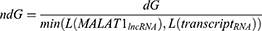Back to Journals » International Journal of General Medicine » Volume 16
MALAT1 in Liquid Biopsy: The Diagnostic and Prognostic Promise for Colorectal Cancer and Adenomas?
Authors Cervena K , Siskova A, Jungwirth J, Volarić M, Kral J, Kohout P, Levy M , Vymetalkova V
Received 5 May 2023
Accepted for publication 27 July 2023
Published 15 August 2023 Volume 2023:16 Pages 3517—3531
DOI https://doi.org/10.2147/IJGM.S420127
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Klara Cervena,1,2 Anna Siskova,1,2 Jiri Jungwirth,3,4 Marin Volarić,5 Jan Kral,6 Pavel Kohout,7 Miroslav Levy,8 Veronika Vymetalkova1,2,9
1Institute of Experimental Medicine, Czech Academy of Sciences, Prague, 142 00, Czech Republic; 2Institute of Biology and Medical Genetics, 1st Medical Faculty, Charles University, Prague, 128 00, Czech Republic; 3Institute of Physiology, 1st Faculty of Medicine Charles University, Prague, 121 08, Czech Republic; 4Department of Surgery, Weiden Clinic, Weiden in der Oberpfalz, 92637, Germany; 5Laboratory for Non-Coding DNA, Ruđer Bošković Institute, Zagreb, 10000, Croatia; 6Department of Hepatogastroenterology, Institute for Clinical and Experimental Medicine, Prague, 140 21, Czech Republic; 7Department of Internal Medicine, 3rd Faculty of Medicine Charles University and Faculty Thomayer Hospital Prague, Prague, 140 00, Czech Republic; 8Department of Surgery, First Faculty of Medicine, Charles University and Thomayer Hospital Prague, Prague, 140 59, Czech Republic; 9Biomedical Centre, Faculty of Medicine in Pilsen, Charles University in Prague, Pilsen, 323 00, Czech Republic
Correspondence: Klara Cervena, Institute of Experimental Medicine, Czech Academy of Sciences, Videnska 1083, Prague, 142 00, Czech Republic, Email [email protected]
Introduction: The development of colorectal cancer (CRC) is a multistep process accompanied by the accumulation of mutations that start from specific precancerous lesion – colorectal adenomas (CA). CRC incidence and mortality can be reduced by the early identification of these neoplasm. Colonoscopy is the most widely used screening method for CRC identification. Nowadays, clinical research interest is shifting to the use of liquid biopsy that may help with the early diagnosis of CA and CRC. In our previous study, we identified long non-coding RNA MALAT1 gene amplification associated with the development of CA.
Methods: This study aimed to describe the potential of MALAT1 expression levels in the adenoma tissue of patients used in the previous study by real-time qPCR. Furthermore, we analysed the plasma samples of an independent group of patients with CA (n=97), CRC (n=101), and cancer-free individuals (CFI, n=48).
Results: There was no difference in the MALAT1 expression level between CA patients with or without MALAT1 amplification. However, the plasma MALAT1 expression levels were significantly upregulated in patients with CRC and CA compared to CFI (for both p< 0.001). Moreover, a correlation between MALAT1 expression and histological types of adenomas was identified– high-CRC-risk adenomas also displayed the highest MALAT1 expression levels. Furthermore, in CRC patients, MALAT1 levels were associated with a response to therapy.
Conclusion: MALAT1 expression levels could serve as a promising circulating biomarker for early CA and CRC diagnosis, and even as a predictor of therapy response in CRC patients.
Keywords: MALAT1, colorectal adenomas, colorectal cancer, plasma, liquid biopsy
Introduction
Most colorectal cancer (CRC) arises from specific precancerous lesion – colorectal adenomas (CA).1,2 CA are neoplastic lesions arising from aberrant cells and some of the adenomas are precursors of CRC.2 Based on their histological characteristics are neoplastic sporadic adenomas divided to benign adenomas (tubular, villous, and tubulovillous), and serrated adenomas (sessile serrated and traditional serrated adenomas). Hyperplastic adenomas are usually identified as non-neoplastic lesion.3 The multistep process that leads to the transformation of small into larger adenomas and, subsequently, carcinomas, is the result of the accumulation of several gene mutations, genome instability and epigenetic factors.4
The early identification and removal of adenomas reduces the incidence of CRC and, consequently, mortality. One of the most frequently used screening methods is colonoscopy, during which the endoscopist can immediately remove an adenoma upon detection.5 Nevertheless, there are several complications (such as extensive patient preparation, risk of intestinal perforations, and bowel bleeding) that deter patients from undergoing this invasive examination.6
Recently, research attention has focused on liquid biopsy, which has many advantages and could compete with traditional biopsy. Liquid biopsy may better reflect the entire genetic profile of tumour clones presented in patients and supports personalized medicine.7 Liquid biopsy may also help with the early diagnosis of adenomas or tumours, prediction of therapy response and resistance, and disease recurrence.8–10 Cell-free nucleic acids (such as DNA, RNA, or non-coding RNAs) can be extracted from many body fluids, analysed by a wide variety of methods, and therefore represent promising biomarkers for cancer and other diseases.10–12
In the past, most studies have focused primarily on short non-coding RNAs, and long non-coding RNAs (lncRNAs) were overlooked. LncRNAs are a class of non-coding RNAs longer than 200 nucleotides and may play an important role in gene regulation and association with the occurrence of many human diseases, such as cancer.13–15 Specifically, in our previous study by Siskova et al, we investigated the structural and numerical chromosomal aberrations in adenoma tissues and adjacent mucosa by array comparative genomic hybridization. A large group of patients displayed the gain of lncRNA, MALAT1. Presented study aimed to investigate whether the MALAT1 amplification may represent an important step for adenoma development.16 MALAT1 expression levels have been already identified as a prognostic biomarker for lung cancer patients however, only a limited number of studies have investigated MALAT1 expression levels in CRC and CA patients.17–19 In the present study we aimed to describe whether i) the amplification of MALAT1 might be reflected at the mRNA expression level, ii) MALAT1 expression levels can be detected in plasma from CRC and CA patients, and iii) MALAT1 expression levels have a potential as a circulating diagnostic biomarker for CA, CRC, and cancer-free individuals (CFI), and iv) MALAT1 levels might be associated with the therapy response.
