Back to Journals » Cancer Management and Research » Volume 12
Long Noncoding RNA FBXL19-AS1 Expedites Cell Growth, Migration and Invasion in Cervical Cancer by miR-193a-5p/PIN1 Signaling
Authors Wan S, Ni G, Ding J, Huang Y
Received 11 May 2020
Accepted for publication 27 August 2020
Published 7 October 2020 Volume 2020:12 Pages 9741—9752
DOI https://doi.org/10.2147/CMAR.S262215
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Su Wan,1 Guantai Ni,1 Jin Ding,1 Yuansheng Huang2
1Department of Obstetrics and Gynecology, Yijishan Hospital of Wannan Medical College, Wuhu, Anhui 241000, People’s Republic of China; 2Department of Orthopedics, Yijishan Hospital of Wannan Medical College, Wuhu, Anhui 241000, People’s Republic of China
Correspondence: Yuansheng Huang
Department of Orthopedics, Yijishan Hospital of Wannan Medical College, No. 2 Zheshan West Road, Jinghu District, Wuhu, Anhui 241000, People’s Republic of China
Tel +86 0553-5739999
Fax +86 0553-5738279
Email [email protected]
Background: Cervical cancer is one of the most prevalent malignancies in gynecology with increasing incidence in recent years. Long noncoding RNAs (lncRNAs) have been reported to regulate human cancers including cervical cancer. F-box and leucine-rich repeat protein 19 antisense RNA 1 (FBXL19-AS1) have been unmasked to exert carcinogenic functions in several cancers except cervical cancer.
Aim: Present study hammered at investigating the function and mechanism of FBXL19-AS1 in cervical cancer.
Methods: RT-qPCR was utilized to test gene expression. EdU staining, colony formation, transwell, flow cytometry and TUNEL assays were applied for measuring the impact of FBXL19-AS1 on cervical cancer cell functions. Moreover, RIP, RNA pull-down and luciferase reporter assays were utilized for detecting the correlations among FBXL19-AS1, miR-193a-5p and PIN1 (peptidylprolyl cis/trans isomerase, NIMA-interacting 1).
Results: FBXL19-AS1 exhibited elevated expression in cervical cancer tissues and cells. Silencing FBXL19-AS1 repressed cell proliferation through arresting cell cycle and stimulating apoptosis, and losing FBXL19-AS1 also restrained cell migration and invasion. Also, we discovered FBXL19-AS1 as a miR-193a-5p sponge, while miR-193a-5p was a tumor inhibitor in cervical cancer. Further, PIN1 was proved as the miR-193a-5p target, and FBXL19-AS1 augmented PIN1 expression in cervical cancer via sequestering miR-193a-5p. Of note, PIN1 accelerated the progression of cervical cancer, and its upregulation counteracted the impacts of depleted FBXL19-AS1 on cervical cancer cell functions.
Conclusion: FBXL19-AS1 contributes to malignant phenotypes in cervical cancer by sponging miR-193a-5p and regulating PIN1.
Keywords: FBXL19-AS1, miR-193a-5p, PIN1, cervical cancer
Introduction
Cervical cancer is one of the most prevalent malignancies in gynecology with serious endangerment to women’s health.1 In recent years, the incidence of cervical cancer is increasing with a younger trend.2 With the development of early screening technology and treatment, the survival rate of early patients is also increasing.3–5 However, the prognosis of those developed into advanced stages is still not good.6 Thus, developing new biomarkers for cervical cancer are urgently demanded.
As a subclass of noncoding RNAs, long noncoding RNAs (lncRNAs) possess a length with longer than 200 nucleotides. LncRNAs cannot translate into proteins, but they can modulate gene expression through diverse manners including post-transcriptional regulation.7,8 An increasing number of reports have indicated that lncRNAs take part in different cellular processes to regulate the development of assorted diseases, even human cancers.9,10 Dysfunctional lncRNAs are always discovered in cancers and they are considered to be associated with cancer development. For example, TUG1 was upregulated and promoted cell proliferation in epithelial ovarian cancer via modulating AURKA.11 A former research also indicated that upregulated PVT1 was associated with low survival rate and it could accelerate the progression of gallbladder cancer via miR-143/HK2 axis.12F-box and leucine-rich repeat protein 19 antisense RNA 1 (FBXL19-AS1) is a kind of lncRNA which has been reported to regulate cancer development by some researches. For examples, FBXL19-AS1 expedited oncogenic cellular behaviors via absorbing miR-346 in osteosarcoma;13 FBXL19-AS1 facilitated cell growth and epithelial–mesenchymal transition in non-small cell lung cancer;14 FBXL19-AS1 acted as a miR-178 sponge to accelerate breast cancer cell proliferation.15 Nonetheless, the specific role and mechanism of FBXL19-AS1 in cervical cancer are still unclear.
