Back to Journals » OncoTargets and Therapy » Volume 13
Long Non-Coding RNA LOXL1-AS1 Enhances Colorectal Cancer Proliferation, Migration and Invasion Through miR-708-5p/CD44-EGFR Axis
Authors Wu X, Cui F, Chen Y, Zhu Y, Liu F
Received 23 April 2020
Accepted for publication 13 July 2020
Published 3 August 2020 Volume 2020:13 Pages 7615—7627
DOI https://doi.org/10.2147/OTT.S258935
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Takuya Aoki
Xiaoyu Wu,1 Facai Cui,1 Yu Chen,2 Ya Zhu,1 Fengzhen Liu1
1Department of Clinical Laboratory, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou, Henan, People’s Republic of China; 2Department of Pathology, Affiliated Tumor Hospital of Zhengzhou University, Zhengzhou, Henan, People’s Republic of China
Correspondence: Xiaoyu Wu Email [email protected]
Introduction: Colorectal cancer (CRC), the third most common cancer worldwide, involves a physiological and pathological long non-coding RNA (lncRNA) paradigm shift. It has been reported that the lncRNA LOXL1-AS1 affects tumor development for many kinds of cancers, but its functions and mechanisms in CRC remain unknown.
Methods: Expression levels of LOXL1-AS1 and miR-708-5p within CRC tissues and cell lines were measured using qRT-PCR. The performance of gain-of-function and loss-of-function assays was aimed at examining the effects of LOXL1-AS1 and miR-708-5p; colony formation and cell viability assays were carried out to measure cell multiplication; and Transwell migration and wound-healing assays were carried out for the measurement of cell migration and invasion. Luciferase reporter assay was used to verify the interactions between LOXL1-AS1 and miR-708-5p and between miR-708-5p and the CD44-EGFR signaling pathway. Finally, expression of CD44 and EGFR proteins was measured by Western blot and immunofluorescence assays.
Results: In this study, we reveal that the regulation of lncRNA LOXL1-AS1 occurs within CRC based on the correlation with poor clinical outcomes. LOXL1-AS1 knockdown along with miR-708-5p overpresentation in CRC cell lines inhibited cell multiplication, migration, and invasion. The inhibiting effect of LOXL1-AS1 knockdown on CRC was reversed by upregulating the CD44-EGFR signal pathway. From the perspective of mechanism, LOXL1-AS1 imposes sponging upon miR-708-5p and thereby promotes the CD44-EGFR signal pathway in CRC cells.
Discussion: This study demonstrated that lncRNA LOXL1-AS1 enhances multiplication, migration, invasion, and progression of CRC by sponging miR-708-5p to regulate the CD44-EGFR signal pathway.
Keywords: colorectal cancer, LOXL1-AS1, CD44-EGFR signal pathway, miR-708-5p, migration
Introduction
Colorectal cancer (CRC), which ranks as the 3rd most commonly diagnosed cancer, showed an increase of 10.2% new cases and 9.2% cancer deaths worldwide in 2018.1 Early-stage CRC patients could be cured by surgery.2 However, 50–60% of CRC patients present with metastases when diagnosed.3 Despite the use of chemotherapy and surgical resection on patients with advanced CRC, the prognosis is poor.4 Previous research implicates a vast array of molecular changes in genes and signal cascades in the development and prognosis of CRC.5,6 Hence, it is urgent for CRC diagnosis and therapy to elucidate the latent molecular mechanisms of CRC.
Long non-coding RNAs (lncRNAs) of >200 nucleotides in length are involved in differentiation, apoptosis, metastasis, and drug-resistance in various cancers. Abundant research has demonstrated that pathological changes in lncRNAs exert critical regulation in all aspects of the initiation and development of CRC.7–9 For example, Ding et al discovered that the lncRNA CASC9 was abnormally upregulation within CRC tissues and promoted the viability, migration, and invasion of CRC cells,8 and Yang et al showed that the lncRNA XIST, a novel diagnostic biomarker in CRC, was associated with poor prognosis through the downregulation of METTL14.9 These data demonstrate that lncRNAs play important roles in tumorigenesis and tumor development.
Recent studies show that the lncRNA lysyl oxidase-like 1 (LOXL1)-AS1 plays crucial roles during the development of many types of cancer, including medulloblastoma,10 prostate cancer,11 and gastric carcinoma.12 Xie et al13 found that LOXL1-AS1 increases non-small-cell lung cancer invasion and proliferation; meanwhile, Li et al14 found that LOXL1-AS1 hindered breast cancer multiplication and migration. In gastric carcinoma, LOXL1-AS1 expression is elevated and accelerates deterioration and poor prognosis.12 Because LOXL1-AS1 has these tissue-specific effects in cancer, its effect on the regulation of CRC biology is unknown.
