Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 16
Microarray Expression Profile of Exosomal circRNAs from High Glucose Stimulated Human Renal Tubular Epithelial Cells
Authors Sha YH, Cao SL, Zhang L, Lai LS, Ke PF, Yu KW, Fang XZ, Deng RT, Wan ZM, Wu XB, Han G, Jie YB, Song LL, Huang XZ, Fu WJ
Received 8 August 2023
Accepted for publication 14 November 2023
Published 5 December 2023 Volume 2023:16 Pages 3937—3951
DOI https://doi.org/10.2147/DMSO.S430131
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Konstantinos Tziomalos
Yan-Hua Sha,1 Song-Ling Cao,1 Lu Zhang,2 Li-Sha Lai,2 Pei-Feng Ke,1 Ke-Wei Yu,1 Xiu-Zhu Fang,2 Ren-Tang Deng,2 Ze-Min Wan,1 Xiao-Bin Wu,1 Guang Han,1 Yu-Bang Jie,2 Lan-Lan Song,2 Xian-Zhang Huang,1 Wen-Jin Fu2
1Department of Laboratory Medicine, The Second Clinical College of Guangzhou University of Chinese Medicine, Guangzhou, Guangdong, People’s Republic of China; 2Clinical Laboratory, Houjie Hospital of Guangdong Medical University, Dongguan, Guangdong, People’s Republic of China
Correspondence: Xian-Zhang Huang, Department of Laboratory Medicine, The Second Clinical College of Guangzhou University of Chinese Medicine, Guangzhou, Guangdong, People’s Republic of China, Email [email protected]; Wen-Jin Fu, Clinical Laboratory, Houjie Hospital of Guangdong Medical University, Dongguan, Guangdong, People’s Republic of China, Email [email protected]
Introduction: Circular RNA (circRNAs) are a type of non-coding RNA (ncRNAs) with a wealth of functions. Recently, circRNAs have been identified as important regulators of diabetic kidney disease (DKD), owing to their stability and enrichment in exosomes. However, the role of circRNAs in exosomes of tubular epithelial cells in DKD development has not been fully elucidated.
Methods: In our study, microarray technology was used to analyze circRNA expression in cell supernatant exosomes isolated from HK-2 cells with or without high glucose (HG) treatment. The small interfering RNAs (siRNA) and plasmid overexpression were used to validate functions of differentially expressed circRNAs.
Results: We found that exosome concentration was higher in HG-stimulated HK-2 cells than in controls. A total of 235 circRNAs were significantly increased and 458 circRNAs were significantly decreased in the exosomes of the HG group. In parallel with the microarray data, the qPCR results showed that the expression of circ_0009885, circ_0043753, and circ_0011760 increased, and the expression of circ_0032872, circ_0004716, and circ_0009445 decreased in the HG group. Rescue experiments showed that the effects of high glucose on regulation of CCL2, IL6, fibronetin, n cadherin, e cadherin and epcam expression can be reversed by inhibiting or overexpressing these circRNAs. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathway analyses indicated that circRNA parental genes are associated with glucose metabolism, lipid metabolism, and inflammatory processes, which are important in DKD development. Further analysis of circRNA/miRNA interactions indicated that 152 differentially expressed circRNAs with fold change (FC) ≥ 1.5 could be paired with 43 differentially expressed miRNAs, which are associated with diabetes or DKD.
Discussion: Our results indicate that exosomal circRNAs may be promising diagnostic and therapeutic biomarkers, and may play a critical role in the progression of DKD.
Keywords: exosome, cicrRNA, diabetic nephropathy, high glucose, microarray
Introduction
Diabetic kidney disease (DKD) is the most frequent microvascular complication leading to end-stage renal disease (ESRD) worldwide.1 Over the years, the incidence of DKD has increased rapidly, resulting in a global economic and medical burden. Renal interstitial fibrosis, a predominant pathological feature of DKD, is characterized by epithelial mesenchymal transformation (EMT), accumulation of extracellular matrix (ECM), and an inflammatory response.2–5 Increasing evidence indicates that interstitial lesions may be mediated through tubular epithelial cell activation and that high glucose levels are an important stimulant of renal interstitial fibrosis.6–8 However, the mechanisms underlying the tubulointerstitial damage induced by high glucose levels have not been fully elucidated. Therefore, additional potential biomarkers are required for the diagnosis of DKD.