Materials and Methods
Design of the Study
MALAT1 expression levels were analysed by real-time quantitative polymerase chain reaction (RT-qPCR) in the tissue of patients with CA. Moreover, the MALAT1 expression levels in plasma from patients with CA, CRC patients, and CFI were measured. The study workflow is depicted in Figure 1.
The Study Population and Collection of Biological Specimens
The pilot cohort comprised adenoma tissue and adjacent mucosa from 14 histologically confirmed patients with CA. Detailed characteristics can be found in Table 1.16
CA patients included in the pilot and liquid biopsy analysis were recruited between March 2017 and May 2022 during the planned colonoscopy, and the collection of samples was carried out in cooperation with the Clinic of Hepatogastroenterology at the Institute for Clinical and Experimental medicine in Prague (Czech Republic). CAs were histologically classified according to the revised Vienna classification by a local pathologist.20
CRC patients were recruited between 2007 and 2018 at the Department of Surgery in the Thomayer Hospital, Prague (Czech Republic) and all patients were followed up until August 2021. Patient characteristics, such as demographics, family history of any cancer including CRC, smoking habits, and body mass index, were collected at the time of diagnosis using a structured questionnaire. Disease characteristics and treatment data including tumour location (colon/rectum), tumour-node-metastasis (TNM) stage, histopathological grade and type of subsequent chemotherapy regimen received were also collected. All information regarding the disease progression, relapse, distant metastasis and date of the last examination/death were also collected along with details of adjuvant chemotherapy (therapy response). CRC patients were also divided into good (those who benefited from chemotherapy and with no relapse) and poor responders (those patients without any therapy response resulting in relapse or death). The exclusion criteria were proven hereditary CRC syndromes, inflammatory bowel disease, histology of hyperplastic polyps, and a size of smaller than 5 mm. Patients with any personal history of previous malignancy, or with colorectal cancer-associated well-defined inherited syndromes (including Lynch syndrome, familial adenomatous, and MUTYH-associated polyposis) were also excluded from the study.
All study individuals signed a written consent to participate in the study and approved the use of their biological samples for genetic analyses according to the Helsinki declaration. The design of the study was approved by the Ethics Committee of the Institute for Clinical and Experimental medicine and Thomayer Hospital in Prague (protocol N. G-17-03-02), Czech Republic. For clinical characteristics of CA and CRC patients, see Table 1 and Table 2.
 |
Table 1 CA Patient’s Clinical Characteristics |
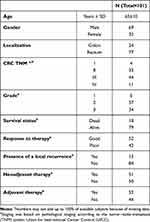 |
Table 2 CRC Patient’s Clinical Characteristics |
Tissue Collection and RNA Isolation
Sample biopsies of adenomas and adjacent tissue were collected into preservative tubes with RNA stabilization solution (RNAlater Stabilization Solution, Invitrogen, USA) and then stored at −80 °C.
All specimens were disrupted by Magna Lyser Green Beads (Roche, Germany) in a homogenizer - MagNaLyser Instrument (Roche, Germany). The RNA from disrupted tissue were extracted using RNA Mini Kit (Qiagen, Germany) and stored at −80 °C. RNA concentration was quantified with Qubit 3.0 Fluorometer using the Qubit RNA HS assay Kit (Thermo Fisher Scientific, USA).
Plasma Collection and RNA Isolation
Peripheral blood was collected from all individuals recruited into the study into EDTA tubes and centrifugated at 1400 rpm at 4°C for 10 min within 2 hours from its collection for plasma separation. The plasma fraction was immediately frozen and preserved at −80 °C.
