Back to Journals » Cancer Management and Research » Volume 12
Long Non-Coding RNA SNHG14 Impedes Viability, Migration and Invasion of Endometrial Carcinoma Cells Through Modulating miR-93-5p/ZBTB7A Axis
Authors Zhang K, Cai Y, Zhou Q, Sun H, Wei J
Received 8 April 2020
Accepted for publication 5 August 2020
Published 1 October 2020 Volume 2020:12 Pages 9515—9525
DOI https://doi.org/10.2147/CMAR.S257419
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Kai Zhang,1,* Yongqin Cai,1,* Qi Zhou,1 Hong Sun,1 Jinying Wei2
1No. 2 Disease Area of Gynaecology, Weifang Yidu Central Hospital, Qingzhou City, Shandong Province 262500, People’s Republic of China; 2No. 1 District of Gynecology, Weifang People’s Hospital, Weifang City, Shandong Province 261041, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jinying Wei Tel +86-18863601086
Email [email protected]
Background: The function of long non-coding RNA small nucleolar RNA host gene 14 (SNHG14) in endometrial carcinoma (EC) has not been thoroughly reported. This research is designed to research the action mechanism of SNHG14 in EC development.
Methods: The expression of SNHG14 was estimated in The Cancer Genome Atlas and was verified by qRT-PCR in EC tissues. The correlation between SNHG14 expression and clinicopathological features of EC patients was analyzed. Cell viability, wound healing rate, and relative invasion rate were examined by MTT, wound healing, and transwell assay. StarBase, TargetScan, RNA pull-down, and dual luciferase reporter gene (DLR) assay were conducted to analyze the relationship among SNHG14, miR-93-5p and ZBTB7A.
Results: SNHG14 was underexpressed in EC. SNHG14 expression was significantly relevant to menstruation, FIGO stage, histological grade and lymphatic metastasis of EC patients. SNHG14 overexpression hampered viability, migration and invasion of EC cells. SNHG14 functioned as a sponge for miR-93-5p, and miR-93-5p inhibition restrained cell viability, migration and invasion in EC. In addition, miR-93-5p directly targeted to ZBTB7A, which was underexpressed in EC. The suppressive action of SNHG14 overexpression on the viability, migration and invasion of EC cells was partly rescued by miR-93-5p overexpression or ZBTB7A silencing.
Conclusion: LncRNA SNHG14 hampered the viability, migration and invasion of EC cells via modulating miR-93-5p/ZBTB7A axis.
Keywords: endometrial carcinoma, invasion, SNHG14, migration, miR-93-5p, ZBTB7A
Introduction
Endometrial carcinoma (EC) is the most common reproductive tract malignancy in women, with an increased incidence in younger women.1 The 5-year survival rate is 90% for EC patients in different stages, and less than 20% for advanced EC patients with distant metastasis. The primary treatment of EC is surgery, especially lymphadenectomy, whereas with unsatisfactory effectiveness.2 Immunotherapy basically consists in stimulating the endogenous immune response specifically against tumor cells and seems the new frontier of the anticancer treatment.3 Recent reportable data have shown that immune checkpoint inhibitors are potentially promising in the treatment of EC.4 The identification of genetic alterations that have a major role in tumorigenesis is leading to the development of new therapeutic options for immunotherapy. Understanding EC at the molecular level and selecting remarkable EC-related biomarkers play important roles in the diagnosis and treatment of EC, and finally improve the survival and cure rate of EC patients.5
Many long non-coding RNAs (lncRNAs), such as small nucleolar RNA host gene 12 (ASLNC04080),6 H19,7 and maternally expressed gene 3 (MEG3),8 have been used as therapeutic biomarkers for EC. LncRNA CCAT2 is found to be overexpressed in EC tissues, and CCAT2 silencing hampers the growth and metastasis of EC cells via targeting miR-216b.9 Overexpression of MEG3 restrains the proliferation, invasion and metastasis, while enhances apoptosis of EC cells through inactivating phosphoinositide 3-kinase (PI3K)/m-TOR pathway.8 LncRNA small nucleolar RNA host gene 14 (SNHG14) has been well researched in many human cancer types, either acts as oncogene or tumor suppressor gene in tumorigenesis of cancers. For instance, SNHG14 promotes the tumorigenesis of patients with breast cancer (BC), and shows potentials as diagnostic and therapeutic target for BC.10 Besides, SNHG14 upregulation contributes to the development of gastric cancer via regulating miR-145/SOX9 axis.11 Conversely, SNHG14 exhibits an anti-cancer function in glioma via sponging miR-92a-3p.12 However, the role of SNHG14 in EC has not been thoroughly reported.
