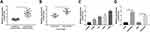Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA NNT-AS1 Contributes to Cisplatin Resistance via miR-1236-3p/ATG7 Axis in Lung Cancer Cells
Authors Wang H, Guo M, Ding D, Yang F, Chen Z
Received 6 November 2019
Accepted for publication 5 April 2020
Published 30 April 2020 Volume 2020:13 Pages 3641—3652
DOI https://doi.org/10.2147/OTT.S237576
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Haifeng Wang,1 Min Guo,2 Dongxiao Ding,3 Feng Yang,4 Zhongjie Chen3
1Department of Hematology and Oncology, Beilun Branch of the First Affiliated Hospital of Medical College of Zhejiang University, Ningbo, Zhejiang, People’s Republic of China; 2Department of Respiratory Medicine, Ningbo Zhenhai Longsai Hospital, Ningbo, Zhejiang, People’s Republic of China; 3Department of Thoracic Surgery, Beilun Branch of the First Affiliated Hospital of Medical College of Zhejiang University, Ningbo, Zhejiang, People’s Republic of China; 4Department of Respiratory Medicine, Beilun Branch of the First Affiliated Hospital of Medical College of Zhejiang University, Ningbo, Zhejiang, People’s Republic of China
Correspondence: Zhongjie Chen
Department of Thoracic Surgery, Beilun Branch of the First Affiliated Hospital of Medical College of Zhejiang University, No. 1288, Lusan East Road, Beilun, Ningbo, Zhejiang 315800, People’s Republic of China
Tel +86 574 86776641
Email [email protected]
Purpose: Lung cancer is one of the most prevailing human cancers worldwide. Emerging evidence implies that long non-coding RNA nicotinamide nucleotide transhydrogenase-antisense RNA1 (NNT-AS1) is implicated in the tumorigenesis of lung cancer. Herein, we aimed to expose the impact of NNT-AS1 on the drug resistance of lung cancer.
Methods: Levels of NNT-AS1, microRNA (miR)-1236-3p and autophagy-related gene 7 (ATG7) were evaluated using quantitative real-time polymerase chain reaction (qRT-PCR) assay. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was implemented to detect cell proliferation and the half maximal inhibitory concentration (IC50) of cisplatin (DDP) in vitro. Moreover, flow cytometry was performed to assess cell apoptosis. Cell migration and invasion were examined utilizing transwell assay in lung cancer cells. Besides, levels of ATG7 and cell behavior-related proteins were determined via Western blot. Dual-luciferase reporter assay was administrated to identify the interaction between miR-1236-3p and NNT-AS1 or ATG7. The biological role of NNT-AS1 in DDP resistance of lung cancer was examined by xenograft tumor model in vivo.
Results: NNT-AS1 and ATG7 were upregulated, whereas miR-1236-3p was curbed in lung cancer tissues and in with or without DDP-resistant cell lines. NNT-AS1 detection significantly constrained cell growth, metastasis, and the IC50 of DDP in A549/DDP and H522/DDP cells. Interestingly, the influence of miR-1236-3p mimic on DDP resistance was overturned via NNT-AS1 upregulation in vitro. Reintroduction of miR-1236-3p inhibitor relieved the effect of ATG7 silencing on DDP sensitivity in A549/DDP and H522/DDP cells. Importantly, NNT-AS1 was a sponge of miR-1236-3p to separate ATG7. Besides, NNT-AS1 silencing enhanced DDP sensitivity of lung cancer in vivo.
Conclusion: NNT-AS1/miR-1236-3p/ATG7 axis regulated DDP resistance in lung cancer cells and might supply a probable target and prognostic marker in lung cancer treatment.
Keywords: NNT-AS1, miR-1236-3p, ATG7, lung cancer
Introduction
Lung cancer is regarded as one of the most common cancers with high incidence and mortality worldwide.1 Non-small cell lung cancer accounts for about 80% of lung cancers and has a high mortality rate due to the metastasis, recurrence, and drug resistance in lung cancer patients.2 Cisplatin (DDP) has been reported to be connected with nuclear DNA binding,3 however, the efficiency of DDP-based chemotherapy is unsatisfactory owing to the chemo-resistance in patients with lung cancer. Until now, the regulatory mechanism of DDP resistance is still cramped because of the utterly complicated processes. Unfortunately, if DDP resistance occurs, the patients may effortlessly develop cross-resistance to another chemo-drug in the clinic.4 Therefore, it is necessary to shed light on the development of DDP resistance via exploring the interaction between DDP and extensive cytoplasmic or nuclear genes in lung cancer patients.