In the current work, we hammered at investigating the function and latent mechanism of FBXL19-AS1 in cervical cancer, which may conduce to improving cervical cancer treatment.
Materials and Methods
Human Cervical Cancer Samples
The paired tumor and non-tumor samples were surgically acquired from 100 cervical cancer patients in the Yijishan Hospital of Wannan Medical College. The patients with no any treatment before operation signed the written informed consents prior to this research. The work was supported by the Ethics Committee of Yijishan Hospital of Wannan Medical College. Specimens were all subjected to quick-frozen via liquid nitrogen and preservation at −80°C.
Cell Culture
Human normal cervical cell line (Ect1/E6E7) and cervical cancer cell lines (C-4-I, SiHa, C-33A and HeLa) were selected for our research. Ect1/E6E7, SiHa, C-33A and HeLa cells were obtained from ATCC (Manassas, VA, USA). C-4-I cell was purchased from CoBioer Biosciences Co., Ltd (Nanjing, China). Keratinocyte Serum-Free Medium (K-SFM; Gibco Laboratories, Grand Island, NY, USA) was utilized to cultivated Ect1/E6E7 cell. Eagle’s Minimum Essential Medium (EMEM) was applied for cultivating SiHa, C-33A and HeLa cells. C-4-I was cultured in DMEM medium with 10% FBS (Gibco). Each kind of culture medium was added with 10% fetal bovine serum (FBS; Gibco). All cells were cultivated under the condition with 5% CO2 at 37°C.
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
The TRIzol reagent was implemented for separating total RNA from indicated tissues or cells. Then, the reverse transcription kit (Takara, Dalian, China) was utilized for reverse transcription of RNA to cDNA. Following, RT-qPCR was conducted via the ABI7500 System (Applied Biosystems, CA, USA). Finally, gene expression relative to GAPDH/U6 was calculated via the 2−ΔΔCt method.
Cell Transfection
For inhibiting FBXL19-AS1 and PIN1, specific short‐hairpin RNAs (sh‐RNAs) were devised and synthesized through GenePharma (Shanghai, China), with a nonsense sequence as the negative control (sh‐NC). For upregulating the expression of PIN1, the sequence of PIN1 cDNA was cloned into pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA) for acquiring pcDNA3.1/PIN1, with empty vector as the negative control. Also, NC mimics/inhibitor and miR-193a-5p mimics/inhibitor were also devised and synthesized through GenePharma. Cell transfection with indicated plasmids for 48 hours was achieved by the utilization of Lipofectamine 3000 (Invitrogen).
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Assay
In accordance with the protocol of EdU labeling/detection kit (RiboBio, Guangzhou, China), EDU experiment was employed for evaluating cell proliferation capability. Cells (1×104) were placed in 96-well plates, followed by the addition of 50 μM of EdU diluent for cultivating cells for 2 hours under the condition of 5% CO2 and 37°C. After fixation by 4% paraformaldehyde and rinsing via PBS, cells were processed with Apollo 567 working solution for 2 hours. Following, DAPI was utilized to counterstain the nuclei for 5 minutes. In the end, cells were observed by fluorescent microscopy (Thermo Fisher Scientific). Bio-repeats were run in triplicate.
Colony Formation Assay
Transfected cells were planted in the 6-well plates (500 cells per well) for two-week incubation. After that, cells were fixated with 4% paraformaldehyde and stained by 0.1% crystal violet. Finally, the number of colonies was monitored manually. Bio-repeats were run in triplicate.
TUNEL Assay
TUNEL assay was operated for assessing cell apoptosis with the in situ Cell Apoptosis Detection Kit (Roche, Basel, Switzerland). Simply put, 1×105 cells were added into 24-well plates. After that, cells were processed consecutively with 4% paraformaldehyde, 0.2% Triton X-100, and DAPI staining solution. Subsequently, the images were captured with a light microscope (CX23, Olympus, Japan). Bio-repeats were run in triplicate.