MicroRNAs (miRNAs) are identified to exert critical functions in diverse biological processes involved in cancer progression.13,14 MiR-93-5p is characterized to play oncogenic effect in diverse types of cancer.15 For instance, miR-93-5p contributes to the tumorigenesis of gastric cancer via blocking the Hippo signaling pathway.16 Additionally, miR-93-5p acts as an important oncogene in non-small cell lung cancer (NSCLC) via targeting PTEN and RB1.17 Recently, lncRNAs could compete with endogenous RNAs (ceRNAs) and sponge miRNAs to regulate the expression of target mRNAs involved in a variety of human cancers.18,19 LncRNA LINC00472 hampers the progression of hepatocellular carcinoma via modulating miR-93-5p/PDCD4 pathway.20 LncRNA H19 serves as an oncogenic function in breast cancer by sponging miR-93-5p to regulating STAT3.21 Nevertheless, the action mechanisms between SNHG14 and miR-93-5p in EC remain elusive.
In this research, we proposed to investigate the mechanism of SNHG14 involved in EC development. Thus, we measured SNHG14 expression in EC tissues and cells, then detected the action of SNHG14 on the progression of EC. Besides, we investigated the correlation among SNHG14, miR-93-5p and Zinc finger and BTB domain containing 7A (ZBTB7A). Our findings demonstrated that SNHG14 might function as a promising therapeutic target for EC.
Methods
Patient Tissue Samples
Fifty-three paired EC tissues and adjacent normal tissues were collected from EC patients via surgical resection at our hospital from 2017 March to 2018 July. EC tissues were histopathologically confirmed. No patient had received preoperative chemotherapy or radiotherapy before tissue collection. Written informed consents were acquired from each patient. This research was permitted by the Ethics Committee of Weifang Yidu Central Hospital.
Cell Culture and Transfection
Human embryonic stem cell (ESC) and EC cell lines (HEC1-A, HEC1-B, AN3CA, Ishikawa) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). ESC cells were maintained in Gibco Essential 8 medium (Thermo Fisher Scientific, Rockford, IL, USA) without fetal bovine serum (FBS). EC cell lines were incubated in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% FBS. All cells were cultured in a humidified 37°C incubator with 5% CO2.
pcDNA-SNHG14 and corresponding control (pcDNA3.1-NC) were constructed in our laboratory. miR-93-5p mimics, miR-93-5p inhibitor, negative control (miR-NC) and si-ZBTB7A were purchased from GenePharma (Shanghai, China). AN3CA and Ishikawa cells were transfected with the above oligonucleotides or plasmids using Lipofectamine 3000 (L3000015, Thermo Fisher Scientific). After transfected for 48h, the cells were harvested for the follow-up assays.
MTT Assay
AN3CA and Ishikawa cells were planted into 96-well plates, and cultured with 20 μL MTT (0.5mg/mL) at 24, 48, 72 h. Then, the precipitated formazan was dissolved in 150 µL of dimethyl sulfoxide (DMSO). The OD values at 450 nm (A450) were assessed using a microplate reader (Bio-Rad, CA, USA).
Wound Healing Assay
AN3CA and Ishikawa cells were seeded in 12-well plates. When cells reached almost 100% confluence, an artificial scratch was created using pipette tip. Then the cells were cultured for 24 h. Finally, photomicrographs of the scratch wounds were captured. The wound healing rate was examined using Image J software.