Exosomes are small extracellular vesicles with diameters ranging between 40 and 160 nm and consist of complex molecular cargos, such as proteins, lipids, DNA, mRNA, and ncRNAs (miRNA, lncRNA, circRNA).9,10 Exosomes mediate intercellular communication both locally and systemically by regulating cellular biological activities, which are strongly associated with the occurrence and development of various diseases.11,12 Many studies have shown that exosomes play an important role in regulating DKD occurrence and progression.13,14 Yang et al showed that serum exosomes extracted from DKD patients may cause dysfunction of human glomerular endothelial cells (HGECs) mainly by mediating the expression of FIBA and 1-MH.15 Studies have indicated that exosomes released by HG-treated GECs can increase α-SMA expression in glomerular mesangial cells through the TGF-β1/Smad3 signaling pathway, resulting in renal fibrosis.16 Tubular epithelial cells also secrete exosomes. Exosomal miR-1269b target Fibulin-1 (FBLN1), thereby inducing EMT in proximal tubular epithelial cells. This suggests that exosomal miR-1269b levels are associated with the severity of renal damage in patients with type 2 diabetes.17
Circular RNA (circRNAs) are ncRNAs that form a unique circular closed-loop structure during RNA splicing.18 Recently, circRNAs have been identified as important regulators of DKD owing to their enrichment and stability in exosomes.19,20 For instance, Li et al confirmed that circTAOK1 expression was upregulated in exosomes extracted from GECs stimulated by high glucose. Mechanistically, exosomal circTAOK1 extracted from GECs can regulate SMAD3 expression by sponging miR-520h to promote the proliferation, fibrosis, and EMT of GMCs.21 Dong et al showed that circNUP98 expression was higher in exosomes isolated from DKD patients. Functionally, circNUP98 knockdown alleviated inflammatory reaction and fibrosis of HMCs treated with high glucose by mediating the miR-151-3p/HMGA2 pathway.22 Furthermore, the diagnostic value of exosomal circNUP98 in DKD was evaluated using ROC curve analysis, with an area under the curve of 0.9146.22 Zhu et al showed that the expression of exosomal circ_0001846 is increased in patients with osteoarthritis.23 In addition, a recent study found that circ_DLGAP4 expression was increased in exosomes isolated from HG-treated mesangial cells, patients with DKD, and DKD rat models.24 Exosomal circ_DLGAP4 promotes an inflammatory response by activating NF-κB signaling, suggesting that deletion of circ_DLGAP4 followed by inhibition of NF-κB signaling may serve as a new therapeutic target for DKD.24 These results suggest that circRNAs may serve as potential diagnostic and therapeutic biomarkers of DKD.
However, current studies exploring the role of exosomal circRNAs in tubular epithelial cells during DKD development remains unclear. In our study, we used microarray technology to analyze circRNA expression in cell supernatant exosomes isolated from HK-2 cells with or without high glucose (HG) treatment to explore the underlying molecular mechanisms of exosomal circRNAs in DKD development. Our study provides new insights into the role of exosomal circRNAs as potential molecular markers for DKD diagnosis.
Materials and Methods
Cell Culture
The human proximal tubular epithelial cell line (HK-2 cells) were purchased from BeNa Culture Collection (Beijing, China). HK-2 cells were cultured in RPMI 1640 medium (Gibco, USA) supplemented with 10% exosome depleted FBS (VivaCell Biosciences) under standard culture conditions (5% CO2, 37°C). In our experiment, 1mL deionized water was used to dissolve 0.18g glucose powder to get 1M glucose solution. And then we add 60μL 1M glucose into 2mL culture medium to make 30mM high glucose condition. The cells were stimulated with 30mM glucose (HG group) or 5.5mM glucose (NG group) for 30h at 37°C. Three duplicates were performed for each group.
Transfection with Small Interfering RNA
The small interfering RNAs (siRNAs) against circ_0009885, circ_0011760, circ_0043753 and an irrelevant 21-nucleotide siRNA, as a negative control, were purchased from Sangon Biotech (Shanghai, China). HK-2 cells were transfected using Lipofectamine 3000 transfection reagent (Invitrogen) for 48h according to the manufacturer’s instructions. After 48h of transfection, real-time RT-PCR performed.
Transfection with Overexpression Plasmid
The plasmid to make circ_0032872, circ_0004716 overexpression were purchased from Geneseed Biotech (Guangzhou, China). HK-2 cells were transfected using Lipofectamine 3000 transfection reagent (Invitrogen) for 48h according to the manufacturer’s instructions. After 48h of transfection, real-time RT-PCR performed.
Exosome Extraction
To exclude cell components, the cell supernatants of the two groups were centrifuged at 300 × g for 10 min. The supernatant was centrifuged at 2000 x g for 10 min and 10,000 × g for 30 min. The supernatant was ultracentrifuged using a P50AT2 rotor (CP80WX; Himac, Tokyo, Japan) at 100,000 × g for 70 min to pellet exosomes. To exclude contaminating proteins, the pellet was washed with PBS and centrifuged at 100,000 g for 70 min. Finally, the exosomes were resuspended in 200µL of PBS. All centrifugation steps were performed at 4 °C.