Currently, long non-coding RNAs (lncRNAs) are recognized as the non-coding transcripts with over than 200 nucleotides in length.5 Accumulating findings demonstrate that lncRNAs can modulate gene expression via epigenetic regulation, transcriptional and post-transcriptional mediation, and the function of lncRNAs in tumor growth and metastasis has been broadly investigated.6–8 Nicotinamide nucleotide transhydrogenase-antisense RNA1 (NNT-AS1) has been manifested to be located in the chromosome 5p12 region with 3304 bp in length.9 Firstly, Wang et al discovered that NNT-AS1 was expressed at a high level in colorectal cancer, and served as an oncogene by reinforcing cell proliferation, migration in colon cancer cells.9 Furthermore, the role of NNT-AS1 in DDP resistance was subsequently uncovered. Emerging evidence showed that the high expression of NNT-AS1 acted as a master mediator for the resistance of DDP via mitogen-activated protein kinase (MAPK)/Slug pathway in lung cancer.10 As mentioned above, we further explored the effect of NNT-AS1 with an ectopic expression on the DDP resistance of lung cancer.
MicroRNAs (miRNAs) are composed of 18–24 nucleotides, which belong to endogenous non-coding transcripts.11 MiRNAs work as the major mediators to regulate the post-transcriptional gene expression.12,13 Growing reports indicate that the dysfunction of miRNAs is involved in cancer pathogenesis and drug resistance in several cancers, such as breast cancer,14 gastric cancer,15 and lung cancer.16 What is more, miR-1236-3p, one of the members of miRNAs, has been proven to be implicated in tumorigenesis, including lung cancer. For example, the upregulation of miR-1236-3p can notably impede cell migration, and invasion, thereby restraining the metastasis of lung cancer cells.17 That is to say, miR-1236-3p may serve as a tumor-suppressive factor in the development of lung cancer. However, the precise effect of miR-1236-3p in cell phenotypes and drug resistance needed to be expounded. Autophagy-related gene 7 (ATG7), a member of ATG family, can regulate the process of autophagy.18 We also explored the molecular function of ATG7 in the resistance of chemotherapy in lung cancer cells induced by DDP.
Herein, we exposed the expression patterns of NNT-AS1, miR-1236-3p, and ATG7 in lung cancer tissues and cell lines. Also, the network between miR-1236-3p and NNT-AS1 or ATG7 was also the objective in the present investigation.
Materials and Methods
Clinical Specimens and Cell Culture
A total of 31 pairs of lung cancer tissues and paracancerous non-tumorous tissues were received from lung cancer patients who underwent surgical resection at Beilun Branch of the First Affiliated Hospital of Medical College of Zhejiang University. The patients who received other treatments were excluded from the present research, and the written informed consents were gained from all donators. The clinical information of the patients included age, gender, tumor size, TNM stage, lymphatic metastasis, vascular invasion and distant metastasis, which are displayed in Table 1. Above all, the study was approved by the Ethics Committee of Beilun Branch of the First Affiliated Hospital of Medical College of Zhejiang University.
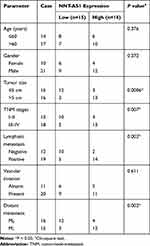 |
Table 1 Correlation Between NNT-AS1 Expression and Clinical Clinicopathological Parameters of the Lung Cancer |
Lung cancer cell lines (H1299, H23, H522, and A549) and Human bronchial epithelioid cells (16-HBE) were purchased from BeNa Culture Collection (Beijing, China). Besides, A549 and H522 cells were induced by DDP with different doses, and the medium gradually increased the DDP concentration until cells could not be tolerated, generating DDP-resistant cells (A549/DDP and H522/DDP). All cells were cultured with Roswell Park Memorial Institute-1640 (RPMI-1640; Gibco, Carlsbad, CA, USA) together with 10% fetal bovine serum (FBS; Gibco), 100 U/mL penicillin (Gibco) and 100 mg/mL streptomycin (Gibco). In the meanwhile, the DDP-resistant cells were supplemented with 1 µg/mL DDP (Sigma-Aldrich, St. Louis, MO, USA) to maintain their drug resistance. The condition for cell incubation was a humidified atmosphere containing 5% CO2 at 37°C.