Transwell Assay
AN3CA and Ishikawa cells were plated in the top chamber coated with 100 μL of Matrigel Matrix. A total of 900 μL of Eagle medium supplemented with 10% FBS was added to the bottom chamber. After incubated for 12 h, the invasive cells were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet for 15 min. Positive stained cells at 6 random fields were pictured under a microscope (Olympus Ckx53).
qRT-PCR
TRIzolTM Plus RNA Isolation Reagents (Invitrogen, Carlsbad, CA, USA) was utilized to extract total RNA from tissues and cells. RNA reverse transcription was performed using reverse transcription kit (Takara, Otsu, Japan). qRT-PCR was carried out using SYBR Premix Ex Taq (Takara) on ABI 7500HT Fast Real-Time PCR System (Applied Biosystems, CA, USA). The amplification program was as follows: 95°C for 3 min, 40 cycles of 95°C for 15 s, 60°C for 30 s and 72°C for 20 s. 2−ΔΔCt method was utilized to calculate the mRNA expression level. The primer sequences were as follows: SNHG14, forward: 5ʹ-AAGGTGGGGTAAGCACACTG-3ʹ and reverse: 5ʹ-CCGAACAAGTGTCCAGGAAT-3ʹ; miR-93-5p, forward: 5ʹ-TCTACAGTGCACGTGTCTCCAG-3ʹ, reverse: 5ʹ-ACCTGCGTAGGTAGTTTCATGT-3ʹ; ZBTB7A, forward: 5ʹ-ATCTGCGAGAAGGTCATCCA-3ʹ, reverse: 5ʹ-CAGCAGCTGTCGCACTGGTA-3ʹ; GAPDH: forward: 5ʹ-GACGGCCGCATCTTCTTGT-3ʹ and reverse: 5ʹ-CACACCGACCTTCACCATTTT-3ʹ; U6, forward:5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ and reverse: 5ʹ-CGCTTCACGAATTTG CGTGTCAT −3ʹ.
Dual Luciferase Reporter (DLR) Gene Assay
According to the predication of StarBase or TargetScan, constructs of the 3ʹ-UTR region fragments of SNHG14 and ZBTB7A interacting with miR-93-5p in pGL3 promoter vectors (Promega, Madison, WI) were generated. Then the recombinant constructs were co-transfected with miR-93-5p mimics or mimics NC into Ishikawa or AN3CA cells with Lipofectamine 3000. At 48 h post-transfection, the luciferase activity was measured by luciferase reporter assay system (YPHBIO, Beijing, China).
RNA Pull-Down Assay
Biotin RNA Labeling Mix (Roche, Basel, Switzerland) and T7/SP6 RNA Polymerase (Roche) were utilized to perform RNA pull-down test. Briefly, miR-93-5p WT, miR-93-5p MUT and miR-NC (GenePharma) were biotinylated to be Bio-miR-93-5p WT, Bio-miR-93-5-MUT and Bio-NC, then were transfected into AN3CA or Ishikawa cells. After cultured for 48 h, cells were lysed with lysis buffer. The mixture of biotinylated RNA and cell lysates (AN3CA and Ishikawa) was incubated with streptavidin agarose beads (Invitrogen) at 37°C for 1 h. After washing, the biotinylated RNAs were assessed by qRT-PCR.
Western Blot
Total proteins were isolated using RIPA lysis buffer (Elabscience, Wuhan, China). The total protein content was quantified with BCA Protein Assay Kit (Thermo Fisher Scientific). Then, protein samples were separated in 10% SDS-PAGE, and transferred to a polyvinylidene fluoride (PVDF) membrane. After blocking in 5% non-fat milk at 37°C for 45 min, the membrane was incubated with primary antibodies (GAPDH, 1:5000, sc-32233, Santa Cruz Biotechnology, Santa Cruz, CA, USA; ZBTB7A, 1:2000, ab175918, Abcam, Cambridge, MA, USA) at 4°C overnight. Then, the membrane was washed with Tris-buffered saline (TBST), and incubated with corresponding secondary antibodies (1:2000, sc-516102, Santa Cruz Biotechnology) for 2 h. The protein bands were visualized using enhanced chemiluminescence reagent (Santa Cruz Biotechnology). The GAPDH level served as an internal control.