Plasma/Serum Circulating and Exosomal RNA Purification Kit (Norgen Biotek, Canada) was used for RNA extraction from plasma. This kit is based on slurry format and was used according to the manufacturer´s protocol. RNA concentration was quantified with Qubit 3.0 Fluorometer (Thermo Fisher Scientific, USA) using the Qubit RNA HS assay Kit (Thermo Fisher Scientific, USA).
LncRNA Expression Analysis by RT-qPCR
Reverse Transcription
Isolated RNA was reverse transcribed by High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, USA). For reverse transcription, the 20 μL reaction (10 μL of RNA sample, 2 μL of 10xRT buffer, 0.8 μL of dNTP, 2 μL of 10xRandom primers, 3.2 μL of water, 1 μL of multiscribe reverse transcriptase and 1 μL of inhibitor) was prepared. Reaction mixtures were incubated for 10 min at 25°C, 120 min at 37°C, 5 min at 85°C and then held at 4°C (MJ Research PTC-200 Thermal Cycler, Marshall Scientific, USA).
The Selection of Reference Genes and Assays for Analysis
GAPDH, TBP, RPLP0 and HPRT1 were chosen (based on literature search) and analysed for the selection of proper reference genes. HPRT1 was further selected as the reference gene by Normfinder (GenEx, MultiD, Sweden). Reference genes with our candidate tested gene were analysed using Taqman Gene expression assays.
Preamplification
Plasma cDNA was pre-amplified prior to use in RT-qPCR using an IQ SuperMix (Bio-Rad, USA). The reaction for preamplification composed of 2 μL of cDNA, 1.5 μL gene expression assay (HPRT1 – assay ID: Hs02800695_m, and MALAT1 – assay ID: Hs00273907_s1), 5 μL of IQ SuperMix and 1.5 μL of RNAse free water. Preamplification was performed on thermal cycle (MJ Research PTC-200 Thermal cycler, Marshal Scientific, USA) in these steps: 95°C for 3 min, 18 cycles of 95°C for 15 sec and 59°C for 4 min and then held at 4°C. Following that, each sample was diluted 200x and stored at −20 °C until the analysis was performed. For the cDNA from tissue, the preamplification was not needed due to the high RNA concentrations.
Real-Time qPCR
RT-qPCR was performed using QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific, USA). This PCR reaction consisted of 5 μL Taqman Universal Mastermix II – no UNG (Thermo Fisher Scientific, USA), 0.5 μL of gene expression assay, 3.5 μL RNAse-free water and 1 μL of 200x diluted pre-amplified cDNA. Reactions were run in a 384-well optical plate at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 10 min.
Statistical Analysis
In this study, the Wilcoxon rank-sum test was used to evaluate the differences in MALAT1 expression levels based on patient data grouped by the variable of interest. Wilcoxon rank-sum test (also known as Mann–Whitney test) is a nonparametric statistical test used to determine if two independent samples come from the same population. This test is particularly useful when the assumptions of normality and equal variances for the parametric t-test are not met. This nonparametric statistical test compares the ranks of observations from the two samples, rather than the raw data, and calculates a test statistic and p-value to determine the significance of the difference between the samples. The p-value obtained was considered statistically significant at a threshold of p<0.05. All statistics and visualizations were performed using the R programming language and the ggplot2 library.
Bioinformatical Analysis
In this study, the interactome data were collected from the ENCORI lncRNA-RNA interaction database21 applying MALAT1 as a search query, using the default database search parameters. This search resulted in a total of 1638 genes. The results were filtered using the standardized free energy approach according to the following Formula no. 1, as stated in.22
Where L() is the mean length of all exons for a certain gene, then either MALAT1 or another genes were obtained from the interactome data. To obtain all the transcript lengths for a certain gene in the interactome gene set, we used the latest Ensembl human genome data GRCh38.p13 downloaded from the Ensembl ftp database (https://ftp.ensembl.org/pub/release-108/gtf/homo_sapiens/). We only kept genes with ndg<-0.1 and the resulting list of 879 genes was used for subsequent analysis.
After filtering the genes of interest, the PANTHER algorithm was used to identify significantly overrepresented biological pathways in the group of genes. The false discovery rate (FDR) was used to correct the p-values, and only the pathways with a corrected p-value of < 0.05 were considered significant.23 In addition, KEGG analysis to demonstrate the biological processes and pathways that may be affected by MALAT1 differential expression was performed.
Results
Tissue-Based Analysis
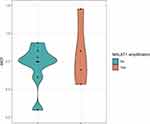
In the pilot study,16 chromosomal instability was studied using array comparative genomic hybridization (aCGH) analysis in adenoma tissue and adjacent mucosa from 16 patients who underwent planned colonoscopy. The MALAT1 amplification was identified in 5 CA patients. These results led us further examine the MALAT1 expression levels in the same CA group of patients. MALAT1 expression levels were successfully analysed by RT-qPCR in CA patients from the pilot phase (n=14). In this analysis, we divided patients according to the presence of MALAT1 amplification. There was no difference in the expression level of MALAT1 between these groups (Figure 2).