Flow Cytometry Analysis
As for cell apoptosis detection, cells were subjected to double-staining via FITC-Annexin V Apoptosis Detection Kit (BD Biosciences) in accordance with protocol of supplier. With respect to cell cycle evaluation, cells were digested and fixed with 75% ethanol, followed by discard of the supernatant and cultivation with PI-contained RNA enzyme (Sigma-Aldrich). Finally, cell apoptotic rate and cell cycle distribution were both determined via flow cytometry (FACScan; BD Biosciences. Bio-repeats were run in triplicate).
Transwell Assay
The capabilities of C-4-I and C-33A cells to invade or migrate were measured by using the transwell chamber with or without Matrigel (Corning Incorporated, Corning, NY), respectively. 2 × 104 cells in serum-free medium were seeded to the upper chamber, while the medium supplementing 10% FBS was poured into the lower chamber. After 24 hours, cells in the lower chamber were subjected to fixation by 4% paraformaldehyde and staining with crystal violet. Afterwards, we observed cells by the microscope (magnification, ×200; Olympus, Tokyo, Japan). Bio-repeats were run in triplicate.
Fluorescent in situ Hybridization (FISH)
Ribo™ Fluorescent in situ Hybridization Kit (Ribobio) was applied for conducted this assay based on the user guides. In short, the FBXL19-AS1 probes labeled with fluorescent dye were incubated with indicated cells. Finally, the signals of fluorescence were analyzed by a confocal laser-scanning microscope (Leica). Bio-repeats were run in triplicate.
Subcellular Fractionation
The nuclear or cytoplasmic Isolation Kit (Biovision, San Francisco, CA, USA) was utilized for the isolation of cytosolic and nuclear fractions in C-4-I or C-33A cells in line with the user guide. The RNA molecules in the cytoplasm and nucleus were then detected via RT-qPCR. U6 and GAPDH served as the controls. Bio-repeats were run in triplicate.
Luciferase Reporter Assay
The synthesized sequences of FBXL19-AS1or PIN1 3ʹUTR covering wild-type (WT) or mutated-type (Mut) of miR-193a-5p interacting sites were inserted into pmirGLO vectors (Promega, Madison, WI, USA) for constructing FBXL19-AS1-WT or PIN1-WT/Mut. Then, cells were subjected to co-transfection with the constructed vectors and miR-193a-5p mimics (or NC mimics) using Lipofectamine™3000 (Invitrogen) for 48 hours. In the end, dual-luciferase reporter gene assay system (Promega) was employed for measuring the luciferase activity. Bio-repeats were run in triplicate.
RNA Immunoprecipitation (RIP) Assay
Magna RIPTM RNA Binding Protein Immunoprecipitation Kit (Millipore) was utilized for conducting this assay. Simply put, cells were lysed via RIP lysis buffer (Solarbio) and then the obtained lysates were cultivated for a whole night in RIP buffer with magnetic beads coating anti-Ago2 (Millipore) or anti-IgG. After that, the precipitated RNAs was isolated and analyzed through RT-qPCR. Bio-repeats were run in triplicate.
RNA Pull-Down Assay
The lysates acquired from C-4-I and C-33A cells via RIPA lysis buffer were cultivated for 1 hour at 4°C with biotinylated miR-193a-5p sequences with wild-type or mutated seed region (Bio-miR-193a-5p-WT/Mut), with the biotin-labelled nonsense sequence as the negative control (Bio-NC). Then, the streptavidin agarose magnetic beads were supplemented into above mixtures, and the enrichment of RNAs captured by beads were analyzed via RT-qPCR. Bio-repeats were run in triplicate.
Statistical Analysis
The data from all three bio-repeats were expressed as mean ± standard deviation (SD) after analysis via SPSS 22.0 software (IBM, Armonk, NY, USA). The group differences were measured via Student’s t-test or one-way ANOVA, with P < 0.05 as statistically significant.