Transient Transfection
Small interfering RNA (siRNA) especially targeting NNT-AS1 (si-NNT-AS1) or ATG7 (si-ATG7), and siRNA negative control (si-NC) were obtained from KeyGEN Biotech (Jiangsu, China). Besides, the full sequences or designed control of NNT-AS1 and ATG7 were sub-cloned into pcDNA3.1, thereby generating the overexpression vector of NNT-AS1 (NNT-AS1), ATG7 (pc-ATG7) and their controls (vector, pc-NC). Apart from that, miR-1236-3p mimic (miR-1236-3p) and inhibitor (anti-miR-1236-3p), as well as their control (miR-NC and anti-miR-NC) were purchased from GenePharma (Shanghai, China). The reagent of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was applied to transfect vectors or oligonucleotides into cells following the producer’s specifications.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Assay
Total RNA from lung tissues and cells was extracted using Trizol reagent (Invitrogen). Then, PrimeScript RT reagent kit (Takara, Dalian, China) was administrated to synthesize complementary DNA (cDNA). Then, the mixtures containing equal RNA, primers and the reagent of the SYBR Premix Ex Taq™ II kit (Takara) were placed in an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Relative levels of genes were standardized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH; for NNT-AS1 and ATG7) or U6 (for miR-1236-3p) via the 2−ΔΔCt method. The primers were as follows: NNT-AS1 (Forward: 5ʹ-TCTCCTAAGTCGAGGACTAGC-3ʹ, Reverse: 5ʹ-AGGCACTCACTAGCATCACGCT-3ʹ); miR-1236-3p (Forward: 5ʹ-CCAATCAGCCTCTTCCCCTT-3ʹ, Reverse: 5ʹ-TATGGTTGTTCACGACTCCTTCAC-3ʹ); ATG7 (Forward: 5ʹ-CCAGTGACGCCAGATTTCC-3ʹ, Reverse: 5ʹ-GGCAGGCACAGATGCTATG-3ʹ); GAPDH (Forward: 5ʹ-AACGTGTCAGTGGTGGACCTG-3ʹ, Reverse: 5ʹ-AGTGGGTGTCGCTGTTGAAGT-3ʹ); U6 (Forward: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, Reverse: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ).
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
A549/DDP and H522/DDP cells were plated into a 96-well plate at a density of 1×104 cells/well. After incubation overnight, cells were treated with different doses of DDP (Sigma-Aldrich; 0 µg/mL, 0.39 µg/mL, 0.78 µg/mL, 1.56 µg/mL, 3.12 µg/mL, 6.25 µg/mL, 12.5 µg/mL, 25 µg/mL, 50 µg/mL, 100 µg/mL) for 48 h to measure the half maximal inhibitory concentration (IC50) of DDP. Apart from that, the A549/DDP and H522/DDP were incubated for 0 h, 24 h, 48 h, or 72 h to measure cell proliferation. The above cells were supplemented with MTT (Sigma-Aldrich; 5mg/mL), the absorbance of lysates was assessed at 490 nm using a microplate reader. The dose of half (50%) inhibitory in cell viability was adopted to represent the IC50 of DDP in lung cells.
Flow Cytometry Assay
In the assay, Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) reagent kit (BD Biosciences, San Diego, CA, USA) was administrated to detect the apoptotic rate of A549/DDP and H522/DDP cells. Firstly, cells (~5×104 cells) were seeded into a 6-well-plate. The cells were collected and washed using ice-cold phosphate buffer saline (PBS; Gibco) at 48 h post-transfection. Subsequently, Annexin V-FITC and PI were employed to stain re-suspended cells for 15 min as per the manuals. The apoptotic cells were recognized via FACSCalibur flow cytometer (BD Biosciences).
Transwell Assay
Transfected cells were harvested with complete mediums and then plated into the upper chamber pre-coated with Matrigel (for cell invasion) or not (for cell migration). At the same time, the lower chamber was filled with medium containing 10% FBS. After incubation for 48 h, the remaining cells on the upper layer were wiped off using cotton swabs, and the migrated and invaded cells were stained with 0.1% crystal violet (Sigma-Aldrich). Ultimately, the migrated and invaded cells were visualized and photographed utilizing a microscope.
Western Blot Assay
The protocol of the Western blot was as per the previous study.19 Briefly, total protein was separated using gel electrophoresis and transfected onto a Polyvinylidene Fluoride (PVDF; Millipore, Bedford, MA, USA). The special protein could bind to the primary antibody, followed by incubation with corresponding secondary antibodies. The combination was visualized via an enhanced chemiluminescence kit (Millipore). The primary antibodies were as follows: ATG7 (1:30,000, ab133528, Abcam, Cambridge, MA, USA), Cleaved-caspase-3 (C-caspase-3; 1:500, ab13847, Abcam), Ki-67 (1:5000, ab92742, Abcam), MMP-9 (1:1000, ab38898, Abcam), and GAPDH (1:7000, ab8245, Abcam).