LncRNAs modulate levels of miRNA and thereby affect numerous biological functions and pathological processes. LOXL1-AS1 regulates progression and poor prognosis through numerous miRNAs, such as miR-541-3p in prostate cancer,11 miR-324-3p in non-small cell lung cancer,13 and miR142-5p in gastric cancer.15 Recently, miR-708-5p, a potential target gene of LOXL1-AS1, was found to be involved in proliferation, metastasis, and development of gastric carcinoma12 and breast cancer.16 It is unknown whether the LOXL1-AS1/miR-708-5p axis is involved in CRC tumorigenesis or progression.
The targets of miR-708-5p may include CD44 and Wnt/β-catenin. CD44 performs the duty of a hyaluronic acid receptor and is an indispensable cell surface glycoprotein that gives rise to activation of stem cell regulatory genes that support the sustainability of CRC stem cells.17–19 Senthil Kumar et al found that miRNA-708 regulates breast cancer tumorigenesis and metastasis by virtue of downregulation of CD44 signaling.20 Wnt/β-catenin signaling exerts a key part in growth, development, and chemoradiotherapy resistance in CRC,21,22 and Liu et al reported that miR-708-5p hinders lung cancer stem cell-like phenotypes by virtue of inhibiting Wnt/β-catenin signaling.23 Interestingly, CD44-EGFR and Wnt-β-catenin promote metastasis, cetuximab resistance, epithelial-to-mesenchymal transition, and progression in CRC.24–26 Whether CD44/EGFR and Wnt/β-catenin signaling are underlying mechanisms of the LOXL1-AS1/miR-708-5p axis demands expounding in detail.
Therefore, this research aims to explore the expression pattern and biological significance of LOXL1-AS1/miR-708-5p in CRC. Additionally, examination was made of CD44/EGFR and Wnt/β-catenin signal pathway’s participation in CRC proliferation and migration. According to our findings, LOXL1-AS1, a known oncogene, was highly expressed in CRC. Mechanistically, LOXL1-AS1 exerted its effect by “sponging” miR-708-5p to promote the CD44/EGFR signal pathway, which provides a theoretical basis for a new treatment strategy in CRC.
Patients and Methods
Patients and Samples
Matched specimens (CRC and adjacent normal tissue) of 40 primary CRC patients who had undergone surgical resection were obtained from People’s Hospital of Zhengzhou University (Zhengzhou, China) between January 2016 and December 2018. After surgical resection, the tissues were immediately frozen in liquid nitrogen and stored at −80°C until further analyses. Records were kept on all characterized aspects of the patients (age, gender, body mass index (BMI), tumor size and differentiation, tumor node metastasis (TNM) phase, lymph node metastasis, liver metastasis, and microsatellite instability (MSI) stage). The written informed consent was obtained from each subject in accordance with the Declaration of Helsinki. All studies were approved by the Ethical Committee of the Henan Provincial People’s Hospital & People’s Hospital of Zhengzhou University.
Cell Culture and Transfection
Human CRC cell lines (HCT8, LoVo, SW620, Caco2 and SW1463) and the normal human intestinal epithelial cell line HIEC were purchased from the ATCC (Manassas, VA, USA). All cell lines were maintained in RPMI-1640 media (Gibco, cat#: 11875-093) supplemented with 10% fetal bovine serum (Life Technologies, Inc., Grand Island, NY, USA), and kept at 37°C in a 5% CO2 incubator.
For knockdown of LOXL1-AS1, small interfering RNA (siRNA) targeting LOXL1-AS1 (si-LOXL1-AS1) and corresponding negative control (si-NC) was purchased from Genechem (Shanghai, China); transfection was made into Lovo and SW480 cells using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s directions. Aiming at upregulation vs downregulation of miR-708-5p, LoVo and SW480 cells were transfected with either miR-708-5p inhibitors or miR-708-5p negative control inhibitors (miR-NC) using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA). Verification of transfection efficiencies was accomplished by the combination of RNA extraction and quantitative real-time PCR (qRT-PCR).
Cell Proliferation Assay
Multiplication of Lovo and SW480 cells was assessed using Cell Counting Kit-8. After relevant transfection for 48 h, cells were transferred at 2000 per well into a 96-well plate with 5 replicates and kept in a humidity-controlled incubator for 4 hours. After that, the addition of 10 μL of CCK-8 solution added to 90 μL of complete medium was made in every well. Proliferation was measured at 0, 12, 24, and 48 hr using Cell Counting Kit-8 (CCK-8; Dojindo, Kyoto, Japan). The measurement at 450 nm absorbance was made with a micro-plate reader.
Colony Formation Assay
Following 24-hour transfection and culture, LoVo and SW480 cells were seeded in 6-well plates for 2 wk culture. After two PBS washes and 30 min fixation in 4% paraformaldehyde, colonies were stained for 30 min with 0.5% crystal violet at room temperature.