Results
Knockdown of FBXL19-AS1 Restrains Cell Proliferation, Migration and Invasion in Cervical Cancer
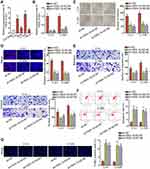
First of all, in order to examine the function ofFBXL19-AS1 in cervical cancer, we tested its expression under malignant conditions. Intriguingly, FBXL19-AS1 presented high expression in cervical cancer tissues relative to matched non-cancerous ones (Supplementary Figure 1A). Consistently, we observed that FBXL19-AS1 was highly expressed in cervical cancer cell lines (C-4-I, SiHa, C-33A and HeLa) in contrast with the normal cell line (Ect1/E6E7), especially in C-4-I and C-33A cells (Figure 1A). Then, for conducting loss-of-function assays, we interfered FBXL19-AS1 expression in C-4-I and C-33A cells using sh-FBXL19-AS1#1/2 (Figure 1B). Subsequently, we discovered that cell proliferative capability was restrained through the lack of FBXL19-AS1, because the quantity of colonies and the rate of EdU positive cells were both reduced after FBXL19-AS1 was knocked down (Figure 1C-D). After that, we measured cell migration and invasion through transwell experiments. Results displayed that the number of migrated and invaded cells was dramatically declined when FBXL19-AS1 was inhibited (Figure 1E), indicating the repressive impact of depleted FBXL19-AS1 on cervical cancer cell migration and invasion. In the end, to further uncover the in-depth influence of FBXL19-AS1 on cervical cancer cell proliferation, we then investigated its impact on cell cycle progression and cell apoptosis. It manifested that the absence of FBXL19-AS1 led to increased percent of cells at G0/G1 phase and decreased proportion of cells at S and G2/M phases (Supplementary Figure 1B), disclosing that FBXL19-AS1inhibition induced cell cycle arrest in cervical cancer cells. Meanwhile, the outcomes of flow cytometry analysis and TUNEL experiment revealed that both the rates of apoptotic cells and TUNEL positive cells were elevated under FBXL19-AS1 deficiency (Figure 1F-G), which demonstrated that cell apoptotic ability could be expedited by FBXL19-AS1 shortage. In a word, the expression of FBXL19-AS1 is extremely high and knockdown of FBXL19-AS1 restrains cell proliferation, migration and invasion in cervical cancer.
FBXL19-AS1 Sponges miR-193a-5p and Overexpression of miR-193a-5p Inhibits the Progression of Cervical Cancer
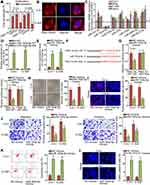
For the sake of exploring the regulatory mechanism of FBXL19-AS1 in cervical cancer, we conducted subcellular fractionation assay and FISH experiment to figure out the distribution of FBXL19-AS1 in cervical cancer cells. The outcomes indicated the major location of FBXL19-AS1 in C-4-I and C-33A cell cytoplasm (Figure 2A-B), which signified that FBXL19-AS1might serve as a miRNA sponge to exert its function at post-transcription level. Thus, we utilized the ENCORI database (http://starbase.sysu.edu.cn/index.php) to search for the latent miRNAs binding to FBXL19-AS1. On the basis of specific conditions (CLIP-Data ≥2; Degradome-Data ≥0; pan-Cancer ≥4), we found seven miRNA candidates (including miR-331-3p,miR-193a-5pmiR-378a-3p, miR-378c, miR-378d, miR-5586-5p and miR-6512-3p). Then, we implemented RT-qPCR experiments to examine the expression of the aforementioned miRNAs in cervical cancer cells. As a result, we discovered that only miR-193a-5p was lowly expressed in cervical cancer cells, while the levels of other miRNAs were relatively high (Figure 2C). Of importance, cervical cancer samples tended to possess higher miR-193a-5p level than paired controls (Supplementary Figure 1C). Hence, we selected miR-193a-5p for further experiments. Through RNA pull-down experiments, we discovered that the enrichment of FBXL19-AS1 was extremely high in biotinylated wild-type miR-193a-5p group rather than the other two groups (Figure 2D). Following, we elevated miR-193a-5p expression in these two cells (Figure 2E), and also acquired the predicted binding sites between miR-193a-5p and FBXL19-AS1 by ENCORI (Figure 2F). As expected, the results of luciferase reporter experiments displayed that overexpressed miR-193a-5p caused a notable reduction in the luciferase activity of FBXL19-AS1-WT, while the luciferase activity of FBXL19-AS1-Mut emerged no visible change (Figure 2G). Furthermore, we examined the role of miR-193a-5p in cervical cancer. Through colony formation and EdU experiments, we discovered that upregulation of miR-193a-5p inhibited cell proliferation (Figure 2H-I). In the same way, cell migration and invasion were also suppressed by miR-193a-5p overexpression according to the results of transwell experiments (Figure 2J). Further, we discovered that miR-193a-5p elevation resulted in cell cycle arrest in both the two cervical cancer cells (Supplementary Figure 1D). Nevertheless, the outcomes of flow cytometry analysis and TUNEL experiments indicated that cell apoptotic ability was accelerated after overexpressing miR-193a-5p (Figure 2K-L). Overall, FBXL19-AS1 sponges miR-193a-5p, a progression-repressor in cervical cancer.