Dual-Luciferase Reporter Assay
LncBase Predicted v.2 was applied to predict the common fragments between miR-1236-3p and NNT-AS1, and potential targets of miR-1236-3p were sought via Targetscan, showing that ATG7 was a probable target of miR-1236-3p. Then, the complementary sequences between miR-1236-3p and NNT-AS1 or the 3ʹ Untranslated Region (3ʹUTR) of ATG7 were inserted into basic vector psiCHECK-2 (Promega, Madison, WI, USA) to create wild-type vectors (WT-NNT-AS1 and WT-ATG7). Similarly, the mutant luciferase reporters (MUT-NNT-AS1 and MUT-ATG7) were simultaneously designed and constructed. For cell transfection, A549/DDP and H522/DDP cells were inoculated in triplicate, and co-transfected with wild-type or mutant reporter together with miR-1236-3p or miR-NC utilizing Lipofectamine 2000 (Invitrogen), followed by measurement with firefly and renilla activities adopting Dual-Luciferase Reporter Assay kit (Promega).
Tumor Xenograft Assay
Lentiviral vector (Lenti-short hairpin) sh-NNT-AS1 for stable NNT-AS1 knockdown was purchased from GeneChem (Shanghai, China), and the lentivirus empty vector acted as a control (sh-NC). Male BALB/C nude mice (five-week-old, n=4 per group) were provided by Shanghai Experimental Animal Center (Shanghai, China). A 5×106 A549/DDP cells transfected with sh-NNT-AS1 or sh-NC were subcutaneously injected on the left flank of the nude mice. After 1 week of injection, mice were intraperitoneally administrated with PBS or 3 mg/kg DDP every 7 days. Tumor volume was detected once a week. Twenty-seven days later, the tumor was excised for subsequent weight, followed by measurement for RT-qPCR assay. This study was approved by the Animal Care and Use Committee of the First Affiliated Hospital of Medical College of Zhejiang University.
Statistical Analysis
The data were statistically analyzed by SPSS 13.0 and presented as mean±standard deviation (SD). The difference between two groups or multiple groups was assayed using Student’s t-test or one-way analysis of variance with Dunnett’s post-test. P-values less than 0.05 were regarded as statistically significant.
Results
Level of NNT-AS1 Was Upregulated in Lung Cancer Tissues, Cell Lines and DDP-Resistant Cells
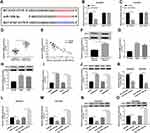
To expose the biological role of NNT-AS1 in the carcinogenesis of lung cancer, we explored the expression level of NNT-AS1 in lung cancer tissues. qRT-PCR analysis indicated that NNT-AS1 was apparently upregulated in tumor tissues as compared with adjacent noncancerous tissues (Figure 1A). Moreover, the expression of NNT-AS1 was notably augmented in advanced lung cancer tissues (III+IV) compared with the lung cancer tissues with the early stages (I+II) (Figure 1B). Meanwhile, our data exhibited that the high expression of NNT-AS1 was significantly related to Tumor size (p=0.0006), advanced TNM stages (p=0.007), lymphatic metastasis (p=0.002) and distant metastasis (p=0.002), implying that NNT-AS1 might affect the clinical prognosis of lung patients. Also, a high expression level of NNT-AS1 was observed in lung cancer cells with DDP resistance or not (Figure 1C and D). The evidence meant that the high expression of NNT-AS1 in DDP-resistant lung cancer cells might go in for the drug resistance in lung cancer cells.
NNT-AS1 Deficiency Could Reinforce DDP Sensitivity in DDP-Resistant Lung Cancer Cells
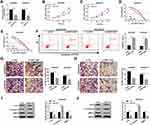
Given the high expression of NNT-AS1 in A549/DDP and H522/DDP cells, the knockdown vector of NNT-AS1 was constructed and transfected into DDP-resistant lung cancer cells, qRT-PCR analysis presented that NNT-AS1 level was specifically downregulated by transfection with si-NNT-AS1 in vitro (Figure 2A). Subsequently, cell proliferation was measured by MTT assay, and the results exhibited that knockdown of NNT-AS1 could markedly block cell proliferation in A549/DDP and H522/DDP cells (Figure 2B and C). Besides, the deficiency of NNT-AS1 could clearly decrease the IC50 of DDP in DDP-resistant lung cancer cells (Figure 2D and E). Next, we further investigated whether the reduction of NNT-AS1 expression could influence cell apoptosis. Flow cytometric analysis suggested that NNT-AS1 detection significantly strengthened cell apoptosis in both A549/DDP and H522/DDP cells (Figure 2F). Simultaneously, the hindered cell migration and invasion were found in DDP-resistant lung cancer cells after transfection with si-NNT-AS1 (Figure 2G and H). Furthermore, cell phenotypes-related proteins were identified by adopting Western blot assay. The decreased Ki-67 indicated the retard of cell proliferation, the improved C-Caspase-3 indicated the enhancement of cell apoptosis, and the reduction of MMP-9 was applied to represent the repression of the mobility in A549/DDP and H522/DDP cells after NNT-AS1 silencing (Figure 2I and J). All the data displayed that NNT-AS1 silencing could retard cell proliferation, migration, invasion, and the IC50 of DDP, and enhance cell apoptosis in A549/DDP and H522/DDP cells.