Transmission Electron Microscopy (TEM)
A 15 µL solution of exosomes was placed in the copper mesh for one min and stained with 2% uranyl acetate solution. The sample was then dried for 10 min under an incandescent light. Finally, TEM (Tecnai G2 Spititi, FEI, USA) was used to observe and photograph the Cu mesh.
Nanoparticle Tracking Analysis (NTA)
Exosomes were diluted with PBS and analyzed using a ZetaViewParticle Metrix instrument, according to the manufacturer’s standard protocols. The particle trajectory was recorded, and the diameter distribution of each sample was computed using the software. The original concentration of exosomes was calculated using multiple dilutions.
Western Blot
Exosomal proteins were extracted using radioimmunoprecipitation assay (RIPA) lysis buffer and separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Antibodies against CD9 (Abcam, USA) and TSG101 (Abcam, USA) were used to detect the presence of exosomes. Chemiluminescence was used to visualize the protein bands using an ECL detection kit (Share-bio).
Exosomal RNA Extraction and Microarray Analysis
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total exosomal RNA. The extracted RNA was quantified using a NanoDrop ND-2000 (Thermo Scientific) and RNA integrity was detected using an Agilent Bioanalyzer 2100 (Agilent Technologies). Then 250ng of total RNA was amplified. cRNA was labeled with Cyanine-3 (Cy3) and purified using the RNeasy Mini Kit (QIAGEN, Germany). Microarray hybridization was performed according to the manufacturer s protocol. After 17 h of hybridization at 65 °C in a rotating Agilent hybridization oven, slides were washed and scanned an Agilent scanner G5761A. Feature Extraction software 12.0 version (Agilent Technologies, USA) was used to extract data. Genespring software 14.8 version (Agilent technologies, USA) was used to normalize raw data. All data were uploaded to the NCBI Gene Expression Omnibus database (accession number GSE207495; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE207495).
Differential expression between the two groups was evaluated using the fold change (FC). The difference in circRNA expression was considered statistically significant according to the filter criteria of FC ≥ 1.5 and p value < 0.05. A t-test was used to assess the statistical significance of the differences.
Quantitative Real-Time PCR (qRT-PCR)
All the primers were synthesized by Sangon Biotech (Shanghai, China). The primer sequences are listed in Supplemental Table S2. After determining the best annealing temperatures, RT-qPCR with SYBR Green detection chemistry to measure the relative circRNA expression was performed on an ABI7500 Fast Real Time PCR system (Applied Biosystems, USA). U6 served as an internal control and the relative expression level of each circRNA was calculated using the 2-Ct method.
Cell Migration Assays
Cell migration assays were performed with transwell chambers (pore size: 8 μm, 24-well; BD Biosciences) following the manufacturer’s protocol. For cell migration assays, cells were detached and washed with PBS, resuspended in serum-free medium, and 200 μL of cell suspensions (a total of 4×104 cells) was added to the upper chamber. Medium with 20% FBS was added to the bottom wells of the chambers. The cells that had not migrated were removed from the upper face of the filters using cotton swabs, and the cells that had migrated to the lower face of the filters were fixed with fixed with methanol, stained with crystal violet solution, photographed under microscope. The mean of triplicate assays for each experimental condition was used.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
GO analysis was performed based on the parental genes of differentially expressed circRNAs. The number distribution of differential genes in GO terms was analyzed based on molecular functions, cellular components, and biological processes. KEGG (http://www.genome.jp/kegg) was used to analyze the molecular pathways enriched by differentially expressed genes.
circRNA/miRNA Network Analysis
TargetScan and miRanda algorithms were used to predict interactions between differentially expressed circRNAs and miRNAs. We searched for miRNA response elements (MREs) in circRNAs and selected miRNAs that matched the MREs. The pair of interaction relations was selected using the criteria of a context score ≥ 90 in TargetScan and a maximum energy ≤ −20 in the miRanda algorithm.
Statistical Analysis
Statistical analyses were performed the SPSS 18.0. All data are expressed as mean ± SD. The Student’s t-test was used to evaluate the differences between the two groups. Statistical significance was determined when p values were less than 0.05.
Results
HG-Treated HK-2 Cells Secrete a Higher Number of Exosomes
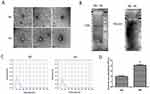
To explore the effect of high glucose on exosome release, HK-2 cells were treated with or without high glucose. First, we identified exosomes by TEM, which revealed the presence of vesicles with a lipid bilayer structure and correct size (Figure 1A). In addition, we detected the protein levels of exosomal markers CD9 and TSG101. As expected, CD9 and TSG101 were detected in the exosomes (Figure 1B). Finally, we analyzed the particle size and concentration of exosomes in the HK-2 cell culture medium using NTA. The results revealed the average particle size of two groups were 130.6 nm and 129.5 nm, respectively (Figure 1C). The concentration of exosomes was increased in the HG group (P<0.05) (Figure 1D). These results indicate that relatively high-quality exosomes were extracted using the ultracentrifugation method, and that high glucose levels promoted exosome secretion in HK-2 cells.
