by traditional culture methods. However, it is often reserved for situations where patients have severe or difficult infections due to factors such as cost. MNGS can offer relatively accurate detection where predicting the patient’s infection status becomes impossible, such as the detection of Balamuthia mandrillaris in cerebrospinal fluid. While eventually diagnosed in the patient as parasitic encephalitis, mNGS still cannot replace the gold standard status of traditional culture methods. The pathogenic microbial spectrum from patients infected with Acinetobacter baumannii, Pseudomonas aeruginosa, Enterococcus faecium, and Enterococcus faecium was collected. These microorganisms were found to be highly detected in both patients and environmental specimens sampled concurrently. It has been suggested that ICU patients are at risk of exposure to microbial transmission from the environment due to the widespread detection of the MecA on various surfaces such as the bed, trolley, workstation, pump, floor, and even the ventilator panel. Additionally, mNGS analysis revealed the presence of ErmX in all sites, which primarily exists in plasmids of microorganisms such as Staphylococcus epidermidis, Acinetobacter baumannii, and Enterococcus faecalis,57 and can widely transfer with the proliferation of microorganisms. Therefore, low-pathogenicity microorganisms may potentially serve as reservoirs for antibiotic-resistant genes (ARGs), posing a risk for transfer to important pathogens.
In recent years, an increasing number of reports have addressed the emergence of CRAB infections.58 All cultured isolates of Acinetobacter baumannii were CRAB, with age being an independent risk factor for multidrug-resistant infections.59 Critically-ill elderly patients carry a higher risk of CRAB-induced sepsis. Longer hospitalization duration correlates with greater infection occurrence risk and poorer prognoses. Floor samples from all three locations within the ICU were positive for metallo-β-lactamase gene IMP-9, whereas work station samples exhibited NDM-6 and OXA-195, both of which are also metallo-β-lactamase genes.32 The abundance of Acinetobacter baumannii in the ICU environment can be attributed to the relatively dry air, which promotes the bacteria’s prolonged survival on ICU surfaces and equipment. Research suggests that roughly 40% of interactions between ICU patients colonized with Acinetobacter baumannii and healthcare workers have contaminated gloves and laboratory coats.60 Thus, the lack of treatment options for multidrug-resistant Acinetobacter baumannii underscores the critical role of hand hygiene and strict environmental infection control in preventing the spread of this almost untreatable pathogen. Due to the relatively low sequencing depth of mNGS in this study, it was not possible to conduct source analysis of Acinetobacter baumannii between patient and environmental samples. However, in the three bed samples, the average relative abundance of Acinetobacter baumannii was higher compared to other sampling points. The relative abundance of Acinetobacter baumannii was also higher in areas near patients, such as infusion pumps and trolleys (Figure 1A). Therefore, it is speculated that the pathogen may be transmitted between the environment and patients.
Due to its high sensitivity and lack of bias, mNGS has proven effective for identifying the species classification and relative abundance of microorganisms in the ICU environment. It has already been widely utilized in detecting clinical pathogenic microorganisms,61 with particular application in critical care medicine and respiratory fields. Moreover, its benefits extend to public health safety and emerging infectious disease detection. As sequencing costs gradually decrease and turn-around time (TAT) shortens, it is expected that mNGS will become an increasingly valuable tool for microbial environment monitoring within hospitals by infection prevention and control departments. Acinetobacter baumannii, and particularly drug-resistant CRAB, pose a prolonged threat to ICU patients while being detected at high abundance within ICU environments. Consequently, pathogenic bacteria with high survival rates on hospital surfaces have the potential to transmit to patients after falling off, resulting in continuous transmission cycles. For geriatric ICU patients, it may therefore be necessary to conduct molecular diagnostic tests upon admission to ascertain whether Acinetobacter baumannii is carrying drug resistance genes, enabling zone-specific treatment and minimizing the spread of CRAB within the geriatric ICU ward. Medical equipment such as trolleys, workstations, infusion pumps, and flooring are highly susceptible to extensive contamination by drug-resistant microorganisms. As such, implementing targeted disinfection measures is essential, utilizing hospital-standard disinfectants and cleaners that generally demonstrate efficacy against prevalent nosocomial pathogens. Subsequently, these sites should undergo more thorough wiping down procedures, with sufficient contact time allowed for the disinfectants to take effect. Whenever possible, disposable parts for respiratory equipment should be used to mitigate the risk of cross-contamination. Furthermore, beds, bed linens, and shared equipment require thorough disinfection between patients. Despite the simplicity and importance of hand hygiene in reducing the spread of drug-resistant bacteria within the ICU, there can be a conflict between patient care responsibilities and devoting adequate time to meticulous hand hygiene.62 Consequently, establishing standards to enhance healthcare workers’ compliance with hand hygiene requirements is necessary.