Statistical Analysis
Data were expressed as the mean ± standard deviation (SD) from at least three independent experiments. The consequences were analyzed using SPSS 22.0 statistical software (SPSS Inc., Chicago, IL) and GraphPad Prism 7.0. Difference. The differences in data between two groups were analyzed by Student’s t-test. The One-way ANOVA test followed by Tukey’s post hoc test was utilized to analyze more than two groups. Differences were considered statistically significant at P < 0.05.
Results
LncRNA SNHG14 Was Underexpressed in EC
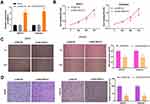
To research the role of SNHG14 in EC, we firstly evaluated SNHG14 expression in The Cancer Genome Atlas (TCGA). As illustrated in Figure 1A, SNHG14 was remarkably downregulated in EC tissues (N = 174) in contrast to normal tissues (N = 91, P < 0.05). To further validate the results obtained from TCGA, qRT-PCR was performed in 53 paired EC tissues and adjacent tissues to examine SNHG14 expression. Consistently, SNHG14 was underexpressed in EC tissues in contrast with adjacent tissues (P < 0.01, Figure 1B). As indicated in Table 1, SNHG14 expression was notably correlated with menstruation, FIGO stage, histological grade and lymphatic metastasis of EC patients (P < 0.05). Kaplan-Meier curve revealed that EC patients with high SNHG14 expression exhibited significant longer overall survival than those with low SNHG14 expression (P = 0.015, Figure 1C). Compared with human embryonic stem cell (ESC), SNHG14 was lowly expressed in EC cell lines (HEC1-A, HEC1-B, AN3CA, Ishikawa) (P < 0.05, Figure 1D). With relatively lower expression of SNHG14, AN3CA and Ishikawa cells were used in subsequent assays.
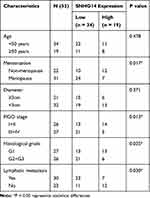 |
Table 1 Correlation Between SNHG14 Expression and Clinicopathological Features in Patients with EC |
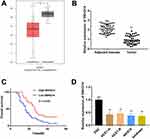 |
Figure 1 LncRNA SNHG14 was underexpressed in endometrial carcinoma (EC) tissues and cells. (A) Relative expression of SNHG14 in The Cancer Genome Atlas (TCGA) (http://gepia.cancer-pku.cn/index.html). *P < 0.05 vs Normal. (B) qRT-PCR was used to examine the relative expression of SNHG14 in EC tissues (Tumor) (n = 53) and adjacent tissues (n = 53). **P < 0.01 vs Adjacent tissue. (C) Kaplan-Meier curve was employed to analyze the correlation between the expression of SNHG14 and overall survival of EC patients. (D) qRT-PCR was used to assess the relative expression of SNHG14 in EC cell lines. **P < 0.01 vs ESC. |
SNHG14 Overexpression Hampered the Viability, Migration and Invasion of EC Cells
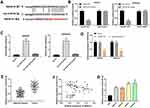
To study the biological effect of SNHG14 in EC, SNHG14 was overexpressed by transfection of pcDNA-SNHG14 into AN3CA and Ishikawa cells. As expected, SNHG14 expression was significantly elevated after transfection of pcDNA-SNHG14 (P < 0.01, Figure 2A). As illustrated in Figure 2B–D, the cell viability, wound healing rate, and relative invasion rate were notably decreased in pcDNA-SNHG14 group in contrast with pcDNA-NC group (P < 0.01).