Overview of circRNA Expression Profiles
In this research, 36,500 circRNAs known in circBase and 48 circRNAs from reported papers were detected by microarray analysis. As shown in Figure 2A, circRNA expression in the culture medium was different between the HG and NG group (Figure 2A). In total, 693 circRNAs were screened as differentially expressed circRNAs by FC ≥1.5 and p value <0.05. Of these, 235 circRNAs were upregulated and 458 circRNAs were downregulated in the HG group (Figure 2B). The differentially expressed circRNAs are shown in a scatter plot (Figure 2C), and the significantly differentially expressed circRNAs are displayed in a volcano plot (Figure 2D). In addition, all circRNAs and differentially expressed circRNAs were distributed across almost all chromosomes (Figure 3A and B). The top 20 upregulated and downregulated circRNAs are shown in Table 1.
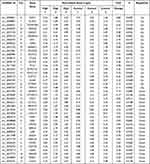 |
Table 1 The Top 20 Up-Regulated and Down-Regulated circRNAs Between HG Group and Control Group |
 |
Figure 3 Chromosomal distribution of circRNAs in exosomes. (A) Chromosomal distribution of all circRNAs. (B) Chromosomal distribution of differentially expressed circRNAs. |
Validation of Differentially Expressed circRNA by qPCR
Based on the relatively high abundance (FC ≥ 2, P<0.05) and their host genes, we selected 10 candidate circRNAs to validate their expression in exosomes from HK-2 cell culture medium of the HG group and NG group, including five up-regulated circRNAs (circ_0009885, circ_0066651, circ_0043753, circ_0041881, and circ_0011760) and five downregulated circRNAs (circ_0032872, circ_0061803, circ_0004716, circ_0009445, and circ_0075792). In parallel with the microarray data, the qPCR results showed that the expression of circ_0009885, circ_0043753, and circ_0011760 increased, and the expression of circ_0032872, circ_0004716, and circ_0009445 decreased in the HG group (Figure 4).
These Differentially Expressed circRNAs Regulate Inflammation and EMT in HK-2 Cells
Many studies have shown that inflammation and epithelial-mesenchymal transition (EMT) play a key role in the development of DKD. Thus, we explored the effects of these differentially expressed circRNAs on regulation of inflammatory factor and EMT related molecule, including CCL2, IL6, fibronetin, n cadherin, e cadherin and epcam. For up-regulated circRNAs, including circ_0009885, circ_0043753 and circ_0011760, we used siRNA (si-0009885, si-0043753 and si-0011760 respectively) to validate their functions. For down-regulated circRNAs, including circ_0032872 and circ_0004716 we used plasmid overexpression (pLC5-ciR0032872 and pLC5-ciR0004716) to validate their functions. The plasmid of circ_0009445 was not successfully constructed because its spliced length was too long. The interference effect and overexpression effect of circRNAs have showen in Figure 5.
We found high glucose increase expression of CCL2, IL6, fibronetin, n cadherin and decrease expression of e cadherin, epcam (Figures 6A and 7A). After si-0009885 treatment, the up-regulation of CCL2, IL6 and fibronetin expression by high glucose were markedly reversed. After si-0011760 treatment, the up-regulation of IL6 and fibronetin expression by high glucose were markedly reversed. After si-0043753 treatment, the down-regulation of epcam expression by high glucose were markedly reversed (Figure 6B–D). After pCL5-0032872 treatment, the up-regulation of IL6 expression by high glucose were markedly reversed. After pCL5-0004716 treatment, the up-regulation of CCL2 and n cadherin expression by high glucose were markedly reversed (Figure 7B–D).
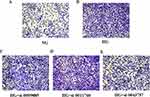
Upregulation of fibronetin, n cadherin and downregulation of e cadherin and epcam are related to EMT occurring. Cell migration is also correlated with EMT occurring. Transwell chamber assays showed that high glucose promote migration in HK-2 cells compared with normal glucose (NG group). Suppression of circ-0043757 resulted in significantly diminished migratory potential (Figure 8).
GO and KEGG Pathway Analysis
The number distribution of differential genes in GO terms was analyzed based on molecular function, cellular component, and biological process (Figure 9). The term with the most genes of molecular function was protein binding (GO:0005515, count=3410), and the most significantly enriched term was wide pore channel activity (GO:0022829, P =3.74E-3). The term with the most genes of cellular component was nucleus (GO:0005634, count =2522), and the most significant enriched term was MutLalpha complex (GO:0032389, P =3.74E-3). The term with the most genes of biological process was transcription, DNA-dependent (GO:0006351, count =887), and the most significant enriched term was retinoid metabolic process (GO:0001523, P = 1.32E-4) (Figure 10).