Conclusions
The microbial composition and abundance in the ICU remained relatively stable over time, although high levels of hospital-acquired infection-related bacteria and fungi persisted in the ICU environment for prolonged durations, such as Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa. Floor exhibited the highest microbial diversity and abundance in the ICU environment, followed by the pump, trolley, and workstation, likely due to frequent medical operations and personnel activity. In contrast, the relatively high cleanliness of the ventilator panel and ventilator inlet outlet, which are single-use items replaced with each patient, may account for their lower microbial diversity. Pathogens, and even drug-resistant bacteria, could potentially spread directly between the environment and patients, as supported by patient cultures and mNGS results. Consequently, particular attentions should be given to disinfecting the pump, trolley, and workstation areas and maintaining a high frequency of hand hygiene. While the detection of drug-resistant genes does not conclusively indicate the presence of drug-resistant bacteria, it enhances our understanding of the resistance situation of ICU surface microorganisms. Considering this, it provides a theoretical foundation for the health management of the geriatric ICU environment.
Acknowledgments
This work was supported by the Clinical Research Center for Geriatric Diseases of Yunnan Province - diagnosis and treatment of geriatric comorbidity and clinical translational research (202102AA310069); the Joint Special Funds for the Department of Science and Technology of Yunnan Province-Kunming Medical University (202001AY070001-205). At the same time, I acknowledged assistance from medical writers, proof-readers and editors. Figure 1 was created with BioRender.com.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation; took part in drafting, revising the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
All authors declare that they have no potential conflicts of interest in this work.
References
1. Li K, Zhu Q, Jiang F, et al. Monitoring microbial communities in intensive care units over one year in China. Sci Total Environ. 2022;811:152353.
2. Kembel SW, Jones E, Kline J, et al. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 2012;6(8):1469–1479. doi:10.1038/ismej.2011.211
3. Vincent JL, Bihari DJ, Suter PM, et al. The prevalence of nosocomial infection in intensive care units in Europe. Results of the European Prevalence of Infection in Intensive Care (EPIC) Study. EPIC International Advisory Committee. JAMA. 1995;274(8):639–644.
4. Esme M, Topeli A, Yavuz BB, et al. Infections in the elderly critically-ill patients. Front Med. 2019;6:118. doi:10.3389/fmed.2019.00118
5. Plowman R. The socioeconomic burden of hospital acquired infection. Euro Surveill. 2000;5(4):49–50. doi:10.2807/esm.05.04.00004-en
6. Manaka A, Tokue Y, Murakami M. Comparison of 16S ribosomal RNA gene sequence analysis and conventional culture in the environmental survey of a hospital. J Pharm Health Care Sci. 2017;3:8. doi:10.1186/s40780-017-0074-y
7. Oberauner L, Zachow C, Lackner S, et al. The ignored diversity: complex bacterial communities in intensive care units revealed by 16S pyrosequencing. Sci Rep. 2013;3:1413. doi:10.1038/srep01413
8. Christoff AP, Sereia AFR, Cruz GNF, et al. One year cross-sectional study in adult and neonatal intensive care units reveals the bacterial and antimicrobial resistance genes profiles in patients and hospital surfaces. PLoS One. 2020;15(6):e0234127. doi:10.1371/journal.pone.0234127
9. Weiner-Lastinger LM, Abner S, Edwards JR, et al. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: summary of data reported to the national healthcare safety network, 2015–2017. Infect Control Hosp Epidemiol. 2020;41(1):1–18.