SNHG14 Acted as a Sponge of miR-93-5p
Using bioinformatics tool StarBase, we discovered that SNHG14 could target miR-93-5p (Figure 3A). DLR assay indicated that miR-93-5p overexpression notably declined the luciferase activity of SNHG14-WT (P < 0.001), while did not influence the luciferase activity of SNHG14-MUT (Figure 3B). The results of RNA pull-down assay confirmed that SNHG14 directly bound with miR-93-5p (P < 0.01, Figure 3C). As revealed in Figure 3D, over expression SNHG14 remarkably declined miR-93-5p expression (P < 0.01). Transfection of pcDNA-NC did not influence miR-93-5p expression. Subsequently, we found that miR-93-5p was dramatically overexpressed in tumor tissues compared with adjacent tissues (P < 0.01, Figure 3E). As demonstrated in Figure 3F, a negative association was observed between SNHG14 and miR-93-5p expression in tumor tissues (r =﹣0.3951, P < 0.01). Additionally, miR-93-5p was upregulated in HEC1-A, HEC1-B, AN3CA and Ishikawa cells in comparison to ESC (P < 0.001, Figure 3G). The above data disclosed that SNHG14 acted as a sponge of miR-93-5p, and miR-93-5p expression was negatively associated with SNHG14 in EC cells.
MiR-93-5p Inhibition Hampered Viability, Migration and Invasion of EC Cells
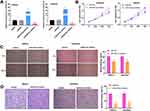
qRT-PCR indicated that miR-93-5p was highly expressed in miR-93-5p mimics group, whereas lowly expressed in miR-93-5p inhibitor group compared to miR-NC group (P < 0.01, Figure 4A). As presented in Figure 4B–D, the cell viability, wound healing rate, and relative invasion rate were all remarkably declined in miR-93-5p inhibitor group in comparison to miR-NC group (P < 0.01).
MiR-93-5p Directly Targeted to ZBTB7A
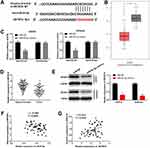
TargetScan predicted a binding site between miR-93-5p and ZBTB7A (Figure 5A). Subsequently, TCGA data indicated that ZBTB7A was remarkably downregulated in EC tissues (N = 174) in contrast to normal tissues (N = 91) (P < 0.05, Figure 5B). As demonstrated in Figure 5C, miR-93-5p mimics notably decreased the relative luciferase activity of ZBTB7A WT, while did not influence the relative luciferase activity of ZBTB7A Mut (P < 0.01). Consistent with the data from TCGA, ZBTB7A expression was dramatically decreased in tumor tissues compared with adjacent tissues (P < 0.01, Figure 5D). The relative protein expression of ZBTB7A was notably decreased by miR-93-5p mimics in AN3CA and Ishikawa cells, while miR-NC did not influence ZBTB7A levels (P < 0.01, Figure 5E). As presented in Figure 5F and G, a negative association was observed between ZBTB7A expression and miR-93-5p (r = −0.3534, P = 0.0093), while a positive association was found between SNHG14 expression and ZBTB7A (r = 0.3313, P = 0.0154). Above all, ZBTB7A was directly targeted to miR-93-5p, and ZBTB7A was lowly expressed in EC.
Overexpression of SNHG14 Impeded Viability, Migration and Invasion of EC Cells by Modulating miR-93-5p/ZBTB7A Axis
To study the regulatory mechanism among SNHG14, miR-93-5p and ZBTB7A, AN3CA cells were co-transfected with pcDNA-SNHG14 and miR-93-5p mimics or si-ZBTB7A. As demonstrated in Figure 6A–C, the cell viability, wound healing rate and relative invasion rate were markedly declined in pcDNA-SNHG14 group compared to pcDNA-NC group (P < 0.01). The inhibitory effect of pcDNA-SNHG14 on the cell viability, wound healing rate and relative invasion rate was partly rescued by miR-93-5p mimics or si-ZBTB7A. Taken together, overexpression of SNHG14 impeded cell viability, migration and invasion in EC via miR-93-5p/ZBTB7A axis.