Similarly, of the top 20 enriched pathways in KEGG pathway analysis, starch and sucrose metabolism (ko00500), fructose and mannose metabolism (ko00051), galactose metabolism (ko00052), type 1 diabetes (ko04940), and type 2 diabetes (ko04930), which are associated with glycometabolism, play key roles in the development of diabetes and DKD (Figure 11).
Prediction of circRNA/microRNA Interaction
The “competing endogenous RNA” hypothesis suggests that circRNAs interact with MRE to mediate target gene expression. Some circRNAs can function as miRNA sponges to affect mRNA stability or translation25 and participate in the development of tumors, diabetes, and cardiovascular disease.26–28
To study the interactions between circRNAs and miRNAs, miRNA sequencing was performed on exosomes from the two groups. TargetScan and miRanda algorithms were used to predict interactions between differentially expressed circRNAs and miRNAs. We found that 152 differentially expressed circRNAs with FC≥1.5 could be paired with 43 differentially expressed miRNAs with the criteria of a context score ≥ 90 and a maximum energy ≤ −20 (Supplemental Table S1).
Discussion
Diabetic kidney disease (DKD) is a severe microvascular complication of diabetes.29–31 Microalbuminuria is a conventional predictor of DKD. However, the microalbumin content in the urine does not change immediately during the early stages of DKD. The sensitivity of microalbumin for early diagnosis of DKD is low. Recent studies have shown that glomerular injury markers SMAD1 and Podocalyxin are strongly associated with the severity of mesangial matrix expansion. The tubular injury markers Netrin-1, L-FABP, and NGAL are significantly higher in patients with diabetes than in controls.32 In addition, many miRNAs and lncRNAs have been considered as biomarkers for the early diagnosis and treatment of DKD.14,33 Nevertheless, it is important to identify more specific and sensitive molecular markers for DKD diagnosis.
Thus, exosomal circRNAs may serve as novel candidate biomarkers for the early diagnosis of DKD. In this study, we constructed circRNA differential expression profiles in HG-stimulated HK-2 cells to examine the correlation between exosomal circRNAs and high glucose conditions. The results showed that the exosome concentration was higher in the culture medium of HG-induced HK-2 cells. More than 36,000 circRNAs in exosomes extracted from the culture medium of the HG group were evaluated, including 693 differentially expressed circRNAs in the HG group compared with the NG group. Based on their relatively high abundance (FC ≥ 2, P<0.05) and their host genes, 10 candidate circRNAs were selected to validate their expression in the exosomes from the HK-2 cell culture medium of the HG group and NG group. In parallel with the microarray data, the qPCR results showed that the expression of circ_0009885, circ_0043753, and circ_0011760 increased, and the expression of circ_0032872, circ_0004716, and circ_0009445 decreased in the HG group. For example, PLOD1, the host gene of circ_0009885, plays an important role in diabetic wound healing.34 EML5 and FHL3, the host genes of circ_0011760 and circ_0032872, respectively, were the most prominently regulated genes in a mouse model of diabetes induced by streptozotocin.35 In addition, many studies have shown that inflammation and EMT play a key role in the development of DKD. Our study found that circ_0009885 can reverse the up-regulation of CCL2, IL6 and fibronetin expression by high glucose. Circ_0011760 reversed the up-regulation of IL6 and fibronetin expression by high glucose. And circ_0043753 reversed the down-regulation of epcam expression by high glucose. These results indicated that circRNAs may play an important role in the occurrence and progression of diabetes and DKD through regulating inflammation and EMT.
The process of DKD is complex and involves multiple pathogeneses such as oxidative stress, inflammation, fibrosis, endothelial dysfunction, hyperglycemia, and dyslipidemia. In addition, signal transduction pathways are also heavily involved and these include NF-κB activation,36 protein kinase C (PKC),37 extracellular signal-regulated kinase (ERK1/2),38 p38 mitogen-activated protein kinases39 and signal transducer and activator of transcription (STAT)40 and so on. GO and KEGG pathway analyses were used to predict the biological functions and potential pathways of differentially expressed circRNAs. GO analysis suggested that exosomal circRNAs are related to lipoprotein metabolic processes, inflammatory cell apoptotic processes, and hexokinase activity, which play important roles in DKD pathogenesis. Recent evidence indicates that lipid accumulation in the kidneys can aggravate DKD progression, including albuminuria, mesangial matrix expansion, and tubulointerstitial lesions.41,42 Abnormal lipid metabolism contributes to DKD progression by promoting tubulointerstitial fibrosis and inflammation.43,44 High glucose promotes apoptosis and inhibits autophagy in renal tubular epithelial cells by increasing cell death-inducing DFF45-like effector C expression. The inhibition of apoptosis may delay DKD.45 In addition, the therapeutic effects of zinc supplementation in DKD were regulated by increasing hexokinase II expression and preserving glucose metabolism-related pathways.46 KEGG pathway analysis revealed that the differentially expressed exosomal circRNAs were related to pathways such as starch and sucrose metabolism, type I diabetes mellitus, type II diabetes mellitus, galactose metabolism, and fructose and mannose metabolism. Among these enriched pathways, long-term hyperglycemia in diabetes mellitus is the primary contributor to DKD development.