10. Strich JR, Palmore TN. Preventing transmission of multidrug-resistant pathogens in the intensive care unit. Infect Dis Clin North Am. 2017;31(3):535–550. doi:10.1016/j.idc.2017.05.010
11. Shams AM, Rose LJ, Edwards JR, et al. Assessment of the overall and multidrug-resistant organism bioburden on environmental surfaces in healthcare facilities. Infect Control Hosp Epidemiol. 2016;37(12):1426–1432. doi:10.1017/ice.2016.198
12. Ohl M, Schweizer M, Graham M, et al. Hospital privacy curtains are frequently and rapidly contaminated with potentially pathogenic bacteria. Am J Infect Control. 2012;40(10):904–906. doi:10.1016/j.ajic.2011.12.017
13. Zhang R, Zhuang Y, Xiao Z-H, et al. Diagnosis and surveillance of neonatal infections by metagenomic next-generation sequencing. Front Microbiol. 2022;13:855988. doi:10.3389/fmicb.2022.855988
14. Zhou H, Ouyang C, Han X, et al. Metagenomic sequencing with spiked-in internal control to monitor cellularity and diagnosis of pneumonia. J Infect. 2022;84(1):e13–e17. doi:10.1016/j.jinf.2021.09.018
15. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi:10.1038/nmeth.1923
16. Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257. doi:10.1186/s13059-019-1891-0
17. Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res. 2018;46(D1):D41–d47. doi:10.1093/nar/gkx1094
18. O’Leary NA, Wright MW, Brister JR, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–45. doi:10.1093/nar/gkv1189
19. Lu J, Breitwieser FP, Thielen P, et al. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;3:e104. doi:10.7717/peerj-cs.104
20. Morganella S, Cerulo L, Viglietto G, et al. VEGA: variational segmentation for copy number detection. Bioinformatics. 2010;26(24):3020–3027. doi:10.1093/bioinformatics/btq586
21. Wickham H. ggplot2: Elegant Graphics for Data Analysis New York. NY: Springer; 2009.
22. Lai MW, Chu Y-D, Hsu C-W, et al. Multi-omics analyses identify signatures in patients with liver cirrhosis and hepatocellular carcinoma. Cancers. 2022;15(1):210. doi:10.3390/cancers15010210
23. Moller AG, Lindsay JA, Read TD. Determinants of phage host range in staphylococcus species. Appl Environ Microbiol. 2019;85(11). doi:10.1128/AEM.00209-19
24. Zeng J, Tang Y, Lin T, et al. Torque-teno virus for the prediction of graft rejection and infection disease after kidney transplantation: a systematic review and meta-analysis. J Med Virol. 2023;95(3):e28677. doi:10.1002/jmv.28677
25. Jump RLP, Crnich CJ, Mody L, et al. Infectious diseases in older adults of long-term care facilities: update on approach to diagnosis and management. J Am Geriatr Soc. 2018;66(4):789–803. doi:10.1111/jgs.15248
26. Castle SC, Uyemura K, Fulop T, et al. Host resistance and immune responses in advanced age. Clin Geriatr Med. 2007;23(3):463–479. doi:10.1016/j.cger.2007.03.005
27. Bokulich NA, Mills DA, Underwood MA. Surface microbes in the neonatal intensive care unit: changes with routine cleaning and over time. J Clin Microbiol. 2013;51(8):2617–2624. doi:10.1128/JCM.00898-13
28. Brooks B, Firek BA, Miller CS, et al. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome. 2014;2(1):1. doi:10.1186/2049-2618-2-1
29. Grice EA. The intersection of microbiome and host at the skin interface: genomic- and metagenomic-based insights. Genome Res. 2015;25(10):1514–1520. doi:10.1101/gr.191320.115
30. Feng GD, Yang S-Z, Wang Y-H, et al. Description of a Gram-negative bacterium, Sphingomonas guangdongensis sp. nov. Int J Syst Evol Microbiol. 2014;64(Pt 5):1697–1702. doi:10.1099/ijs.0.056853-0