Discussion
In recent years, lncRNAs are reported to be closely correlated with human cancers, and become one of the hotspots in cancer research.22 Among them, SNHG14 is frequently dysregulated in various human tumors. For instance, SNHG14 expression is remarkably enhanced in ovarian cancer tissues, and silenced SNHG14 hampers cell proliferation, migration and invasion in ovarian cancer.23 Liu et al11 have demonstrated that SNHG14 might act as an oncogenic role in gastric cancer. In contrast with the above studies, Zhang et al24 have illustrated that SNHG14 is markedly downregulated in colorectal cancer (CRC) tissues, and overexpression of SNHG14 restrains the proliferation, migration, and invasion of CRC cells. Wang et al12 have disclosed that SNHG14 is lowly expressed in glioma, and overexpressed SNHG14 hampers proliferation and invasion, and enhances apoptosis of glioma cells. Similar with the previous studies, our findings revealed that SNHG14 was remarkably downregulated in EC, and SNHG14 overexpression hampered the viability, migration and invasion of EC cells, which suggested that SNHG14 might act as an anti-tumor role in EC progression. Moreover, Zhang et al24 also indicated that high expressed SNHG14 indicates a better overall survival of CRC patients by Kaplan-Meier curve. Similarly, we discovered that EC patients with higher SNHG14 expression had a longer overall survival, which indicated that SNHG14 might function as a biomarker for EC prognosis.
Mounting researches show that lncRNAs compete with endogenous RNAs (ceRNAs) via targeting miRNAs to modulate the development of cancers, including EC.9,25,26 For example, the upregulated lncRNA MIR22 host gene remarkably promotes the cell apoptosis and suppresses cell proliferation through regulating miR-141-3p/DAPK1 axis in EC.27 LncRNA TUG1 is overexpressed in EC tissues, and promotes the EC evolution and progression by suppressing miR-299 and miR-34a-5p.28 Here, we validated that SNHG14 acted as a sponge for miR-93-5p. MiR-93-5p is identified to play a regulatory role in the progression of various tumors. For example, miR-93-5p is reported to be overexpressed in cervical cancer, and contributes to the proliferation and metastasis of cervical cancer cells.29 MiR-93-5p is highly expressed in NSCLC, and miR-93-5p silencing impedes the proliferation, migration and invasion of NSCLC cells.17 Notably, Chen et al30 have demonstrated that miR-93 is highly expressed in EC tissues, and overexpressed miR-93 facilitates the proliferation, migration and invasion of EC cells. Consistently, we detected that miR-93-5p was markedly upregulated in EC tissues, and miR-93-5p expression was negatively associated with SNHG14 in EC cells. Further functional experiments indicated that miR-93-5p inhibition hampered the viability, migration and invasion of EC cells. Our findings indicated that SNHG14 might exert its anti-tumor effect on EC progression via functioning as a sponge inhibiting miR-93-5p.
To explore the mechanism between SNHG14 and miR-93-5p in EC, we focused on the downstream target of miR-93-5p. ZBTB7A exerts a crucial role in the tumorigenesis of human malignancies, and the promotion or inhibition effect is associate with tumor type. ZBTB7A is overexpressed in breast cancer tissues, and facilitates the progression of breast cancer.31 Conversely, ZBTB7A is underexpressed in melanoma, and restrains adhesion, invasion of melanoma cells via inhibiting MCAM.32 Similarly, we detected that ZBTB7A was downregulated in EC, and further ascertained that ZBTB7A was a direct target of miR-93-5p. Previous studies reveal that ZBTB7A acts as target gene for different miRNAs in various cancer types. For instance, recent studies have revealed that ZBTB7A serves as a functional target for miR-106b in hepatocellular carcinoma and ovarian carcinoma.33,34 Zhu et al35 have illustrated that miR-520e impedes cell growth, invasion and migration in NSCLC by targeting ZBTB7A. Furthermore, we demonstrated that the inhibiting action of SNHG14 overexpression on viability, migration and invasion of EC cells was partially rescued by miR-93-5p upregulation or ZBTB7A inhibition, suggesting that SNHG14 may exert an anti-tumor role in EC progression via regulating miR-93-5p/ZBTB7A axis.