Plasma-Based Analysis
The potential of the expression level of MALAT1 as a non-invasive diagnostic biomarker was examined by liquid biopsy approach. The MALAT1 expression levels in the plasma of patients with CA (n=97) or CRC (n=101) and CFI (n=48) were studied. Plasma MALAT1 levels were significantly upregulated in the patients with CA (p<0.001, 5.25- fold change) and CRC (p<0.001, 5.16-fold change) compared to CFIs, but MALAT1 was unable to distinguish between CA and CRC patients (Figure 3). CA patients were further divided into adenomas groups according to histology type. MALAT1 levels could distinguish patients with hyperplastic adenomas and tubulovillous/villous adenomas (p=0.002, 6.14-fold change, Figure 4). Tubulovillous and villous adenomas that have the highest risk of the cancer development, were characterized by higher levels of MALAT1 expression. We further divided the CRC patients into good and poor responders based on their therapy response and found a significant difference between them: MALAT1 expression levels were upregulated in poor responders compared to good responders (p=0.04, 1.86-fold change, Figure 5). No association between MALAT1 expression level and the patient´s other characteristics (such as sex, localization and CRC stage) were observed.
Bioinformatical Analysis
A functional analysis was performed in silico to describe the target pathways that are influenced by the increased MALAT1 expression. A Gene Ontology (GO) analysis of the validated and predicted MALAT1 targets was conducted, and several biological processes (BP), molecular functions (MF), and cellular components (CC) terms were identified (Tables 3-5). Among them, the most affected biological processes were transmembrane transport (GO:0015988 and GO:0015990) and the antigen processing and presentation of endogenous peptide antigen via MHC class I (GO:0002486). The molecular functions were related to Major Histocompatibility Complex (MHC) class I and II proteins (GO:0042612, GO:0023026), while the cellular components were again related to MHC Class I protein complex (GO:0042612).
 |
Table 3 The List of MALAT1 Targets Related to Biological Processes |
 |
Table 4 The List of MALAT1 Targets Related to Molecular Functions |
 |
Table 5 The List of MALAT1 Targets Related to Cellular Components |
Gene set enrichment analysis on KEGG terms uncovered significant terms (Supplementary Table S1) again related to the MHC signaling pathway (padjust=0.039) and to ribosome-related pathway (padjust<0.0001) that was also presented in the top pathways in GO analysis. Schematic pathway map of affected gene associated with antigen processing and presentation by MALAT1 is illustrated in Supplementary Figure 1.
Discussion
The early identification and removal of adenomas during colonoscopy is the key to improving the CRC diagnosis and survival. However, some patients avoid the procedure due to the discomfort and invasiveness of the examination itself. The last decade has shown progress in the identification of new circulating cancer biomarkers that may allow the earlier diagnosis of tumours or precancerous lesions with minimal invasiveness. Increasing evidence shows that non-coding RNA plays an important role in the CRC development and aspirate as CRC biomarkers.16,24
LncRNA MALAT1 was first connected with cancer in 2003.17 The authors discovered a higher MALAT1 expression level associated with a worse prognosis for metastatic non-small cell lung cancer. From this date, the number of studies focusing on MALAT1 and cancer has rapidly increased and in addition to lung cancer, MALAT1 was aberrantly expressed in many other cancers, such as gastric, breast, and pancreatic cancer.25–27 Generally, MALAT1 has a significant role in the transcriptional processes involving interactions with transcriptional and splicing factors.27–29 MALAT1 has a posttranscriptional role in chromatin regulation and as competing endogenous RNAs (ceRNAs) in microRNA (miRNA) regulation.30 This interaction is between the miRNA and RNA that own the same miRNA recognition elements (MRE) and thus inhibit the function of miRNAs.31,32 MiRNAs that have the same MRE as MALAT1 and have also been marked as those with significant clinical relevance for CRC are miR-15 family, miR-101, or miR-363-3p.15,33–35
In our previous study, we identified the occurrence of MALAT1 gain in adenoma tissue, compared to adjacent mucosa by aCGH.16 This study aimed to examine the association between the presence of MALAT1 gain and the level of MALAT1 expression in adenoma tissues in the same group of CA patients, and, simultaneously, in the plasma samples of an enlarged cohort of CA, CRC patients, and CFI. We observed no significant difference in the level of MALAT1 expression in the tissue of patients with MALAT1 gain and patients without gain. We assume that CA patients with MALAT1 gain have this aberration in their adenoma tissue to compensate for the need of elevated MALAT1 levels in their tissue as a possible deviation from healthy tissue and possibly CRC development. Our claim is supported by the fact of increased MALAT1 levels in CA patients as well as in CRC patients compared to CFI. Additionally, our bioinformatical analysis has shown the involvement of MALAT1 in an adaptive immune response due to the relation to MHC proteins. Besides, we carried out a literature search and several other research articles suggested the role of MALAT1 in the immune pathway as well.36–38 In the study of Liu et al, the authors showed that a high level of MALAT1 in lung cancer lead to the inhibition of dendritic cells, the most powerful antigen-presenting cells, and the inhibition of T-cells proliferation.36 T-cells activation dependent on MALAT1 inhibition has been reported in another study by Hewitson et al.37 The authors observed negative regulation of MALAT1 by interleukin IL-10 and macrophage-activating factor. MALAT1 can be involved in suppressing antigen presentation through the inhibition of TNF-α and IL-6.38 Another interaction that may be MALAT1 involved in the immune system is through sponging of miR-195 and thus the upregulation of PD-L1, which serves as an important checkpoint in immune response and has already been described to be related to immune escape in