Wound-Healing Assay
For the wound-healing assay, Lovo and SW480 cells were transferred to 24-well plates at a concentration of 5 × 104 cells per well. Cells were harvested at 48 hr after transfection upon reaching 90% confluence. After a scratch wound was made through the cell layer with a pipette tip, the cell layer was gently washed three times with culture media to remove cell debris. After 48 hr further incubation, the scratch wounds were examined under the microscope to view cells that had migrated; the extent of healed area of the scratch was used as a measure of cell migration ability.
Transwell Assay
The evaluation of cell migration or invasion ability was made using Transwell chambers. LoVo and SW480 cells were seeded into the upper chamber of an 8 μm pore size insert (BD Biosciences, Franklin Lakes, NJ, USA). Following 24-hour culture starvation with serum-free medium, the cell suspension was shifted to the apical chamber. After 24 h incubation at 37°C, migration or invasion of cells to the lower surface of the insert membrane was assessed following 4% paraformaldehyde fixation for 30 min and staining with 0.1% crystal violet. Photography and analysis of cells that had migrated or invaded was performed using Image Pro-Plus software.
Dual-Luciferase Reporter Assay
Identification of the LOXL1-AS1 target was achieved with TargetScan online software (http://www.targetscan.org). To assess the LOXL1-AS1/miR-708-5p binding specificity, sub-clones of the wild type (wt) or the mutant type (mut) 3′-untranslated regions (3′-UTR) attached to LOXL1-AS1 binding sites were made on the pGL3 luciferase reporter vector (Promega, Madison, WI). Synthesis was made on the CD44, together with Wnt 3ʹ-UTR-luciferase reporter plasmid which include wild-type and mutant-type reporter 3ʹ-UTR of Wnt (Wnt-WT and Wnt-MUT) and CD44 (CD44-WT and CD44-MUT). pGL3 control vectors (Invitrogen, Waltham, MA, USA) were inserted in the plasmid. After culturing, 48-hour measurement to luciferase activity was made in virtue of a FLUO star device (Omega Engineering, Deckenpfronn, Germany)
Western Blot Assay
Protein was extracted from SW480 and Lovo cells using RIPA lysis buffer (Beyotime, Shanghai, China). Briefly, the BCA (Beyotime, Shanghai, China) method was used to determine the protein density. Total protein (30 μg/sample) was separated using SDS-PAGE electrophoresis, then transferred to a PVDF membrane. Following blocking with 5% skimmed milk, the membrane was exposed to primary antibodies against CD44, EGFR, or β-catenin (all from Abcam, Cambridge, MA, USA) for overnight at 4°C, then washed and exposed to the second antibody, goat anti-mouse IgG-HRP (Bioworld) for one hour at room temperature. The 350 μL ECL luminous substrate was added and images were formed in the darkroom. Antibodies to β-actin and GAPDH-HRP (Beyotime, Shanghai, China) were used as internal controls for the experiment.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Extraction of RNA from cells was made using TRIzol (Yaji Mall, Minhang, Shanghai, China). Analysis of purity and amount was achieved with a NanoDrop ND-2000 spectrophotometer (Thermo Fisher, Scientific Inc., Waltham, MA, USA). Reverse transcription used the TaqMan RNA Reverse Transcription kit as well as the TaqMan MicroRNA Reverse Transcription kit (both from Applied Biosystems). Reverse transcription of total RNA to cDNAs used the PrimeScript RT Reagent Kit (Takara Bio, Inc., Japan) according to the manufacturer’s specifications. The sequences of the primers were as follows: LOXL1-AS1 (forward): 5′-TTCCCATTTACCTGCCCGAAG-3′, LOXL1-AS1 (reverse): 5′-GT CAGCAAACACATGGCAAC-3′; miR-708-5p (forward): 5′-GGCGC GCAAGGAGCTTACAATC-3′, miR-708-5p (reverse): 5′-GTGCAGGGTCCGAGGTAT-3′; U6 (for-ward): 5′-GCTTCGGCAGCACATATACTAAAAT-3′, U6 (reverse): 5′-CGCTTCACGAATTTGCGTGTCAT-3′. Detection of miR-708-5p expression used the Prime Script miRNA RT-PCR Kit (Takara Bio Inc., Japan). Calculation of relative gene expression was assessed using the 2–ΔΔCt method.
Immunofluorescence Assay
Cells on glass coverslips were fixed with pre-cooled acetone, rinsed 3 times with PBS, incubated in 0.3% hydrogen peroxide in 10% goat serum, and then incubated overnight at 4°C with primary antibodies to CD44, EGFR, Wnt, and β-catenin (all from Abcam). After rinsing, the cells were incubated at room temperature for 45 min with HRP-conjugated secondary antibody (ZSGB-BIO, Xicheng, Beijing, China). Finally, nuclei were stained with DAPI (Boster Biotechnology, Wuhan, China).