PIN1 is the Target of miR-193a-5p in Cervical Cancer
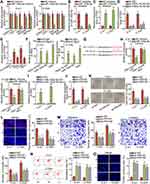
In order to further investigate the downstream molecules, we began to find the target genes of miR-193a-5p. After searching on ENCORI database based on following conditions: CLIP-Data ≥2, Degradome-Data≥1, pan-Cancer ≥10 and programNum ≥1), we discovered 5 potential genes (CARD19, FDFT1,PIN1, INO80Eand REPIN1) targeted by miR-193a-5p. Thereafter, RT-qPCR was conducted to measure the influence of upregulated miR-193a-5p on the expression of above targets. The results presented that the expression of PIN1 (peptidylprolyl cis/trans isomerase, NIMA-interacting 1) was visibly inhibited by miR-193a-5p enhancement, while that of others almost unchanged (Figure 3A). Then, we intended to inhibit miR-193a-5p expression in the two cells by transfecting with miR-193a-5p inhibitor, and examined the interference efficiency using RT-qPCR. The results indicated that miR-193a-5p expression was evidently declined by an miR-193a-5p inhibitor (Figure 3B). Further, we discovered that the expression of PIN1 was ascended in response to miR-193a-5p deficiency (Figure 3C), but declined in face of FBXL19-AS1 knockdown (Figure 3D). In addition, it was unveiled that the expression of PIN1 was heightened in cervical cancer specimens compared to adjacent non-tumor ones (Supplementary Figure 1E), as well as in cervical cancer cells relative to normal controls (Figure 3E). Following, RIP experiments were implemented and the outcomes displayed that FBXL19-AS1, miR-193a-5p and PIN1 were co-precipitated by anti-Ago2, proving their existence in RNA-induced silencing complexes (RISCs) (Figure 3F). In Figure 3G, we displayed the binding sites between miR-193a-5p and PIN1 which were predicted by ENCORI. Furthermore, luciferase reporter experiment results showed that only the luciferase activity of PIN1 3ʹUTR-WT was evidently declined when miR-193a-5p was upregulated (Figure 3H). Moreover, the outcomes of RNA pull-down experiments also proved the combining relationship between miR-193a-5p and PIN1 (Figure 3I). Subsequently, we evaluated the function of PIN1 in cervical cancer cellular behaviors after validating the successful decline of PIN1 expression by sh-PIN1#1/2 (Figure 3J). As anticipated, we found that loss of PIN1 could hinder cell proliferation, migration and invasion (Figure 3K-M). In depth, knockdown of PIN1 in cervical cancer cells could induce cell cycle arrest (Supplementary Figure 1F), and boost apoptosis (Figure 3N-O). In sum, PIN1 that serves as a tumor-accelerator is the downstream of FBXL19-AS1/miR-193a-5p signaling in cervical cancer.
FBXL19-AS1 Accelerates Cervical Cancer Progression via Regulating PIN1
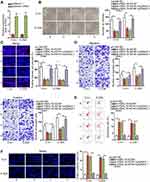
In order to examine whether FBXL19-AS1 accelerated cervical cancer progression via regulating PIN1, we conducted following rescue experiments. Firstly, we overexpressed PIN1 in C-4-I and C-33A cells and tested the overexpression efficiency of PIN1 through RT-qPCR (Figure 4A). Then, we implemented colony formation and EdU experiments to evaluate the influence of overexpressed PIN1 on the proliferation of FBXL19-AS1-silenced cells. The outcomes indicated that cell proliferation restrained by silenced FBXL19-AS1 was then reversed by overexpression of PIN1 (Figure 4B-C). Also, transwell experiments were carried out and the outcomes demonstrated that the inhibited cell migration and invasion caused by the lack of FBXL19-AS1 could be recovered when PIN1 was upregulated (Figure 4D). Additionally, we revealed that cell cycle arrested by depleted FBXL19-AS1 was propelled under PIN1 elevation (Supplementary Figure 1G). Furthermore, it was proved that the promoting influence of FBXL19-AS1suppression on cell apoptosis was offset when augmenting PIN1(Figure 4E-F). Taken together,FBXL19-AS1 accelerates cervical cancer progression via regulating PIN1.