cancer cells.39 The potential targets of MALAT1 in association with CRC, including miRNAs, were summarized in our review article.15 Briefly, potential targets of MALAT1 include proteins that have been already described as proto-oncogenes, for example, SOX9 or YAP1.19,40 SOX9 has been found overexpressed in CRC and its expression correlated with tumor progression, advanced stages of tumor, and lower overall survival.41–43 A positive correlation was found between MALAT1 and SOX9 expression levels and another constituent of this pathway is represented by miR-145.19 Specifically, MALAT1 can upregulate SOX9 and downregulate miR-145 and promote cell proliferation, migration, and invasion. Reverse (upstream) regulation of MALAT1 was described for transcriptional factor YAP1 which is a possible oncogene in many cancers.44 YAP1 in complex with other proteins binds to the MALAT1 promoter which can sponge miR-126-5p and thus promote expression of angiogenic factors (such as VEGFA, SLUG, or TWIST) that are direct targets of this miRNA. The process facilitates angiogenesis and epithelial-mesenchymal transition (EMT).40
Furthermore, in plasma, MALAT1 was upregulated in CRC and CA patients compared to CFI but MALAT1 levels were not able to distinguish patients with CA and CRC. In CA patients, we also observed an association of MALAT1 expression level with the histological type of adenomas – the type with the highest risk of CRC development displayed the highest MALAT1 expression level. Moreover, we observed that the patients characterized as poor responders showed a higher level of MALAT1 expression.
There are many studies analysing the expression level of MALAT1 in CRC tumour tissue and adjacent mucosa, but only a small number of studies have focused on CA. A study by Chaleshi et al included not only CRC patients (n=26) but also 46 tissue samples from CA patients.18 Nevertheless, they observed no significant difference between neoplastic tissue and non-neoplastic tissue. However, in our opinion, it is optimal to separate these two groups and preferably analyse them separately. Surprisingly, they described a high MALAT1 expression level in hyperplastic adenomas, which present the group of adenomas with the lowest risk of CRC development. This is not in concordance with our results, as hyperplastic adenomas were characterized by the lowest MALAT1 expression level. This dissimilarity may be due to the different RNA sources (tissue in the Chaleshi study vs plasma in our study) and the different way of the grouping of adenomas. In Chaleshi et al hyperplastic, adenomatous, and inflammatory adenomas were studied while in our study we used hyperplastic, tubular, and tubulovillous/villous adenomas. According to Jass,45 adenoma subdivisions include villous, tubulovillous or tubular, and serrated polyps, which are then subdivided into hyperplastic polyps, sessile serrated polyps or traditional serrated adenomas. The histological discrimination of hyperplastic polyps from sessile serrated lesions is rather difficult. Sessile serrated lesions and hyperplastic polyps are both types of serrated polyps that carry the different risk of malignancy, have different surveillance intervals, and are sometimes difficult to distinguish. In our study, purely hyperplastic polyps were used, however, no information about the sessile serrated polyps is mentioned in the study of Chaleshi.18
In CRC tissue, all studies observed MALAT1 upregulation in tumour tissue compared to adjacent mucosa, and a high expression level was also associated with lower overall survival.15,19,46–48 Moreover, in the study of Li et al, the authors divided the patients according to their response to oxaliplatin therapy and described how poor responders had higher MALAT1 expressions compared with good responders. This result was confirmed in tumour tissue and even in serum.49 A significant MALAT1 upregulation in patients resistant to oxaliplatin was observed in a study by Fan et al.50 These results are in concordance with our results. In the study of Li et al, the authors suggested the potential pathway for how MALAT1 may influence the therapy response in CRC patients.49 High MALAT1 expression levels led to a decreased levels of CDH1, an important protein for intercellular junctions and evasion to EMT. EMT causes high invasivity and migration, the most frequent causes of chemoresistance The interaction of MALAT1 with CDH1 is mediated by interaction with the EZH2 protein. MALAT1 binds it to the gene promoter CDH1 (encoding cadherin) thereby repressing cadherin expression and stimulating EMT. The authors also found that miR-218, containing a binding site for MALAT1, could also be part of this regulatory pathway.49 Another potential regulatory pathway is the MALAT1-miR-324-3p-ADAM17 axis that contains a disentegrin gene that has already been reported to be associated with the drug resistance in CRC.50,51
Circulating MALAT1 levels in association with CA or CRC have rarely been studied. To the best of our knowledge, no study has analysed MALAT1 levels in plasma samples. Siddique at al. analysed MALAT1 in the blood of CRC patients and CFI. MALAT1 levels were upregulated in the CRC patients compared to CFI.52 Another study focusing on the circulating MALAT1 levels in CRC also comprised CA patients.53 The authors reported higher serum MALAT1 expression levels in patients with CRC and CA, compared to CFI. These data were consistent with and reinforce our results. Most studies analysing MALAT1 in the blood were focused on the identification of single-nucleotide polymorphisms that were associated with a higher risk of CRC.54–57 The potential of lncRNAs as a circulating cancer biomarker was further supported by ongoing and completed clinical trials. As a results, one of these trials is focused on lncRNA ELNAT1 in exosomes in repeated urine samplings (before and after radical cystectomy) as a predictive cancer biomarker for the presence of lymphatic metastasis from bladder cancer (NCT05270174). An Egyptian study aims to evaluate the diagnostic value of lncRNA CCAT1 in the blood of CRC and CA patients (NCT04269746). Other lncRNA tested in clinical trials included H19 (hepatocellular cancer) and HOTAIR (thyroid cancer) (NCT04767750, NCT03469544).