Statistical Analysis
Statistical analysis used SPSS 23.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad Software, Inc., CAPrism6, USA). Data are presented as mean ± standard deviation (SD). Repeated-measures analysis of variance (ANOVA) was used to compare means across one or more variables based on repeated observations. Pairwise comparison of means used Student’s t-test; multiple group comparisons were made using ANOVA with LSD test. P values <0.05 were taken as indicating statistical significance.
Results
LOXL1-AS1 is Highly Expressed in CRC and Related to Poor Clinicopathologic Characteristics
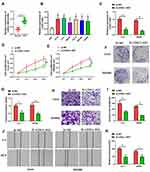
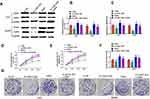
The level of LOXL1-AS1 is significantly higher in CRC tissue than in adjacent normal tissue (P < 0.05, Figure 1A). Correlations between LOXL1-AS1 levels and CRC clinicopathologic characteristics were analyzed and revealed that increased LOXL1-AS1 expression is significantly related to tumor size (P < 0.001), differentiation (P < 0.001), TNM stage (P < 0.001), liver metastasis (P < 0.05), and MSI stage (P < 0.05) (Table 1). Similarly, LOXL1-AS1 levels in CRC cell lines (HCT8, LoVo, SW620, Caco2, SW1468, and SW480) were significantly higher than in the normal human colorectal mucosa cell lines (HIEC) (P < 0.05, Figure 1B). Thus, LOXL1-AS1 is upregulated in both CRC tissue and CRC cell lines, and CRC tissue levels are positively correlated with poor clinicopathologic characteristics.
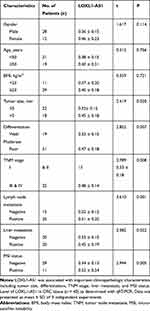 |
Table 1 Correlation Between LOXL1-AS1 Level (X ± SD) and Clinicopathological Characteristics |
LOXL1-AS1 Contributes to Multiplication, Colony Formation, Migration, and Invasion in CRC Cells
After transfection with the pcDNA-LOXL1-AS1 vector, levels of LOXL1-AS1 in Lovo and SW480 cells were lower (P < 0.01; Figure 1C), indicating that LOXL1-AS1 knockdown was effective. CCK-8 assay showed that proliferation of si-LOXL1-AS1 cells was obviously depressed compared to si-NC cells (P < 0.01; Figure 1D and E). In contrast with the si-NC, downregulation of LOXL1-AS1 by si-LOXL1-AS1 greatly reduced the capacity for colony formation in CRC cells (P < 0.01, Figure 1F; P < 0.01, Figure 1G). Transwell assays showed that LOXL1-AS1 knockdown dramatically inhibited migration and invasion of Lovo and SW480 cells (Figure 1H and I). Likewise, the wound-healing assays showed that migration of Lovo and SW480 cells was reduced when the transfection of cells was made with the pcDNA-LOXL1-AS1 vector (P < 0.01, Figure 1J and K). Taken together, these findings demonstrated that LOXL1-AS1 can contribute to colorectal carcinogenesis.
LOXL1-AS1 Acts as a Sponge for miR-708-5p in CRC Cells
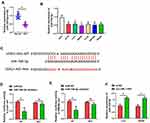
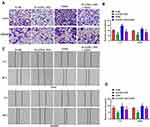
Significant downregulation of miR-708-5p expression occurred in both CRC tissue (P < 0.01, Figure 2A) and CRC lines (P < 0.01, Figure 2B). More interestingly, the expression of miR-708-5p in Lovo and SW480 cells was significantly upregulated after transfection with pcDNA-LOXL1-AS1 vector (P < 0.01, Figure 2F), which indicated a potential association between LOXL1-AS1 and miR-708-5p. To determine whether miR-708-5p could interact with LOXL1-AS1, StarBase v3.0 (http://starbase.sysu.edu.cn/panCancer.php) was used and showed that LOXL1-AS1 has a putative binding site of miR-708-5p (Figure 2C). To confirm whether miR-708-5p is a LOXL1-AS1 target, we created LOXL1-AS1 mutant (Mut) and wild type (WT) luciferase constructs. The luciferase reporter assays showed that down-expression of miR-708-5p suppressed luciferase activity in the LOXL1-AS1-WT group but not the LOXL1-AS1-Mut group when compared with miR-NC group, respectively (Figure 2D and E). Those findings indicate miR-708-5p bound to the transcript position of LOXL1-AS1.