NNT-AS1 Was a Sponge of miR-1236-3p
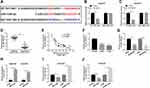
LncBase Predicted v.2 was applied to predict the probable targets of NNT-AS1, the binding sites between miR-1236-3p and NNT-AS1 were found (Figure 3A). Then, dual-luciferase reporter analysis validated that the luciferase of WT-NNT-AS1 was reduced by more than 50% after co-transfection with miR-1236-3p in A549/DDP and H522/DDP cells. However, the luciferase of MUT-NNT-AS1 had no evident alteration after transfection with miR-1236-3p in vitro (Figure 3B and C). As shown in Figure 3D, a low expression of NNT-AS1 was observed in lung cancer tissues in comparison with matched control. Of course, the passive correlation between miR-1236-3p level and NNT-AS1 level was manifested via qRT-PCR analysis (Figure 3E). In addition, we also disclosed that the level of miR-1236-3p was particularly weakened in lung cancer cells no matter with or without DDP resistance (Figure 3F and G). After that, we constructed the overexpression vector of NNT-AS1, the efficiency of NNT-AS1 overexpression is shown in Figure 3H. Lastly, the NNT-AS1 or si-NNT-AS1 was introduced into A549/DDP and H522/DDP cells so as to analyze the expression of miR-1236-3p. Expectantly, the level of miR-1236-3p was decreased in cells transfected with NNT-AS1, and the miR-1236-3p level was increased in cells transfected with si-NNT-AS1 in vitro (Figure 3I and J). These findings confirmed that NNT-AS1 was able to target miR-1236-3p in DDP-resistant lung cancer cells.
The Effect of miR-1236-3p on Cell Phenotypes and DDP Resistance Was Abrogated by NNT-AS1 Overexpression in vitro
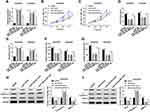
As mentioned above, the relevance between miR-1236-3p and NNT-AS1 was verified at the molecular aspect, rescue assays were conducted to explore the functional mechanism between them. Firstly, miR-1236-3p alone or along with NNT-AS1 was transfected into A549/DDP and H522/DDP cells, and qRT-PCR analysis proved that the acceleratory effect of miR-1236-3p mimic on the level of miR-1236-3p was abrogated via co-transfection with NNT-AS1 in vitro (Figure 4A). Additionally, the inhibitory impact of miR-1236-3p upregulation on cell proliferation in DDP-resistant lung cancer cells was relieved after the synchronous introduction of NNT-AS1 (Figure 4B and C). Similarly, reintroduction of NNT-AS1 could sharply attenuate the repressive role of miR-1236-3p mimic in the IC50 of DDP in both A549/DDP and H522/DDP cells (Figure 4D). Meanwhile, cell apoptosis expedited as a result of miR-1236-3p overexpression, and such promotion could be restored by the co-introduction of NNT-AS1 in both A549/DDP and H522/DDP cells (Figure 4E). As the results of transwell assay, the capacity of the mobility was illustrated at the aspects of cell migration and invasion, reintroduction of NNT-AS1 could remarkably regain the inhibiting influence of miR-1236-3p overexpression on the mobility of DDP-resistant lung cancer cells (Figure 4F and G). The alterations of Ki-67, C-Caspase-3, and MMP-9 expression were also proved the above conclusion about cell behaviors in vitro (Figure 4H and I). According to all the above results, we corroborated that NNT-AS1 upregulation could abolish the effect of miR-1236-3p mimic on DDP resistance in A549/DDP and H522/DDP cells.