Recent studies have shown that circRNAs can serve as ceRNAs that sponge miRNAs and subsequently increase mRNA transcription, thereby regulating gene expression. For instance, exosomal circ_DLGAP4 induces proliferation and fibrosis in HG-treated mesangial cells (MCs) by sponging miR-143, ultimately promoting DKD progression.24 In addition, circNUP98 knockdown inhibited the inflammatory reaction and fibrosis of HMCs induced by high glucose levels by mediating the miR-151-3p/HMGA2 pathway.22 In our study, in order to study the interaction between circRNAs and miRNAs, miRNAs sequencing was performed in exosomes of two groups. Differentially expressed circRNAs (FC≥1.5 were selected to analyze the interaction with miRNAs expressed in HK-2 cells. Some differentially expressed circRNAs, combined with one or more miRNA-binding sites, are associated with diabetes or DKD. For example, hsa-miR-15a-5p expression is decreased in the plasma of patients with moderate and severe diabetic renal disease.47 Moreover, hsa-miR-497-5p relieved fibrosis and inflammation in HG-treated HK-2 cells by inhibiting CCL19 expression.48 Our results showed that miR-15a-5p and miR-497-5p matched circ_0012339, indicating that exosomal circ_0012339 may play a role in DKD development. Therefore, we speculated that circRNAs participate in DKD progression through regulating miRNA target genes. However, further investigation of the detailed mechanism of circRNA-miRNA-mRNA interactions is needed.
The findings of this study indicate a significant difference in exosomal circRNA expression between high glucose-stimulated HK-2 cells and the control group. Comprehensive network analysis indicated that certain circRNAs and miRNAs may play critical roles in DKD pathogenesis. These data offer novel insights into DKD pathogenesis and could be a promising strategy for elucidating the molecular mechanisms of DKD.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Consent for Publication
The authors grant publisher permission to publish this work in Inflammation.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The authors gratefully acknowledge financial support from the Guangdong Basic and Applied Basic Research Foundation Project-key Project of Regional Joint Fund (2019B1515120004), Dongguan Science and Technology of Social Development Program (201950715023190), Administration of Traditional Chinese Medicine Bureau of Guangdong Province (20221184), and Key Projects of Social Science and Technology Development in Dongguan City, Guangdong Province (202050715023190).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Gnudi L, Coward RJM, Long DA. Diabetic nephropathy: perspective on novel molecular mechanisms. Trends Endocrinol Metab. 2016;27(11):820–830. doi:10.1016/j.tem.2016.07.002
2. Liu DW, Liu FX, Li ZY, et al. HNRNPA1-mediated exosomal sorting of miR-483-5p out of renal tubular epithelial cells promotes the progression of diabetic nephropathy-induced renal interstitial fibrosis. Cell Death Dis. 2021;12(3):255. doi:10.1038/s41419-021-03460-x
3. Xiang E, Han B, Zhang Q, et al. Human umbilical cord-derived mesenchymal stem cells prevent the progression of early diabetic nephropathy through inhibiting inflammation and fibrosis. Stem Cell Res Ther. 2020;11(1):336. doi:10.1186/s13287-020-01852-y
4. Wang S, Zhou Y, Zhang Y, et al. Roscovitine attenuates renal interstitial fibrosis in diabetic mice through the TGF-β1/p38 MAPK pathway. Biomed Pharmacother. 2019;115:108895. doi:10.1016/j.biopha.2019.108895
5. Yao Y, Li Y, Zeng X, et al. Losartan alleviates renal fibrosis and inhibits Endothelial-to-Mesenchymal Transition (EMT) under high-fat diet-induced hyperglycemia. Front Pharmacol. 2018;9:1213. doi:10.3389/fphar.2018.01213
6. Du L, Qian X, Li Y, et al. Sirt1 inhibits renal tubular cell epithelial-mesenchymal transition through YY1 deacetylation in diabetic nephropathy. Acta Pharmacol Sin. 2021;42(2):242–251. doi:10.1038/s41401-020-0450-2