miR-708-5p Represses the Occurrence of Proliferation, Colony Formation, Migrating and Invading to CRC Cells
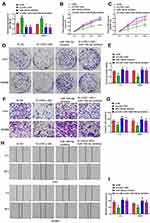
Next, in order to determine whether LOXL1-AS1-dependent malignant behaviors are present after downregulation of miR-708-5p, cells down-expressing LOXL1-AS1 were transfected with the miR-708-5p inhibitor. As shown with qRT-PCR, miR-708-5p was downregulated in Lovo and SW480 cells after transfection of miR-708-5p inhibitor (P < 0.01; Figure 3A). Evaluating the effects of miR-708-5p on CRC cell biology, it was concluded that the downregulation of miR-708-5p completely blocked the effect of si-LOXL1-AS1 in reducing viability (Lovo: P < 0.01, Figure 3B; SW480: P < 0.01, Figure 3C) and colony formation (P < 0.01, Figure 3D and E). Moreover, wound-healing and Transwell invasion assays showed that the inhibitory effect of si-LOXL1-AS1 on CRC migration (P < 0.01, Figure 3F and G) and invasion (P < 0.01, Figure 3H and I) was reversed by miR-708-5p inhibitor. These findings indicate that decreasing the level of miR-708-5p is crucial to the acceleration of cell proliferation, colony formation, migration, and invasion after LOXL1-AS1 downregulation.
CD44-EGFR is the Target Pathway of miR-708-5p
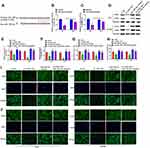
TargetScan 7.2 was used for predicting the targets of miR-708-5p and showed that miR-708-5p targeted CD44 (Figure 4A). We next used luciferase reporter assays to verify the repression of miR-708-5p on the activity of pGL3-CD44-WT both in Lovo (P < 0.01, Figure 4B) and SW480 (P < 0.01, Figure 4C) cells. Western blot assay showed that LOXL1-AS1 knockdown dramatically inhibited the protein levels of CD44 and EGFR both in Lovo (P < 0.01, Figure 4D and E) and SW480 (P < 0.01, Figure 4D and F) which could be rescued by miR-708-5p inhibitor. Meanwhile, immunofluorescence assay showed that CD44 and EGFR expression were increased with miR-708-5p inhibitor when compared with si-LOXL1-AS1 (Lovo: P < 0.01, Figure 4G, I and J; SW480: P < 0.01, Figure 4H–J).
Although the TargetScan analysis also indicated that miR-708-5p of might target β-catenin (Figure S1A), luciferase reporter assays indicated that miR-708-5p exerted no effect upon the activity of pGL3- β-catenin-WT in either Lovo (P > 0.05, Figure S1B) or SW480 cells (P > 0.05, Figure S1C). Moreover, Western blotting showed no significant changes in the level of β-catenin protein after knockdown of LOXL1-AS1 or co-transfection with the miR-708-5p inhibitor (P >0.05, Figure S1D–F), and β-catenin expression remained unchanged in different treatments group both in Lovo (P > 0.05, Figure S2A) and SW480 (P > 0.05, Figure S2B) cells.
CD44 Overexpression Blocks the Effect of LOXL1-AS1 Downregulation in CRC Cells
To determine the involvement of CD44-EGFR following LOXL1-AS1 downregulation in Lovo and SW480, CD44 expression was upregulated by transfection with a plasmid of pLVX-CD44 encoding the full-length coding sequence without the 3ʹUTR region, which resulted in upregulation of the levels of CD44 and EGFR (P < 0.01, Figure 5A–C). The proliferation (Lovo: P < 0.01, Figure 5D; SW480: P < 0.01, Figure 5E) and viability (P < 0.01, Figure 5F and G) of both cell lines, which was significantly decreased after knockdown of LOXL1-AS1, was reversed by CD44 overexpression. Meanwhile, upregulation of CD44 completely blocked the inhibitory effects of LOXL1-AS1 downregulation on cell migration capability (P < 0.01, Figure 6A and B) and invasion capability (P < 0.01, Figure 6C and D) of the Lovo and SW480 cells. These data indicated that CD44 overexpression contributes to the enhancement of cell multiplication, migration, and invasion after LOXL1-AS1 downregulation.
Discussion
This study has first demonstrated that upregulation of LOXL1-AS1 occurs in CRC tissues as well as CRC cell lines, and that this upregulation is relevant to poor clinicopathologic features of CRC. LOXL1-AS1 acts as a sponge for miR-708-5p in CRC cells, thereby contributing to proliferation, colony formation, migration, and invasion of CRC cells. Meanwhile, the CD44-EGFR pathway was shown to be the direct target gene of miR-708-5p, and expression of CD44 and EGFR could be increased by downregulating miR-708-5p. In addition, the suppressive effects of LOXL1-AS1 downregulation upon CRC cell progression were restored by CD44 overexpression. Collectively, these findings suggest that LOXL1-AS1 enhances the development of CRC by virtue of sponging miR-708-5p and thus promoting the CD44-EGFR signal pathway.