MiR-1236-3p Directly Targeted ATG7
To further investigate the role of miR-1236-3p in drug resistance in lung cancer cells, Targetsacn software was employed to predict the possible target of miR-1236-3p and the results are exhibited in Figure 5A. Whereafter, dual-luciferase reporter assay acted as a suitable assistant to verify miR-1236-3p and ATG7 could bind to each other in both A549/DDP and H522/DDP cells. Results of the assays discovered a truth that WT-ATG7 could interact with miR-1236-3p unlike the mutant luciferase reporter (Figure 5B and C). In the meanwhile, ATG7 was expressed at a high level in lung cancer tissues in comparison with matched control (Figure 5D). Simultaneously, qRT-PCR analysis revealed that ATG7 was negatively related to miR-1236-3p in lung cancer tissues (Figure 5E). Also, the mRNA and protein levels of ATG7 were dramatically intensified in lung cancer tissues (Figure 5F). Expectantly, the level of ATG7 was substantially facilitated in lung cancer cells no matter with or without DDP resistance (Figure 5G–J). Subsequently, the efficiency of anti-miR-1236-3p was analyzed, and the level of miR-1236-3p was notably reduced after transfection with anti-miR-1236-3p in vitro (Figure 5K). To further research the relevance between miR-1236-3p and ATG7, miR-1236-3p or anti-miR-1236-3p was transfected into DDP-resistant lung cancer cells, we found that the expression level of ATG7 was opposite with miR-1236-3p at the aspects of mRNA and protein expression (Figure 5L–O). The above descriptions corroborated that ATG7 was a target of miR-1236-3p in DDP-resistant lung cancer cells.
MiR-1236-3p Inhibition Could Overturn the Impact of ATG7 Detection on Cell Behaviors and DDP Resistance in A549/DDP and H522/DDP Cells
Considering the interaction between miR-1236-3p and ATG7, we deeply explored the functional regulation between them. Firstly, A549/DDP and H522/DDP cells were transfected with si-ATG7 or si-ATG7 plus anti-miR-1236-3p, respectively. Seen from Figure 6A–C, the curb impact of ATG7 silencing on the mRNA and protein levels of ATG7 was recovered by miR-1236-3p inhibition in DDP-resistant lung cancer cells. Synchronously, MTT analysis demonstrated that the hindering effect of ATG7 reduction on cell proliferation and the IC50 of DDP was reversed via co-transfection with miR-1236-3p inhibitor in A549/DDP and H522/DDP cells (Figure 6D–F). Moreover, miR-1236-3p repression could overturn the boosting role of ATG7 deficiency in cell apoptosis in vitro (Figure 6G). Besides, transwell analysis exposed that introduction of si-ATG7 could efficiently retard cell migration and invasion, such reduction was rescued by the co-transfection of miR-1236-3p inhibitor in both A549/DDP and H522/DDP cells (Figure 6H and I). Interestingly, the changes of Ki-67, C-Caspase-3, and MMP-9 levels also concluded the interaction between miR-1236-3p and ATG7 in regulating DDP-resistance in vitro (Figure 6I and J). In sum, the miR-1236-3p inhibitor could abrogate the impact of ATG7 silencing on DDP resistance in A549/DDP and H522/DDP cells.
ATG7 Was Co-Regulated by miR-1236-3p and NNT-AS1 in DDP-Resistant Lung Cancer Cells
According to the above instructions, the regulatory mechanism between ATG7 and NNT-AS1 or miR-1236-3p was highlighted. Firstly, si-NC, miR-1236-3p, miR-1236-3p+vector, or miR-1236-3p+NNT-AS1 were introduced into A549 and H522 cells under DDP resistance, respectively. QRT-PCR analysis illustrated that the mRNA level of ATG7 was impeded due to miR-1236-3p upregulation, which was overturned by the synchronous transfection with NNT-AS1 in both A549/DDP and H522/DDP cells (Figure 7A and B). Besides, a similar tendency was observed in the alteration of ATG7 protein expression in vitro (Figure 7C and D). Meanwhile, our data exhibited that NNT-AS1 silencing repressed the ATG7 level, while the upregulation of ATG7 relieved the effect in A549/DDP (Figure S1A) and H522/DDP (Figure S1B) cells. Namely, ATG7 was co-modulated by miR-1236-3p and NNT-AS1 in lung cancer cells under DDP resistance in vitro. In addition, a xenograft tumor mouse model was performed to verify the role of NNT-AS1 on DDP-resistance in vivo. As displayed in Figure S2, tumor volume (Figure S2A) and weight (Figure S2B) were distinctly reduced due to DDP treatment or NNT-AS1 downregulation, suggesting that DDP treatment or NNT-AS1 knockdown inhibited lung tumor growth in vivo. Interestingly, combined sh-NNT-AS1 and DDP resulted in more overt suppression on tumor growth. Meanwhile, our data exhibited that the NNT-AS1 level was remarkably decreased in tumor tissues from the sh-NNT-AS1 group relative to the sh-NC group (Figure S2C). In a word, NNT-AS1 deficiency could enhance DDP sensitivity of lung cancer in vivo.