The major advantage of our study is the topic of liquid biopsy and the focus on colorectal adenomas, since most of the studies analyzing lncRNA consisted only of CRC patients and their tumour tissue. Our study is the only one to investigate plasma MALAT1 levels in CA patients - it even includes almost 100 plasma samples from CA patients. The main potential of this study is based on the use of plasma as a source of lncRNA. Our plasma results are strengthened by concordance with the results of other studies focusing on serum/blood MALAT1 studies, which also describe the higher expression in CRC and CA patients. The main limitation of our study is that we did not observe an association between MALAT1 DNA amplification in and gene expression. Nevertheless, this could be explained by the small number of tissues, which was further reduced by two patients (from 16 to 14) for whom we no longer had tissue for RNA isolation. One of these patients even displayed MALAT1 amplification. The second limitation is the lack of patient tissues used in our liquid biopsy analysis.
Conclusions
In conclusion, we did not confirm an association between DNA amplification and gene expression. However, plasma MALAT1 might be able to distinguish CRC and CA patients from CFI and in addition, its expression level might reflect the patient´s therapy response. MALAT1 expression levels could serve as a circulating biomarker for the early diagnosis of CRC and even CA patients, and as a predictor of therapy response in CRC patients.
Clinical Trials
Changhao M Chen. A prospective, Multicenter Cohort Study of Urinary Exosome lncRNAs for Preoperative Diagnosis of Lymphatic Metastasis in Patients With Bladder Cancer. ClinicalTrials.gov. NCT05270174.
Alyaa Abd El-Rasoul Sayed Refae. Assessment Of Long Noncoding RNA CCAT1 In Colorectal Cancer Patients, 2020. Clinical Trials.gov. NTC04269746.
Shehalbeldin MS N. Role of LncRNA H19 in the regulation of IGF-1R Expression. ClinicalTrials.gov. NCT04767750.
Abdelghafour HS. Long Non Coding RNA HOTAIR and Midkine as Biomarkers in Thyroid Cancer. ClinicalTrials.gov. NCT03469544.
Acknowledgment
KC, AS and JJ acknowledge support by Charles University, First Faculty of Medicine at the project “Grant Schemes at CU” (reg. no. CZ.02.2.69/0.0/ 0.0/19_073/0016935). VV was supported by the Grant Agency of the Czech Republic (grant no. 22-05942S). Thanks to Frances Zatrepalkova for his kind assistance in proofreading and editing this manuscript.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi:10.3322/caac.21708
2. Siskova A, Cervena K, Kral J, et al. Colorectal Adenomas-Genetics and Searching for New Molecular Screening Biomarkers. Int J Mol Sci. 2020;21(9):3260. doi:10.3390/ijms21093260
3. Rubio CA, Jaramillo E, Lindblom A, et al. Classification of colorectal polyps: guidelines for the endoscopist. Endoscopy. 2002;34(3):226–236. doi:10.1055/s-2002-20296
4. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi:10.1016/0092-8674(90)90186-I
5. Allescher HD, Weingart V. Optimizing Screening Colonoscopy: strategies and Alternatives. Visc Med. 2019;35(4):215–225. doi:10.1159/000501835
6. Kim SY, Kim HS, Park HJ. Adverse events related to colonoscopy: global trends and future challenges. World J Gastroenterol. 2019;25(2):190–204. doi:10.3748/wjg.v25.i2.190
7. Gonzalez-Kozlova EE. Molecular Profiling of Liquid Biopsies for Precision Oncology. Adv Exp Med Biol. 2022;1361:235–247.
8. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18(5):297–312. doi:10.1038/s41571-020-00457-x
9. Lone SN, Nisar S, Masoodi T, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022;21(1):79. doi:10.1186/s12943-022-01543-7
10. Cervena K, Pardini B, Urbanova M, et al. Mutational landscape of plasma cell-free DNA identifies molecular features associated with therapeutic response in patients with colon cancer. A pilot study. Mutagenesis. 2021;36(5):358–368. doi:10.1093/mutage/geab024
11. Suraj S, Dhar C, Srivastava S. Circulating nucleic acids: an analysis of their occurrence in malignancies. Biomed Rep. 2017;6(1):8–14. doi:10.3892/br.2016.812
12. Szilagyi M, Pös O, Márton É, et al. Circulating Cell-Free Nucleic Acids: main Characteristics and Clinical Application. Int J Mol Sci. 2020;21(18):6827. doi:10.3390/ijms21186827
13. Brannan CI, Dees EC, Ingram RS, et al. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10(1):28–36. doi:10.1128/mcb.10.1.28-36.1990
14. Kosinska-Selbi B, Mielczarek M, Szyda J. Review: long non-coding RNA in livestock. Animal. 2020;14(10):2003–2013. doi:10.1017/S1751731120000841
15. Cervena K, Vodenkova S, Vymetalkova V. MALAT1 in colorectal cancer: its implication as a diagnostic, prognostic, and predictive biomarker. Gene. 2022;843:146791. doi:10.1016/j.gene.2022.146791
16. Siskova A, Kral J, Drabova J, et al. Discovery of Long Non-Coding RNA MALAT1 Amplification in Precancerous Colorectal Lesions. Int J Mol Sci. 2022;23(14):7656. doi:10.3390/ijms23147656
17. Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi:10.1038/sj.onc.1206928
18. Chaleshi V, Irani S, Alebouyeh M, et al. Association of lncRNA-p53 regulatory network (lincRNA-p21, lincRNA-ROR and MALAT1) and p53 with the clinicopathological features of colorectal primary lesions and tumors. Oncol Lett. 2020;19(6):3937–3949. doi:10.3892/ol.2020.11518
19. Xu Y, Zhang X, Hu X, et al. The effects of lncRNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol Med. 2018;24(1):52. doi:10.1186/s10020-018-0050-5
20. Schlemper RJ, Riddell RH, Kato YE, et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut. 2000;47(2):251–255. doi:10.1136/gut.47.2.251
21. Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. doi:10.1093/nar/gkt1248
22. Li J, Ma W, Zeng P, et al. LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Brief Bioinform. 2015;16(5):806–812. doi:10.1093/bib/bbu048
23. Mi H, Thomas P. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol Biol. 2009;563:123–140.
24. He J, Wu F, Han Z, et al. Biomarkers (mRNAs and Non-Coding RNAs) for the Diagnosis and Prognosis of Colorectal Cancer - From the Body Fluid to Tissue Level. Front Oncol. 2021;11:632834. doi:10.3389/fonc.2021.632834
25. Okugawa Y, Toiyama Y, Hur K, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35(12):2731–2739. doi:10.1093/carcin/bgu200
26. Pang EJ, Yang R, Fu X-B, et al. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2015;36(4):2403–2407. doi:10.1007/s13277-014-2850-8
27. Yue X, Wu W-Y, Dong M, et al. LncRNA MALAT1 promotes breast cancer progression and doxorubicin resistance via regulating miR-570-3p. Biomed J. 2021;44(6 Suppl 2):S296–S304. doi:10.1016/j.bj.2020.11.002
28. Arun G, Aggarwal D, Spector DL. MALAT1 Long Non-Coding RNA: functional Implications. Noncoding RNA. 2020;6(2). doi:10.3390/ncrna6020022
29. Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi:10.1016/j.molcel.2010.08.011
30. Yang L, Lin C, Liu W, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–788. doi:10.1016/j.cell.2011.08.054
31. Su X, Xing J, Wang Z, et al. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chin J Cancer Res. 2013;25(2):235–239. doi:10.3978/j.issn.1000-9604.2013.03.08
32. Tang Y, Xiao G, Chen Y, et al. LncRNA MALAT1 promotes migration and invasion of non-small-cell lung cancer by targeting miR-206 and activating Akt/mTOR signaling. Anticancer Drugs. 2018;29(8):725–735. doi:10.1097/CAD.0000000000000650
33. Xie JJ, Li WH, Li X, et al. LncRNA MALAT1 promotes colorectal cancer development by sponging miR-363-3p to regulate EZH2 expression. J Biol Regul Homeost Agents. 2019;33(2):331–343.