31. Prakash O, Lal R. Description of Sphingobium fuliginis sp. nov., a phenanthrene-degrading bacterium from a fly ash dumping site, and reclassification of Sphingomonas cloacae as Sphingobium cloacae comb. nov. Int J Syst Evol Microbiol. 2006;56(Pt 9):2147–2152. doi:10.1099/ijs.0.64080-0
32. Álvarez C, Almuzara M, Tosello C, et al. Actinomyces europaeus (Gleimia europaea) asociado con absceso cerebral: comunicación de tres casos. Rev Argent Microbiol. 2023. doi:10.1016/j.ram.2022.07.003
33. Deveci A, Coban AY. Optimum management of Citrobacter koseri infection. Expert Rev Anti Infect Ther. 2014;12(9):1137–1142. doi:10.1586/14787210.2014.944505
34. Ward HH, Lauber P, Laubach LT, et al. Citrobacter koseri meningitis with cerebral edema and pneumocephalus in a neonate. Radiol Case Rep. 2021;16(3):528–530. doi:10.1016/j.radcr.2020.12.039
35. Kwaees TA, Hakim Z, Weerasinghe C, et al. Musculoskeletal infections associated with Citrobacter koseri. Ann R Coll Surg Engl. 2016;98(7):446–449. doi:10.1308/rcsann.2016.0209
36. Licata G, De Rosa A, Gambardella A, et al. Bullous erysipelas caused by citrobacter koseri. J Clin Aesthet Dermatol. 2021;14(5):12.
37. Hatta J, Anzawa K, Kubota K, et al. A Case of recalcitrant phaeohyphomycosis of the face caused by exophiala lecanii-corni. Med Mycol J. 2021;62(2):35–39. doi:10.3314/mmj.20-00018
38. Miyakubo T, Todokoro D, Satake Y, et al. Exophiala lecanii-corni keratitis presenting as a serpiginous pigmented superficial lesion: a case report. Medicine. 2020;99(36):e22121. doi:10.1097/MD.0000000000022121
39. Brancaccio M, Legendre GG. Megasphaera elsdenii endocarditis. J Clin Microbiol. 1979;10(1):72–74. doi:10.1128/jcm.10.1.72-74.1979
40. Maľučká LU, Uhrinová A, Lysinová P. Medicinal mushrooms Ophiocordyceps sinensis and Cordyceps militaris. Ceska Slov Farm. 2022;71(6):259–265.
41. Kämpfer P, Lodders N, Falsen E. Luteibacter anthropi sp. nov., isolated from human blood, and reclassification of Dyella yeojuensis Kim et al. 2006 as Luteibacter yeojuensis comb. nov. Int J Syst Evol Microbiol. 2009;59(Pt 11):2884–2887. doi:10.1099/ijs.0.009100-0
42. Mittal S, Sharma M, Yadav A, et al. Acinetobacter lwoffii an emerging pathogen in neonatal ICU. Infect Disord Drug Targets. 2015;15(3):184–188. doi:10.2174/1871526515666150826114745
43. Rathinavelu S, Zavros Y, Merchant JL. Acinetobacter lwoffii infection and gastritis. Microbes Infect. 2003;5(7):651–657. doi:10.1016/S1286-4579(03)00099-6
44. Hosoya S, Kutsuna S, Shiojiri D, et al. Leuconostoc lactis and Staphylococcus nepalensis Bacteremia. Japan Emerg Infect Dis. 2020;26(9):2283–2285. doi:10.3201/eid2609.191123
45. Lepargneur JP. Lactobacillus crispatus as biomarker of the healthy vaginal tract. Ann Biol Clin. 2016;74(4):421–427. doi:10.1684/abc.2016.1169
46. Boisrenoult P. Cutibacterium acnes prosthetic joint infection: diagnosis and treatment. Orthop Traumatol Surg Res. 2018;104(1s):S19–s24. doi:10.1016/j.otsr.2017.05.030
47. Gharamti AA, Kanafani ZA. Cutibacterium (formerly Propionibacterium) acnes infections associated with implantable devices. Expert Rev Anti Infect Ther. 2017;15(12):1083–1094. doi:10.1080/14787210.2017.1404452
48. Achermann Y, Goldstein EJC, Coenye T, et al. Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin Microbiol Rev. 2014;27(3):419–440. doi:10.1128/CMR.00092-13
49. Abdullah HM, Waqas Q, Abdalla A, et al. Cutibacterium acnes pneumonia in an immunocompromised patient: a case report and review of the literature. S D Med. 2021;74(11):523–526.