Discussion
In the paper, we excavated that the expression of NNT-AS1 and ATG7 was specifically enhanced, but the miR-1236-3p level was distinctly hindered in lung cancer tissues and cell lines, as well as DDP-resistant lung cancer cells than matched controls. In addition, the high expression of NNT-AS1 was implicated in the histological grade of lung cancer. Functional analysis demonstrated that NNT-AS1 silencing could remarkably retard the progression, migration, invasion, and the IC50 of DDP, and trigger the apoptosis of DDP-resistant lung cells (A549/DDP and H522/DDP). Subsequently, rescue assays were implemented to verify the interaction between miR-1236-3p and NNT-AS1 or ATG7 in vitro.
NNT-AS1, a newly discovered lncRNA, has been implied to express in obvious tumors aberrantly, and NNT-AS1 with the ectopic expression participates in the pathogenesis and tumorigenesis in human cancers. For instance, the high expression of NNT-AS1 could expedite the progression of cervical cancer via regulating the Wnt/β-catenin signaling pathway, showing as the acceleration of the proliferation and invasion in vitro.20 Besides, the overexpression of NNT-AS1 in osteosarcoma was implicated in tumor growth and poor prognosis in patients.21 It was worth noting that the uncovered lncRNAs were relative to chemo-resistance, including DDP resistance. For example, homeobox transcript antisense intergenic RNA (HOTAIR) deficiency could regain the sensitivity by causing cell-cycle arrest in DDP-resistant lung cells.22 Based on the previous researches, we further investigated the function of NNT-AS1 in chemo-resistance of lung cancer. Functional assays exhibited that NNT-AS1 deficiency could enhance the drug sensitivity in DDP-resistant A549 and H522 cells in vitro, presenting the repression of cell proliferation, migration, invasion and the IC50 of DDP, and the promotion of cell apoptosis in vitro. Furthermore, chemo-resistance of NNT-AS1 was also verified on lung cancer xenografts in nude mice. The above conclusion was in keeping with previous studies.10 The work pathway of NNT-AS1 in mediating DDP resistance was the subsequent gist.
As we all know, lncRNAs participate in the development, pathogenesis and metastasis of diverse human carcinomas.23 More importantly, lncRNAs act as the sponges of miRNAs to modulate tumorigenesis.24 miRNA expression is extremely specific in tumors, and many miRNAs function as oncogenic or tumor-suppressive factors.25 MiR-1236-3p, one of the common miRNAs, was manifested to be downregulated in lung cancer tissues and DDP-resistant cells in our study. Not only that, subsequent assays elucidated that miR-1236-3p was a target of NNT-AS1, the role of miR-1236-3p mimic on cell phenotypes was relieved via co-transfection with NNT-AS1 in A549/DDP and H522/DDP cells. Emerging findings also indicated that miR-1236-3p was connected with the DDP resistance of lung cancer cells.26 Also, miR-1236-3p exerted its tumor-suppressive role by targeting Krueppel-like factor 8 (KLF8) in lung cancer cells.17 In the present investigation, we further clarified miR-1236-3p was strictly associated with DDP resistance in lung carcinoma cells. Next, the binding sites between miR-1236-3p and potential targets were predicted by Targetscan software.
With the help of dual-luciferase reporter assay, we corroborated that ATG7 was a target of miR-1236-3p. In regard to ATG7, it is part of the ATGs family and is deemed as autophagy-related genes. Autophagy is an evolutional conversed process, leading to orderly degradation and recycling in cellular organelles and proteins.27 Over the past decades, autophagy has been suggested to play an important function in the resistance of chemotherapy in multiple tumors.28,29 Referring to previous evidence, the autophagy activity of A549/DDP cells was higher than A549 cells, the inhibition of autophagy has been expounded to be related to the promotion of DDP sensitivity in lung cancer cells.30 Currently, ATG7 was particularly upregulated in lung cancer tissues and lung cancer cells no matter with or without DDP resistance. Subsequently, in vitro experiments were implemented to illustrate the relationship between miR-1236-3p and ATG7, the result showed that the impact of ATG7 silencing on cell behaviors was recovered via miR-1236-3p inhibition. The conclusion about ATG7 in DDP resistance was in accord with earlier findings.31