Conclusion
In conclusion, SNHG14 was underexpressed in EC, and overexpressed SNHG14 hampered viability, migration and invasion of EC cells. SNHG14 served as a sponge for miR-93-5p. Furthermore, ZBTB7A was directly targeted to miR-93-5p. Overexpression of SNHG14 impeded viability, migration and invasion of EC cells by modulating miR-93-5p/ZBTB7A axis. Our findings demonstrated that SNHG14 might act as an anti-tumor role in EC progression, and may be utilized as a novel therapeutic target for EC treatment.
Data Sharing Statement
All data in this study may be obtained from the corresponding author upon reasonable request.
Ethics Approval and Informed Consent
Written informed consents were acquired from each patient. This research was permitted by the Ethics Committee of Weifang Yidu Central Hospital.
Consent for Publication
All patients agreed to the study being published.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Schouten LJ, Goldbohm RA, Van Den Brandt PA. Anthropometry, physical activity, and endometrial cancer risk: results from the Netherlands Cohort Study. J Natl Cancer Inst. 2004;96(21):1635–1638.
2. Wright JD, Medel NIB, Sehouli J, Fujiwara K, Herzog TJ. Contemporary management of endometrial cancer. Lancet. 2012;379(9823):1352–1360.
3. Di Tucci C, Capone C, Galati G, et al. Immunotherapy in endometrial cancer: new scenarios on the horizon. J Gynecol Oncol. 2019;30(3):e46.
4. Makker V, Green AK, Wenham RM, Mutch D, Davidson B, Miller DS. New therapies for advanced, recurrent, and metastatic endometrial cancers. Gynecol Oncol Res Pract. 2017;4:19.
5. Yang L, Zhang J, Jiang A, et al. Expression profile of long non-coding RNAs is altered in endometrial cancer. Int J Clin Exp Med. 2015;8(4):5010–5021.
6. Wen Z, Xu L, Shouzhen W, Yan Z, Huan P, Wei C. Microarray expression profile of lncRNAs and the upregulated ASLNC04080 lncRNA in human endometrial carcinoma. Int J Oncol. 2015;46(5):2125–2137.
7. Zhang L, Wang DL, Yu P. LncRNA H19 regulates the expression of its target gene HOXA10 in endometrial carcinoma through competing with miR-612. Eur Rev Med Pharmacol Sci. 2018;22(15):4820–4827.
8. Sun KX, Wu DD, Chen S, Zhao Y, Zong ZH. LncRNA MEG3 inhibit endometrial carcinoma tumorigenesis and progression through PI3K pathway. Apoptosis. 2017;22(12):1543–1552.
9. Xie P, Cao H, Li Y, Wang J, Cui Z. Knockdown of lncRNA CCAT2 inhibits endometrial cancer cells growth and metastasis via sponging miR-216b. Cancer Biomarkers. 2017;21(1):123–133.
10. Dong H, Wang W, Mo S, et al. Long non-coding RNA SNHG14 induces trastuzumab resistance of breast cancer via regulating PABPC1 expression through H3K27 acetylation. J Cell Mol Med. 2018;22(10):4935–4947.
11. Liu Z, Yan Y, Cao S, Chen Y. Long non-coding RNA SNHG14 contributes to gastric cancer development through targeting miR-145/SOX9 axis. J Cell Biochem. 2018;119(8):6905–6913.
12. Wang Q, Teng Y, Wang R, et al. The long non-coding RNA SNHG14 inhibits cell proliferation and invasion and promotes apoptosis by sponging miR-92a-3p in glioma. Oncotarget. 2018;9(15):12112–12124.
13. Esquela-Kerscher A, Slack FJ. Oncomirs — microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269.
14. Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol. 2007;171(3):728–738.
15. Wang CZ, Deng F, Li H, et al. MiR-101: a potential therapeutic target of cancers. Am J Transl Res. 2018;10(11):3310–3321.
16. Li L, Zhao J, Huang S, et al. MiR-93-5p promotes gastric cancer-cell progression via inactivation of the Hippo signaling pathway. Gene. 2018;641:240–247.
17. Yang W, Bai J, Liu D, et al. MiR-93-5p up-regulation is involved in non-small cell lung cancer cells proliferation and migration and poor prognosis. Gene. 2018;647:13–20.