34. Guo J, Ding Y, Yang H, et al. Aberrant expression of lncRNA MALAT1 modulates radioresistance in colorectal cancer in vitro via miR-101-3p sponging. Exp Mol Pathol. 2020;115:104448. doi:10.1016/j.yexmp.2020.104448
35. Ji Q, Cai G, Liu X, et al. MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis. Cell Death Dis. 2019;10(6):378. doi:10.1038/s41419-019-1598-x
36. Liu Y, Yin Z, Lu P, et al. Lung Carcinoma Cells Secrete Exosomal MALAT1 to Inhibit Dendritic Cell Phagocytosis, Inflammatory Response, Costimulatory Molecule Expression and Promote Dendritic Cell Autophagy via AKT/mTOR Pathway. Onco Targets Ther. 2020;13:10693–10705. doi:10.2147/OTT.S256669
37. Hewitson JP, West KA, James KR, et al. Malat1 Suppresses Immunity to Infection through Promoting Expression of Maf and IL-10 in Th Cells. J Immunol. 2020;204(11):2949–2960. doi:10.4049/jimmunol.1900940
38. Zhao G, Su Z, Song D, et al. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-kappaB. FEBS Lett. 2016;590(17):2884–2895. doi:10.1002/1873-3468.12315
39. Wang QM, Lian G-Y, Song Y, et al. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. 2019;231:116335. doi:10.1016/j.lfs.2019.03.040
40. Sun Z, Ou C, Liu J, et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38(14):2627–2644. doi:10.1038/s41388-018-0628-y
41. Matheu A, Collado M, Wise C, et al. Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 2012;72(5):1301–1315. doi:10.1158/0008-5472.CAN-11-3660
42. Lu B, Fang Y, Xu J, et al. Analysis of SOX9 expression in colorectal cancer. Am J Clin Pathol. 2008;130(6):897–904. doi:10.1309/AJCPW1W8GJBQGCNI
43. Lizarraga-Verdugo E, Carmona T, Ramos-Payan R, et al. SOX9 is associated with advanced T-stages of clinical stage II colon cancer in young Mexican patients. Oncol Lett. 2021;22(1):497. doi:10.3892/ol.2021.12758
44. Szulzewsky F, Holland EC, Vasioukhin V. YAP1 and its fusion proteins in cancer initiation, progression and therapeutic resistance. Dev Biol. 2021;475:205–221. doi:10.1016/j.ydbio.2020.12.018
45. Jass JR. Gastrointestinal polyposes: clinical, pathological and molecular features. Gastroenterol Clin North Am. 2007;36(4):927–46, viii. doi:10.1016/j.gtc.2007.08.009
46. Zheng HT, Shi D-B, Wang Y-W, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174–3181.
47. Wu X, Li R, Song Q, et al. JMJD2C promotes colorectal cancer metastasis via regulating histone methylation of MALAT1 promoter and enhancing beta-catenin signaling pathway. J Exp Clin Cancer Res. 2019;38(1):435. doi:10.1186/s13046-019-1439-x
48. Li Q, Dai Y, Wang F, et al. Differentially expressed long non-coding RNAs and the prognostic potential in colorectal cancer. Neoplasma. 2016;63(6):977–983. doi:10.4149/neo_2016_617
49. Li P, Zhang X, Wang H, et al. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol Cancer Ther. 2017;16(4):739–751. doi:10.1158/1535-7163.MCT-16-0591
50. Fan C, Yuan Q, Liu G, et al. Long non-coding RNA MALAT1 regulates oxaliplatin-resistance via miR-324-3p/ADAM17 axis in colorectal cancer cells. Cancer Cell Int. 2020;20(1):473. doi:10.1186/s12935-020-01549-5
51. Kyula JN, Van Schaeybroeck S, Doherty J, et al. Chemotherapy-induced activation of ADAM-17: a novel mechanism of drug resistance in colorectal cancer. Clin Cancer Res. 2010;16(13):3378–3389. doi:10.1158/1078-0432.CCR-10-0014
52. Siddique H, Al-Ghafari A, Choudhry H, et al. Long Noncoding RNAs as Prognostic Markers for Colorectal Cancer in Saudi Patients. Genet Test Mol Biomarkers. 2019;23(8):509–514. doi:10.1089/gtmb.2018.0308
53. Radwan AF, Shaker OG, El-Boghdady NA, et al. Association of MALAT1 and PVT1 Variants, Expression Profiles and Target miRNA-101 and miRNA-186 with Colorectal Cancer: correlation with Epithelial-Mesenchymal Transition. Int J Mol Sci. 2021;22(11):6147. doi:10.3390/ijms22116147
54. Li Y, Bao C, Gu S, et al. Associations between novel genetic variants in the promoter region of MALAT1 and risk of colorectal cancer. Oncotarget. 2017;8(54):92604–92614. doi:10.18632/oncotarget.21507
55. Zhao K, Jin S, Wei B, et al. Association study of genetic variation of lncRNA MALAT1 with carcinogenesis of colorectal cancer. Cancer Manag Res. 2018;10:6257–6261. doi:10.2147/CMAR.S177244
56. Yang Q, Zheng W, Shen Z, et al. MicroRNA Binding Site Polymorphisms of the Long-Chain Noncoding RNA MALAT1 are Associated with Risk and Prognosis of Colorectal Cancer in Chinese Han Population. Genet Test Mol Biomarkers. 2020;24(5):239–248. doi:10.1089/gtmb.2020.0013
57. Wu S, Sun H, Wang Y, et al. MALAT1 rs664589 Polymorphism Inhibits Binding to miR-194-5p, Contributing to Colorectal Cancer Risk, Growth, and Metastasis. Cancer Res. 2019;79(20):5432–5441. doi:10.1158/0008-5472.CAN-19-0773
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.