According to an increasing number of studies, LOXL1-AS1 participates in the growth and development of several tumors but with different characteristics in different tissues.10,13,14 In this study, LOXL1-AS1 was found to be higher in CRC tumor tissue samples and associated with important clinicopathologic characteristics including tumor size, differentiation, TNM stage, liver metastasis, and MSI. These findings suggest that miR-708-5p, acting as a positive regulator, might be an important part in the CRC progression and prognosis of CRC patients. As for our in vitro research, LOXL1-AS1 presentation was shown to be significantly upregulated in several CRC cell lines, and knockdown of LOXL1-AS1 could inhibit tumor cell vitality, colony formation, migration and invasion in both Lovo and SW480 cells. This showed that LOXL1-AS1 promoted tumor development and migration in CRC, which is supported by previous studies.10,11 On the other hand, Li et al found that LOXL1-AS1 inhibited proliferation and apoptosis in breast cancer through regulation of miR-143-3p,14 which is clearly indicative of tissue specificity regarding LOXL1-AS1 function.
LOXL1-AS1 is one of the most prominent regulators in human cancer. However, the mechanism through which LOXL1-AS1 alters the host’s noncoding RNAs, especially in CRC, remains poorly understood. Previous studies showed that LOXL1-AS1 facilitates tumorigenesis and stemness via regulation of miR-708-5p in gastric carcinoma and breast cancer.12,16 Our study demonstrates significant miR-708-5p downregulation expression in CRC tissues (P < 0.01, Figure 2A) and CRC lines, which implies an interaction between LOXL1-AS1 and miR-708-5p. To further verify the targeting reaction between LOXL1-AS1 and miR-708-5p in CRC, TargetScan was performed to predict the targets of LOXL1-AS1, and luciferase reporter assay revealed that miR-708-5p acted as a direct target of LOXL1-AS1. Functional experiments suggested that the reversal of the inhibition of cell proliferation, colony formation, migrating, and invading under the influence of LOXL1-AS1 knockdown can be made by virtue of a decrease in miR-708-5p. This suggests that miR-708-5p can act as a negative regulator in the pathogenesis of CRC and that malignant behaviors depending on LOXL1-AS1 are associated with miR-708-5p downregulation in CRC. Similar relationships have been found in gastric carcinoma and breast cancer.12,16
CD44 has multiple spliced isoforms and heavy glycosylation, which is involved in tumor cell migration in prostate cancer27 and breast cancer.28 Kim et al found that hepatocellular carcinoma patients with low expression of CD44 expressed a significantly better disease-free survival rate compared to those with higher CD4429 expression. Jing et al examined 36 CRC patients with hepatic metastasis and found that CD44 expression was an independent factor associated with survival.30 Lina et al demonstrated that upregulation of CD44 as well as the variant isoforms can promote the activation induced by EGFR downstream signaling pathways. They also held the view that CD44 suppression held back EGFR presentation and the activation featured by downstream signaling pathways among CRC cells, which is closely associated with CRC cell migration and drug resistance. Luciferase reporter assays in the current study showed that CD44 was a direct target of miR-708-5p. Surprisingly, downregulating miR-708-5p increased the level of CD44 and EGFR protein as shown with Western blot and immunofluorescence assays. In summary, miR-708-5p-mediated repression of the CD44-EGFR signaling pathway may underlie its role in affecting the tumor-initiating subpopulation of CRC.
To further investigate the role CD44-EGFR signaling pathway exerts in the CRC cell progression facilitated by LOXL1-AS1, we transfected a plasmid of pLVX-CD44 to upregulate expression of CD44. CD44 overexpression blocked the suppressive effect of LOXL1-AS1 in CRC cell multiplication, migration, and invasion, which demonstrates the biological connection between LOXL1-AS1 expression and the CD44-EGFR signaling pathway in CRC. Previous studies have shown that EGFR-mediated signaling by way of the Ras-MAPK and PI3K-AKT pathways calls for the CD44 cytoplasmic tail in relevance to the receptor tyrosine kinases which promote tumor progress.31–33 Mutation of KRAS should lead to constitutive stimulation of the Erk1/2 pathway, which gives rise to these cells in resistance to EGFR-issued signals.34 Meanwhile, molecules functionally interacting with CD44 such as receptor tyrosine kinases, matrix proteinases, and ERM proteins could involve the close connection betwixt cell migration and miR-708-5p expression.35
It is often reported that aberrant Wnt/β-catenin signaling in CRC is implicated in increased resistance to radiotherapy.36,37 Stimulation of the Wnt/β-catenin signaling pathway drives CRC growth by acting on downstream target genes like mTOR and c-myc proto-oncogene.36 In the current study, TargetScan analysis indicated that miR-708-5p might target β-catenin, but we discovered that miR-708-5p had no effect on Wnt/β-catenin signaling. Thus, Wnt/β-catenin signaling may not participate in biological effects of the LOXL1-AS1/miR-708-5p axis in CRC.