18. Chen SC, Xiao BX, Guo JM. Roles of interaction between miRNA and lncRNA in tumorigenesis. Chin J Biochem Mol Biol. 2014.
19. Chaofeng TU. The interaction between lncRNA and microRNA contributes to tumor. Onco Targets Ther. 2013;29(11):1029–1034.
20. Chen C, Zheng Q, Kang W, Yu C. Long non-coding RNA LINC00472 suppresses hepatocellular carcinoma cell proliferation, migration and invasion through miR-93-5p/PDCD4 pathway. Clin Res Hepatol Gastroenterol. 2019;43(4):436–445.
21. Li J-P, Xiang Y, Fan L-J, Yao A, Li H, Liao X-H. Long noncoding RNA H19 competitively binds miR-93-5p to regulate STAT3 expression in breast cancer. J Cell Biochem. 2019;120(3):3137–3148.
22. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981.
23. Li L, Zhang R, Li S-J. Long noncoding RNA SNHG14 promotes ovarian cancer cell proliferation and metastasis via sponging miR-219a-5p. Eur Rev Med Pharmacol Sci. 2019;23(10):4136–4142.
24. Zhang W, Duan W, Mo Z, et al. Upregulation of SNHG14 suppresses cell proliferation and metastasis of colorectal cancer by targeting miR-92b-3p. J Cell Biochem. 2020;121(2):1998–2008.
25. Ke J, Shen Z, Hu W, et al. LncRNA DCST1-AS1 was upregulated in endometrial carcinoma and may sponge miR-92a-3p to upregulate Notch1. Cancer Manag Res. 2020;12:1221–1227.
26. Shi F, Wang T, Liu Z, et al. LncRNA miR143HG up-regulates p53 in endometrial carcinoma by sponging miR-125a. Cancer Manag Res. 2019;11:10117–10123.
27. Cui Z, An X, Li J, Liu Q, Liu W. LncRNA MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression and inhibits endometrial carcinoma cells proliferation. Biomed Pharmacother. 2018;104:223–228.
28. Liu L, Chen X, Zhang Y, Hu Y, Shen X, Zhu W. Long non-coding RNA TUG1 promotes endometrial cancer development via inhibiting miR-299 and miR-34a-5p | liu. Oncotarget. 2017;8(19):31386–31394.
29. Sun XY, Han XM, Zhao XL, Cheng XM, Zhang Y. MiR-93-5p promotes cervical cancer progression by targeting THBS2/MMPS signal pathway. Eur Rev Med Pharmacol Sci. 2019;23(12):5113–5121.
30. Chen S, Xi C, Kai-Xuan S, et al. MicroRNA-93 promotes epithelial–mesenchymal transition of endometrial carcinoma cells. PLoS One. 2016;11(11):e0165776.
31. Mao A, Chen M, Qin Q, et al. ZBTB7A promotes migration, invasion and metastasis of human breast cancer cells through NF-κB-induced epithelial–mesenchymal transition in vitro and in vivo. J Biochem. 2019;166(6):485–493.
32. Liu X-S, Genet MD, Haines JE, et al. ZBTB7A suppresses melanoma metastasis by transcriptionally repressing MCAM. Mol Cancer Res. 2015;13(8):1206–1217.
33. Liang X, Zhao Q, Geng T, Luo S, He Q. MiR-106b regulates the apoptosis and tumorigenesis of hepatocellular carcinoma via targeting Zinc finger and BTB domain-containing protein 7A (Zbtb7a). J Biochem Mol Toxicol. 2018;32(8):e22169.
34. Yue R, Chen Y, Lai W, Wei W. miR-106b exerts tumor suppressive functions in ovarian carcinoma by directly targeting ZBTB7A. Minerva Med. 2020;12(10):06475–06477.
35. Zhijun Z, Jingkang H. MicroRNA-520e suppresses non-small-cell lung cancer cell growth by targeting Zbtb7a-mediated Wnt signaling pathway. Biochem Biophys Res Commun. 2017;486(1):49–56.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.