50. Harden ME, Munger K. Human papillomavirus molecular biology. Mutat Res Rev Mutat Res. 2017;772:3–12. doi:10.1016/j.mrrev.2016.07.002
51. Whitley RJ, Kimberlin DW. Herpes simplex encephalitis: children and adolescents. Semin Pediatr Infect Dis. 2005;16(1):17–23. doi:10.1053/j.spid.2004.09.007
52. Han D, Yu F, Zhang D, et al. The real-world clinical impact of plasma mNGS testing: an observational study. Microbiol Spectr. 2023;11(2):e0398322. doi:10.1128/spectrum.03983-22
53. Rachmadi AT, Torrey JR, Kitajima M. Human polyomavirus: advantages and limitations as a human-specific viral marker in aquatic environments. Water Res. 2016;105:456–469. doi:10.1016/j.watres.2016.09.010
54. Del Valle L, Piña-Oviedo S. Human polyomavirus JCPyV and its role in progressive multifocal leukoencephalopathy and oncogenesis. Front Oncol. 2019;9:711. doi:10.3389/fonc.2019.00711
55. van Doremalen N, Bushmaker T, Morris D, et al. Aerosol and surface stability of HCoV-19 (SARS-CoV-2) compared to SARS-CoV-1. medRxiv. 2020;2020:1.
56. Mullis L, Saif LJ, Zhang Y, et al. Stability of bovine coronavirus on lettuce surfaces under household refrigeration conditions. Food Microbiol. 2012;30(1):180–186. doi:10.1016/j.fm.2011.12.009
57. Li B, Chen D, Lin F, et al. Genomic island-mediated horizontal transfer of the erythromycin resistance geneerm (X) among bifidobacteria. Appl Environ Microbiol. 2022;88(10):e0041022. doi:10.1128/aem.00410-22
58. Rossi I, Royer S, Ferreira ML, et al. Incidence of infections caused by carbapenem-resistant Acinetobacter baumannii. Am J Infect Control. 2019;47(12):1431–1435. doi:10.1016/j.ajic.2019.07.009
59. Fang EF, Scheibye-Knudsen M, Jahn HJ, et al. A research agenda for aging in China in the 21st century. Ageing Res Rev. 2015;24(Pt B):197–205. doi:10.1016/j.arr.2015.08.003
60. Morgan DJ, Liang SY, Smith CL, et al. Frequent multidrug-resistant acinetobacter baumannii contamination of gloves, gowns, and hands of healthcare workers. Infect Control Hosp Epidemiol. 2010;31(7):716–721. doi:10.1086/653201
61. Liu H, Zhang Y, Yang J, et al. Application of mNGS in the etiological analysis of lower respiratory tract infections and the prediction of drug resistance. Microbiol Spectr. 2022;10(1):e0250221. doi:10.1128/spectrum.02502-21
62. Stahmeyer JT, Lutze B, von Lengerke T, et al. Hand hygiene in intensive care units: a matter of time? J Hosp Infect. 2017;95(4):338–343. doi:10.1016/j.jhin.2017.01.011
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.