Conclusion
To sum up, the research shows that LOXL1-AS1 is a critical mediator in CRC patients and cell lines, clarifying the regulatory mechanisms underlying the function of LOXL1-AS1 in CRC cell lines. For the first time, this report shows that LOXL1-AS1 regulates the CD44-EGFR signaling pathway for the execution of its oncogenic influence in CRC by sponging miR-708-5p. This is an important avenue for further research to explore therapeutic treatments for CRC.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
This research was supported by Ethics Committee of Henan Provincial People’s Hospital & People’s Hospital of Zhengzhou University; the written informed consent of each patient was obtained.
Disclosure
The authors have no financial interests or conflicts of interest to declare.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi:10.1016/S0140-6736(13)61649-9
3. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–117. doi:10.3322/caac.21220
4. Sun L, Jiang C, Xu C, et al. Down-regulation of long non-coding RNA RP11-708H21.4 is associated with poor prognosis for colorectal cancer and promotes tumorigenesis through regulating AKT/mTOR pathway. Oncotarget. 2017;8(17):27929–27942. doi:10.18632/oncotarget.15846
5. Xiao R, Li C, Chai B. miRNA-144 suppresses proliferation and migration of colorectal cancer cells through GSPT1. Biomed Pharmacother. 2015;74:138–144. doi:10.1016/j.biopha.2015.08.006
6. Han C, Song Y, Lian C. MiR-769 inhibits colorectal cancer cell proliferation and invasion by targeting HEY1. Med Sci Monit. 2018;24:9232–9239. doi:10.12659/MSM.911663
7. Liu Y, Chen X, Chen X, et al. Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer. Cell Death Dis. 2020;11(3):175. doi:10.1038/s41419-020-2268-8
8. Ding Y, Li X, Zhang Y, Zhang J. Long non-coding RNA cancer susceptibility 9 (CASC9) up-regulates the expression of ERBB2 by inhibiting miR-193a-5p in colorectal cancer. Cancer Manag Res. 2020;12:1281–1292. doi:10.2147/CMAR.S234620
9. Yang X, Zhang S, He C, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19(1):46. doi:10.1186/s12943-020-1146-4
10. Gao R, Zhang R, Zhang C, Liang Y, Tang W. LncRNA LOXL1-AS1 promotes the proliferation and metastasis of medulloblastoma by activating the PI3K/AKT pathway. Anal Cell Pathol (Amst). 2018;2018:9275685.
11. Long B, Li N, Xu XX, et al. Long noncoding RNA LOXL1-AS1 regulates prostate cancer cell proliferation and cell cycle progression through miR-541-3p and CCND1. Biochem Biophys Res Commun. 2018;505(2):561–568. doi:10.1016/j.bbrc.2018.09.160
12. Sun Q, Li J, Li F, et al. LncRNA LOXL1-AS1 facilitates the tumorigenesis and stemness of gastric carcinoma via regulation of miR-708-5p/USF1 pathway. Cell Prolif. 2019;52(6):e12687. doi:10.1111/cpr.12687
13. Xie N, Fei X, Liu S, Liao J, Li Y. LncRNA LOXL1-AS1 promotes invasion and proliferation of non-small-cell lung cancer through targeting miR-324-3p. Am J Transl Res. 2019;11(10):6403–6412.