Taken together, the NNT-AS1 and ATG7 with high expression, and the miR-1236-3p with low level played pivotal roles in the initiation and progression of lung cancer cells with DDP resistance. Knockdown of NNT-AS1 could elevate cell apoptosis, and curb proliferation, migration, invasion and the IC50 of DDP in A549/DDP and H522/DDP cells. Collectively, NNT-AS1 exerted its function via miR-1236-3p/ATG7 axis in mediating DDP resistance in vitro. However, the potential role of NNT-AS1 still needed to be further highlighted.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi:10.3322/caac.20107
2. Heist RS, Engelman JA. SnapShot: non-small cell lung cancer. Cancer Cell. 2012;21:
3. Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378. doi:10.1016/j.ejphar.2014.07.025
4. Piskareva O, Harvey H, Nolan J, et al. The development of cisplatin resistance in neuroblastoma is accompanied by epithelial to mesenchymal transition in vitro. Cancer Lett. 2015;364:142–155. doi:10.1016/j.canlet.2015.05.004
5. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(1):R17–29. doi:10.1093/hmg/ddl046
6. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi:10.1186/s13059-017-1348-2
7. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi:10.1016/j.ccell.2016.03.010
8. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi:10.1038/onc.2017.184
9. Wang Q, Yang L, Hu X, et al. Upregulated NNT-AS1, a long noncoding RNA, contributes to proliferation and migration of colorectal cancer cells in vitro and in vivo. Oncotarget. 2017;8:3441–3453. doi:10.18632/oncotarget.13840
10. Cai Y, Dong ZY, Wang JY. LncRNA NNT-AS1 is a major mediator of cisplatin chemoresistance in non-small cell lung cancer through MAPK/Slug pathway. Eur Rev Med Pharmacol Sci. 2018;22:4879–4887. doi:10.26355/eurrev_201808_15624
11. Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi:10.1007/s10555-009-9188-5
12. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi:10.1038/nrg1379
13. Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. doi:10.1038/35103511
14. Kovalchuk O, Filkowski J, Meservy J, et al. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152–2159. doi:10.1158/1535-7163.mct-08-0021
15. Xia L, Zhang D, Du R, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi:10.1002/ijc.23501
16. Qiu T, Zhou L, Wang T, et al. miR-503 regulates the resistance of non-small cell lung cancer cells to cisplatin by targeting Bcl-2. Int J Mol Med. 2013;32:593–598. doi:10.3892/ijmm.2013.1439
17. Bian T, Jiang D, Liu J, et al. miR-1236-3p suppresses the migration and invasion by targeting KLF8 in lung adenocarcinoma A549 cells. Biochem Biophys Res Commun. 2017;492:461–467. doi:10.1016/j.bbrc.2017.08.074
18. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi:10.1016/j.cell.2011.10.026
19. Zhao Y, Liu Y, Lin L, et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. 2018;17:69. doi:10.1186/s12943-018-0820-2
20. Hua F, Liu S, Zhu L, et al. Highly expressed long non-coding RNA NNT-AS1 promotes cell proliferation and invasion through Wnt/beta-catenin signaling pathway in cervical cancer. Biomed Pharmacother. 2017;92:1128–1134. doi:10.1016/j.biopha.2017.03.057
21. Ye H, Lin J, Yao X, et al. Overexpression of long non-coding RNA NNT-AS1 correlates with tumor progression and poor prognosis in osteosarcoma. Cell Physiol Biochem. 2018;45:1904–1914. doi:10.1159/000487966
22. Liu Z, Sun M, Lu K, et al. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregulation of p21(WAF1/CIP1) expression. PLoS One. 2013;8:e77293. doi:10.1371/journal.pone.0077293
23. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi:10.1186/1476-4598-10-38
24. Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA-lncRNA interactions. Methods Mol Biol. 2016;1402:271–286. doi:10.1007/978-1-4939-3378-5_21
25. Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–355. doi:10.1038/cgt.2008.8
26. Wang Z, Liu L, Guo X, et al. microRNA-1236-3p regulates DDP resistance in lung cancer cells. Open Med. 2018;14:41–51. doi:10.1515/med-2019-0007
27. Kobayashi S. Choose delicately and reuse adequately: the newly revealed process of autophagy. Biol Pharm Bull. 2015;38:1098–1103. doi:10.1248/bpb.b15-00096
28. Wu WK, Coffelt SB, Cho CH, et al. The autophagic paradox in cancer therapy. Oncogene. 2012;31:939–953. doi:10.1038/onc.2011.295
29. Sui X, Chen R, Wang Z, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4:e838. doi:10.1038/cddis.2013.350
30. Zhang N, Yang GQ, Shao XM, et al. GAS5 modulated autophagy is a mechanism modulating cisplatin sensitivity in NSCLC cells. Eur Rev Med Pharmacol Sci. 2016;20:2271–2277. PMID: 27338051.
31. Pan X, Chen Y, Shen Y, et al. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 2019;10:429. doi:10.1038/s41419-019-1660-8
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.