Discussion
Cervical cancer is one of the most common gynecological malignant tumors, and it is a major hidden danger to women’s health in the world. So far, a large number of researches have confirmed that assorted cancers are associated with the maladjusted lncRNAs, which means that the variation on the expression of lncRNAs influences the process of tumor formation. Similarly, cervical cancer, as a cancer with high incidence, has been discovered to be tightly associated with lncRNAs by a crowd of scholars. For instance, GAS5 restrains the development of cervical cancer through sponging miR-196a and miR-205.16 PVT1 epigenetically silences miR-195 and regulates EMT in cervical cancer.17 Also, SNHG20 strengthens the proliferative capability of cervical cancer cells by miR-140-5p/ADAM10 axis.18 Although lots of lncRNAs have been searched in cervical cancer, the functions of many other lncRNAs in cervical cancer still need to be further unearthed. FBXL19-AS1 has been unveiled to be highly expressed in osteosarcoma cells and to facilitate malignancy in this cancer.13 Nevertheless, the specific function and mechanism of FBXL19-AS1 in cervical cancer are still in an unknown state. In our research, we discovered that the expression of FBXL19-AS1 was singularly strong in cervical cancer tissues and cells. In recent years, lncRNAs have been reported to be regulated at transcriptional and post-transcriptional levels.19 Further, FBXL19-AS1 was proved to be post-transcriptionally stabilized by LIN28A in breast cancer.20 Nevertheless, the mechanism whereby FBXL19-AS1 was upregulated in cervical cancer remains unknown, and this needs to be further focused on in future studies.
Thereupon we took the functional experiments to search the influence of FBXL19-AS1 silence on the behaviors of cervical cancer cells. Interestingly, we discovered that cervical cancer cell proliferation could be restrained by the lack of FBXL19-AS1, which was proved to be attributed to arrested cell cycle and accelerated cell apoptosis under the context of FBXL19-AS1 inhibition. Meanwhile, we also elucidated that cell migration and invasion capabilities were suppressed when FBXL19-AS1 was subjected to inhibition. Consequently, we deduced that FBXL19-AS1 exerted a carcinogenic function in cervical cancer.
In recent years, lncRNAs have been proposed to serve as the sponge of miRNAs, so as to release the controlled mRNAs.21 For example, HOXD-AS1acts as a ceRNA for SOX4 to accelerate the development of liver cancer.22 Besides, UICLM expedites colorectal cancer progression through serving as a sponge for miR-215 to boost ZEB2 expression.23 Importantly, whether certain lncRNA could function as a ceRNA primarily depends on its cellular position. In other words, nuclear lncRNAs always regulate pre-transcription or transcription, while cytoplasmic lncRNAs often function as a ceRNA by sponging miRNAs to indirectly regulate target mRNA expression at the post-transcriptional level. In our research, we found that the distribution of FBXL19-AS1 was mostly in the cytoplasm of cervical cancer cells. Consequently, we conjectured that FBXL19-AS1 worked by the ceRNA mechanism. Hence, we discovered miR-193a-5p as the downstream sponged by FBXL19-AS1 in cervical cancer. Further, miR-193a-5p was indicated to hamper cell proliferation, migration and invasion in cervical cancer, consistently as it did in other cancer types like colon cancer,24 osteosarcoma,25 and gastric cancer.26
Subsequently, we discovered that PIN1 was the target of miR-193a-5p. PIN1 has been reported to expedite the progression of hepatocellular carcinoma,27 esophageal squamous-cell carcinoma28 and prostate cancer.29 In our study, we proved that PIN1 was the downstream of FBXL19-AS1/miR-193a-5p pathway in cervical cancer. Besides, silencing PIN1 mitigated the oncogenic behaviors of cervical cancer cells, and the findings supporting PIN1 as a cancer-promoter in this tumor was consistent with a formerstudy.30 Moreover, rescue experiments demonstrated that overexpressing PIN1 could reverse the influences of silenced FBXL19-AS1on cervical cancer cellular functions. Previously, PIN1 has been suggested to have roles in cell cycle control,31 regulation of PI3K/AKT and Wnt/β-catenin pathways,32 or modulation of Hippo signaling.33 More importantly, several reports revealed that PIN1 also affects cyclin D1 expression and Wnt/β-catenin pathway in cervical cancer,34,35 which might be the downstream mechanism underlying the contribution of FBXL19-AS1/miR-193a-5p/PIN1 axis to cervical cancer development.