14. Li GH, Yu JH, Yang B, Gong FC, Zhang KW. LncRNA LOXL1-AS1 inhibited cell proliferation, migration and invasion as well as induced apoptosis in breast cancer via regulating miR-143-3p. Eur Rev Med Pharmacol Sci. 2019;23(23):10400–10409. doi:10.26355/eurrev_201912_19679
15. Li M, Cai O, Tan S. LOXL1-AS1 drives the progression of gastric cancer via regulating miR-142-5p/PIK3CA axis. Onco Targets Ther. 2019;12:11345–11357. doi:10.2147/OTT.S223702
16. Dong HT, Liu Q, Zhao T, et al. Long non-coding RNA LOXL1-AS1 drives breast cancer invasion and metastasis by antagonizing miR-708-5p expression and activity. Mol Ther Nucleic Acids. 2020;19:696–705. doi:10.1016/j.omtn.2019.12.016
17. Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4(1):33–45. doi:10.1038/nrm1004
18. Wahab SMR, Islam F, Gopalan V, Lam AK. The identifications and clinical implications of cancer stem cells in colorectal cancer. Clin Colorectal Cancer. 2017;16(2):93–102. doi:10.1016/j.clcc.2017.01.011
19. Mikami S, Mizuno R, Kosaka T, Saya H, Oya M, Okada Y. Expression of TNF-alpha and CD44 is implicated in poor prognosis, cancer cell invasion, metastasis and resistance to the sunitinib treatment in clear cell renal cell carcinomas. Int J Cancer. 2015;136(7):1504–1514. doi:10.1002/ijc.29137
20. Senthil Kumar KJ, Gokila Vani M, Hsieh HW, et al. MicroRNA-708 activation by glucocorticoid receptor agonists regulate breast cancer tumorigenesis and metastasis via downregulation of NF-kappaB signaling. Carcinogenesis. 2019;40(2):335–348. doi:10.1093/carcin/bgz011
21. Emons G, Spitzner M, Reineke S, et al. Chemoradiotherapy resistance in colorectal cancer cells is mediated by Wnt/beta-catenin signaling. Mol Cancer Res. 2017;15(11):1481–1490. doi:10.1158/1541-7786.MCR-17-0205
22. Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16(1):9. doi:10.1186/s12943-017-0583-1
23. Liu T, Wu X, Chen T, Luo Z, Hu X. Downregulation of DNMT3A by miR-708-5p inhibits lung cancer stem cell-like phenotypes through repressing Wnt/beta-catenin signaling. Clin Cancer Res. 2018;24(7):1748–1760. doi:10.1158/1078-0432.CCR-17-1169
24. Sun L, Fang Y, Wang X, et al. miR-302a inhibits metastasis and cetuximab resistance in colorectal cancer by targeting NFIB and CD44. Theranostics. 2019;9(26):8409–8425. doi:10.7150/thno.36605
25. Sun J, Zhang T, Cheng M, et al. TRIM29 facilitates the epithelial-to-mesenchymal transition and the progression of colorectal cancer via the activation of the Wnt/beta-catenin signaling pathway. J Exp Clin Cancer Res. 2019;38(1):104. doi:10.1186/s13046-019-1098-y
26. Pan S, Liu Y, Liu Q, et al. HOTAIR/miR-326/FUT6 axis facilitates colorectal cancer progression through regulating fucosylation of CD44 via PI3K/AKT/mTOR pathway. Biochim Biophys Acta Mol Cell Res. 2019;1866(5):750–760. doi:10.1016/j.bbamcr.2019.02.004
27. Saini S, Majid S, Shahryari V, et al. miRNA-708 control of CD44(+) prostate cancer-initiating cells. Cancer Res. 2012;72(14):3618–3630. doi:10.1158/0008-5472.CAN-12-0540
28. Tan W, Tang H, Jiang X, et al. Metformin mediates induction of miR-708 to inhibit self-renewal and chemoresistance of breast cancer stem cells through targeting CD47. J Cell Mol Med. 2019;23(9):5994–6004. doi:10.1111/jcmm.14462
29. Kim J, Jiang J, Badawi M, Schmittgen TD. miR-221 regulates CD44 in hepatocellular carcinoma through the PI3K-AKT-mTOR pathway. Biochem Biophys Res Commun. 2017;487(3):709–715. doi:10.1016/j.bbrc.2017.04.121
30. Jing F, Kim HJ, Kim CH, Kim YJ, Lee JH, Kim HR. Colon cancer stem cell markers CD44 and CD133 in patients with colorectal cancer and synchronous hepatic metastases. Int J Oncol. 2015;46(4):1582–1588. doi:10.3892/ijo.2015.2844
31. Dhar D, Antonucci L, Nakagawa H, et al. Liver cancer initiation requires p53 inhibition by CD44-enhanced growth factor signaling. Cancer Cell. 2018;33(6):1061–1077 e1066. doi:10.1016/j.ccell.2018.05.003
32. Zoller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11(4):254–267. doi:10.1038/nrc3023
33. Prochazka L, Tesarik R, Turanek J. Regulation of alternative splicing of CD44 in cancer. Cell Signal. 2014;26(10):2234–2239. doi:10.1016/j.cellsig.2014.07.011
34. Lake D, Correa SA, Muller J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol Life Sci. 2016;73(23):4397–4413. doi:10.1007/s00018-016-2297-8
35. Zhu Y, Ailane N, Sala-Valdes M, et al. Multi-factorial modulation of colorectal carcinoma cells motility – partial coordination by the tetraspanin Co-029/tspan8. Oncotarget. 2017;8(16):27454–27470. doi:10.18632/oncotarget.16247
36. Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. doi:10.1016/j.ctrv.2017.11.002
37. Liu L, Zhang Y, Wong CC, et al. RNF6 promotes colorectal cancer by activating the wnt/beta-catenin pathway via ubiquitination of TLE3. Cancer Res. 2018;78(8):1958–1971. doi:10.1158/0008-5472.CAN-17-2683
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.