7. Lu Q, Chen YB, Yang H, et al. Inactivation of TSC1 promotes epithelial-mesenchymal transition of renal tubular epithelial cells in mouse diabetic nephropathy. Acta Pharmacol Sin. 2019;40(12):1555–1567. doi:10.1038/s41401-019-0244-6
8. Zhong X, Chung ACK, Chen HY, et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia. 2013;56(3):663–674. doi:10.1007/s00125-012-2804-x
9. Kalluri R, LeBleu VS. The biology function and biomedical applications of exosomes. Science. 2020;367(6478):eaau6977. doi:10.1126/science.aau6977
10. He C, Zheng S, Luo Y, Wang B. Exosome theranostics: biology and translational medicine. Theranostics. 2018;8(1):237–255. doi:10.7150/thno.21945
11. Gurunathan S, Kang M-H, Jeyaraj M, et al. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells. 2019;8(4):307. doi:10.3390/cells8040307
12. Tikhomirov R, Donnell BR, Catapano F, et al. Exosomes: from potential culprits to new therapeutic promise in the setting of cardiac fibrosis. Cells. 2020;9(3):592. doi:10.3390/cells9030592
13. Chen J, Zhang Q, Liu D, et al. Exosomes: advances, development and potential therapeutic strategies in diabetic nephropathy. Metabolism. 2021;122:154834. doi:10.1016/j.metabol.2021.154834
14. Xu YX, Pu SD, Li X, et al. Exosomal ncRNAs: novel therapeutic target and biomarker for diabetic complications. Pharmacol Res. 2022;178:106135. doi:10.1016/j.phrs.2022.106135
15. Yang J, Liu D, Liu Z. Integration of metabolomics and proteomics in exploring the endothelial dysfunction mechanism induced by serum exosomes from diabetic retinopathy and diabetic nephropathy patients. Front Endocrinol. 2022;13:830466. doi:10.3389/fendo.2022.830466
16. Wu XM, Gao YB, Cui FQ, Zhang N. Exosomes from high glucose-treated glomerular endothelial cells activate mesangial cells to promote renal fibrosis. Biol Open. 2016;5(4):484–491. doi:10.1242/bio.015990
17. Tsai YC, Hung -W-W, Chang W-A, et al. Autocrine exosomal fibulin-1 as a target of MiR-1269b induces epithelial-mesenchymal transition in proximal tubule in diabetic nephropathy. Front Cell Dev Biol. 2021;9:789716. doi:10.3389/fcell.2021.789716
18. Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7(17):4183–4191. doi:10.7150/thno.21299
19. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi:10.1038/cr.2015.82
20. Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. doi:10.1186/s12943-019-1041-z
21. Li B, Sun G, Yu H, Meng J, Wei F. Exosomal circTAOK1 contributes to diabetic kidney disease progression through regulating SMAD3 expression by sponging miR-520h. Int Urol Nephrol. 2022;54(9):2343–2354. doi:10.1007/s11255-022-03139-y
22. Dong Q, Dong L, Wang YX, et al. Circular ribonucleic acid nucleoporin 98 knockdown alleviates high glucose-induced proliferation, fibrosis, inflammation and oxidative stress in human glomerular mesangial cells by regulating the microribonucleic acid −151-3p–high mobility group AT-hook 2 axis. J Diabetes Investig. 2022;13(8):1303–1315. doi:10.1111/jdi.13821
23. Zhu C, Shen K, Zhou W, Wu H, Lu Y. Exosome-mediated circ_0001846 participates in IL-1β-induced chondrocyte cell damage by miR-149-5p-dependent regulation of WNT5B. Clin Immunol. 2021;232:108856. doi:10.18632/oncotarget.8589
24. Bai S, Xiong X, Tang B, et al. Exosomal circ_DLGAP4 promotes diabetic kidney disease progression by sponging miR-143 and targeting ERBB3/NF-κB/MMP-2 axis. Cell Death Dis. 2020;11(11):1008. doi:10.1038/s41419-020-03169-3
25. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
26. Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. doi:10.18632/oncotarget.8589
27. Peng F, Gong W, Li S, et al. circRNA_010383 acts as a sponge for miR-135a, and its downregulated expression contributes to renal fibrosis in diabetic nephropathy. Diabetes. 2021;70(2):603–615. doi:10.2337/db20-0203
28. Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun. 2017;487(4):769–775. doi:10.1016/j.bbrc.2017.04.044