Conclusions
All in all, our research unmasked that FBXL19-AS1 accelerates cell growth, migration and invasion in cervical cancer by sponging miR-193a-5p to augment PIN1, which provides a new direction for treating patients with cervical cancer.
Data Sharing Statement
Research data have been presented within this manuscript and the Additional file.
Ethics Approval and Informed Consent
This work was conducted under the support of the Ethics Committee of Yijishan Hospital of Wannan Medical College. All cervical cancer patients enrolled in signed the informed consents before this research.
Acknowledgments
We thank each partner in our team and the kind people who gave us a hand during this work.
Author Contributions
Conceptualization: Su Wan
Methodology: Guantai Ni
Software: Yuansheng Huang
Validation: Jin Ding
Formal Analysis: Su Wan
Investigation: Jin Ding
Data Curation: Guantai Ni
Writing – Original Draft Preparation: Su Wan
Writing – Review & Editing: Yuansheng Huang
Visualization: Guantai Ni
Supervision: Yuansheng Huang
Project Administration: Su Wan.
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
2. Small W
3. Galic V, Herzog TJ, Lewin SN, et al. Prognostic significance of adenocarcinoma histology in women with cervical cancer. Gynecol Oncol. 2012;125(2):287–291. doi:10.1016/j.ygyno.2012.01.012
4. Lee YY, Choi CH, Kim TJ, et al. A comparison of pure adenocarcinoma and squamous cell carcinoma of the cervix after radical hysterectomy in stage IB-IIA. Gynecol Oncol. 2011;120(3):439–443. doi:10.1016/j.ygyno.2010.11.022
5. Saei Ghare Naz M, Kariman N, Ebadi A, Ozgoli G, Ghasemi V, Rashidi Fakari F. Educational interventions for cervical cancer screening behavior of women: a systematic review. Asian Pac J Cancer Prev. 2018;19(4):875–884.
6. Park JY, Kim DY, Kim JH, Kim YM, Kim YT, Nam JH. Outcomes after radical hysterectomy in patients with early-stage adenocarcinoma of uterine cervix. Br J Cancer. 2010;102(12):1692–1698. doi:10.1038/sj.bjc.6605705
7. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi:10.1038/nature10887
8. Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. doi:10.1073/pnas.0904715106
9. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
10. Enfield KS, Pikor LA, Martinez VD, Lam WL. Mechanistic roles of noncoding RNAs in lung cancer biology and their clinical implications. Genet Res Int. 2012;2012:737416. doi:10.1155/2012/737416
11. Li T, Chen Y, Zhang J, Liu S. LncRNA TUG1 promotes cells proliferation and inhibits cells apoptosis through regulating AURKA in epithelial ovarian cancer cells. Medicine. 2018;97(36):e12131. doi:10.1097/MD.0000000000012131
12. Chen J, Yu Y, Li H, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 2019;18(1):33. doi:10.1186/s12943-019-0947-9
13. Pan R, He Z, Ruan W, et al. lncRNA FBXL19-AS1 regulates osteosarcoma cell proliferation, migration and invasion by sponging miR-346. Onco Targets Ther. 2018;11:8409–8420. doi:10.2147/OTT.S160963
14. Yu DJ, Li YH, Zhong M. LncRNA FBXL19-AS1 promotes proliferation and metastasis via regulating epithelial-mesenchymal transition in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2019;23(11):4800–4806.