29. Webster AC, Nagler EV, Morton RL, Masson P. Chronic kidney disease. Lancet. 2017;389(10075):1238–1252. doi:10.1016/S0140-6736(16)32064-5
30. Umanath K, Lewis JB. Update on diabetic nephropathy: core curriculum 2018. Am J Kidney Dis. 2018;71(6):884–895. doi:10.1053/j.ajkd.2017.10.026
31. Alicic RZ, Rooney MT, Tuttle KR. Diabetic kidney disease: challenges, progress, and possibilities. Clin J Am Soc Nephrol. 2017;12(12):2032–2045. doi:10.2215/CJN.11491116
32. Liu H, Feng J, Tang L. Early renal structural changes and potential biomarkers in diabetic nephropathy. Front Physiol. 2022;13:1020443. doi:10.3389/fphys.2022.1020443
33. Gu YY, Lu FH, Huang XR, et al. Non-coding RNAs as biomarkers and therapeutic targets for diabetic kidney disease. Front Pharmacol. 2021;11:583528. doi:10.3389/fphar.2020.583528
34. Hu M, Wu Y, Yang C, et al. Novel long noncoding RNA lnc-URIDS delays diabetic wound healing by targeting Plod1. Diabetes. 2020;69(10):2144–2156. doi:10.2337/db20-0147
35. Friedrichs P, Schlotterer A, Sticht C, et al. Hyperglycaemic memory affects the neurovascular unit of the retina in a diabetic mouse model. Diabetologia. 2017;60(7):1354–1358. doi:10.1007/s00125-017-4254-y
36. Wang H, Huang X, Xu P, et al. Apolipoprotein C3 aggravates diabetic nephropathy in type 1 diabetes by activating the renal TLR2/NF-κB pathway. Metabolism. 2021;119:154740. doi:10.1016/j.metabol.2021.154740
37. Li X, Xu L, Hou X, et al. Advanced oxidation protein products aggravate tubulointerstitial fibrosis through protein kinase C-dependent mitochondrial injury in early diabetic nephropathy. Antioxid Redox Signal. 2019;30(9):1162–1185. doi:10.1089/ars.2017.7208
38. Wang L, Chang JH, Buckley AF, Spurney RF. Knockout of TRPC6 promotes insulin resistance and exacerbates glomerular injury in Akita mice. Kidney Int. 2019;95(2):321–332. doi:10.1016/j.kint.2018.09.026
39. Liu F, Guo J, Qiao Y, et al. MiR-138 plays an important role in diabetic nephropathy through SIRT1-p38-TTP regulatory axis. J Cell Physiol. 2021;236(9):6607–6618. doi:10.1002/jcp.30238
40. Zhang X, Zhao Y, Zhu X, et al. Active vitamin D regulates macrophage M1/M2 phenotypes via the STAT-1-TREM-1 pathway in diabetic nephropathy. J Cell Physiol. 2019;234(5):6917–6926. doi:10.1002/jcp.27450
41. Wu L, Liu C, Chang D, et al. The Attenuation of diabetic nephropathy by annexin A1 via regulation of lipid metabolism through the AMPK/PPARα/CPT1b pathway. Diabetes. 2021;70(10):2192–2203. doi:10.2337/db21-0050
42. Lucas O, Mas S, Marín-Royo G, et al. Lipotoxicity and diabetic nephropathy: novel mechanistic insights and therapeutic opportunities. Int J Mol Sci. 2020;21(7):2632. doi:10.3390/ijms21072632
43. Jankowski E, Wulf S, Ziller N, Wolf G, Loeffler I. MORG1-A negative modulator of renal lipid metabolism in murine diabetes. Biomedicines. 2021;10(1):30. doi:10.3390/biomedicines10010030
44. Yang W, Luo Y, Yang S, et al. Ectopic lipid accumulation: potential role in tubular injury and inflammation in diabetic kidney disease. Clin Sci. 2018;132(22):2407–2422. doi:10.1042/CS20180702
45. Zheng GS, Tan YM, Shang YY, et al. CIDEC silencing attenuates diabetic nephropathy via inhibiting apoptosis and promoting autophagy. J Diabetes Investig. 2021;12(8):1336–1345. doi:10.1111/jdi.13534
46. Sun W, Wang Y, Miao X, et al. Renal improvement by zinc in diabetic mice is associated with glucose metabolism signaling mediated by metallothionein and Akt, but not Akt2. Free Radic Biol Med. 2014;68:22–34. doi:10.1016/j.freeradbiomed.2013.11.015
47. Dieter C, Assmann TS, Costa AR, et al. MiR-30e-5p and MiR-15a-5p expressions in plasma and urine of type 1 diabetic patients with diabetic kidney disease. Front Genet. 2019;10:563. doi:10.3389/fgene.2019.00563
48. Zhang D, Chen X, Zheng D. A novel MIR503HG/miR-497-5p/CCL19 axis regulates high glucose-induced cell apoptosis, inflammation, and fibrosis in human HK-2 cells. Appl Biochem Biotechnol. 2022;194(5):2061–2076. doi:10.1007/s12010-021-03776-6
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.