15. Ding Z, Ye P, Yang X, Cai H. LncRNA FBXL19-AS1 promotes breast cancer cells proliferation and invasion via acting as a molecular sponge to miR-718. Biosci Rep. 2019;39:4. doi:10.1042/BSR20182018
16. Yang W, Hong L, Xu X, Wang Q, Huang J, Jiang L. LncRNA GAS5 suppresses the tumorigenesis of cervical cancer by downregulating miR-196a and miR-205. Tumour Biol. 2017;39(7):1010428317711315. doi:10.1177/1010428317711315
17. Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J Drug Target. 2017;25(7):637–644. doi:10.1080/1061186X.2017.1307379
18. Guo H, Yang S, Li S, Yan M, Li L, Zhang H. LncRNA SNHG20 promotes cell proliferation and invasion via miR-140-5p-ADAM10 axis in cervical cancer. Biomed Pharmacother. 2018;102:749–757. doi:10.1016/j.biopha.2018.03.024
19. Wu Z, Liu X, Liu L, et al. Regulation of lncRNA expression. Cell Mol Biol Lett. 2014;19(4):561–575. doi:10.2478/s11658-014-0212-6
20. Zhang Y, Xiao X, Zhou W, Hu J, Zhou D. LIN28A-stabilized FBXL19-AS1 promotes breast cancer migration, invasion and EMT by regulating WDR66. In Vitro Cell Dev Biol Anim. 2019;55(6):426–435. doi:10.1007/s11626-019-00361-4
21. Qu J, Li M, Zhong W, Hu C. Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int J Clin Exp Med. 2015;8(10):17110–17116.
22. Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. doi:10.1186/s12943-017-0680-1
23. Chen DL, Lu YX, Zhang JX, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7(19):4836–4849.
24. Shirafkan N, Shomali N, Kazemi T, et al. microRNA-193a-5p inhibits migration of human HT-29 colon cancer cells via suppression of metastasis pathway. J Cell Biochem. 2018.
25. Pu Y, Zhao F, Cai W, Meng X, Li Y, Cai S. MiR-193a-3p and miR-193a-5p suppress the metastasis of human osteosarcoma cells by down-regulating Rab27B and SRR, respectively. Clin Exp Metastasis. 2016;33(4):359–372. doi:10.1007/s10585-016-9783-0
26. Chou NH, Lo YH, Wang KC, Kang CH, Tsai CY, Tsai KW. MiR-193a-5p and −3p play a distinct role in gastric cancer: miR-193a-3p suppresses gastric cancer cell growth by targeting ETS1 and CCND1. Anticancer Res. 2018;38(6):3309–3318. doi:10.21873/anticanres.12596
27. Leong KW, Cheng CW, Wong CM, Ng IO, Kwong YL, Tse E. miR-874-3p is down-regulated in hepatocellular carcinoma and negatively regulates PIN1 expression. Oncotarget. 2017;8(7):11343–11355. doi:10.18632/oncotarget.14526
28. Chen M, Xia Y, Tan Y, Jiang G, Jin H, Chen Y. Downregulation of microRNA-370 in esophageal squamous-cell carcinoma is associated with cancer progression and promotes cancer cell proliferation via upregulating PIN1. Gene. 2018;661:68–77.
29. Lee KH, Lin FC, Hsu TI, et al. MicroRNA-296-5p (miR-296-5p) functions as a tumor suppressor in prostate cancer by directly targeting Pin1. Biochim Biophys Acta. 2014;1843(9):2055–2066. doi:10.1016/j.bbamcr.2014.06.001
30. Ma JQ, Yang Y, Juan J, et al. Over-expression of prolyl isomerase Pin1 promotes cervical tumorigenesis and metastasis. Int J Clin Exp Pathol. 2018;11(2):664–674.
31. Cheng C-W, Tse E. PIN1 in cell cycle control and cancer. Front Pharmacol. 2018;9:1367. doi:10.3389/fphar.2018.01367
32. Zhang Z, Yu W, Zheng M, et al. Pin1 inhibition potently suppresses gastric cancer growth and blocks PI3K/AKT and Wnt/β-catenin oncogenic pathways. Mol Carcinog. 2019;58(8):1450–1464. doi:10.1002/mc.23027
33. Khanal P, Yeung B, Zhao Y, Yang X. Identification of Prolyl isomerase Pin1 as a novel positive regulator of YAP/TAZ in breast cancer cells. Sci Rep. 2019;9(1):6394.
34. Li H, Wang S, Zhu T, et al. Pin1 contributes to cervical tumorigenesis by regulating cyclin D1 expression. Oncol Rep. 2006;16(3):491–496.
35. Wang T, Liu Z, Shi F, Pin WJ. 1 modulates chemo-resistance by up-regulating FoxM1 and the involvements of Wnt/β-catenin signaling pathway in cervical cancer. Mol Cell Biochem. 2016;413(1):179–187. doi:10.1007/s11010-015-2651-4
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.