Back to Journals » OncoTargets and Therapy » Volume 14
LncRNA MCTP1-AS1 Regulates EMT Process in Endometrial Cancer by Targeting the miR-650/SMAD7 Axis
Authors Gao Q, Huang Q, Li F, Luo F
Received 25 November 2019
Accepted for publication 26 February 2020
Published 3 February 2021 Volume 2021:14 Pages 751—761
DOI https://doi.org/10.2147/OTT.S240010
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Qin Gao,* Qin Huang,* Fangbing Li, Fang Luo
Obstetrics and Gynecology of Pu Ren Hospital in Wuhan, Wuhan, 430081, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Fang Luo Tel +86-13507157935
Email [email protected]
Background: Long noncoding RNAs (lncRNAs) play critical roles in the pathogenesis of several diseases, especially some kinds of cancer. This study aimed to investigate the expression of MTCP1-AS1 and its effects on endometrial cancer (EC).
Methods: MTCP1-AS1 expression level was determined in human EC tissues and cell lines by qRT-PCR. The role of MTCP1-AS1 on EC cell proliferation, migration, invasion and epithelial to mesenchymal transition (EMT) was detected by CCK8, wound-healing assay, transwell assay and Western blot, respectively. Moreover, luciferase reporter assay and RNA-binding protein immunoprecipitation (RIP) assay were performed to verify the targeting relationship between miR-650, MCTP1-AS1 and SMAD7 in EC cells.
Results: Our data showed that MCTP1-AS1 expression was downregulated in EC tissues and cell lines. Overexpression of MCTP1-AS1 inhibited cell proliferation, migration, invasion and EMT process of EC cells. Moreover, MCTP1-AS1 was proved to be the target of miR-650 and reversely correlated with its expression. In addition, MCTP1-AS1 reversed the effect of miR-650 on the EC cells, which might be associated with the role of SMAD7. Moreover, Western blot showed siRNA-SMAD7 transfection could rescue the repressed TGF-β/SMAD pathway induced by MCTP1-AS1 in EC cells.
Conclusion: Taken together, these data suggested that lncRNA MCTP1-AS1 inhibited cell proliferation, migration, invasion and EMT process of EC cells via targeting the miR-650/SMAD7 axis and it has the potential to be explored as a therapeutic target for the treatment of EC in the future.
Keywords: endometrial cancer, EC, lncRNAs, MCTP1-AS1, miR-650, SMAD7, EMT
Background
Endometrial cancer (EC) is the fourth most common gynecological cancer in women.1,2 Standard treatment for EC consists of surgery and radiation therapy depending on risk of disease recurrence.2–4 However, the prognosis of EC is still not satisfactory, and the underlying mechanisms remain to be fully understood. Therefore, there is an urgent demand to develop predictive biomarkers for treatment selection.
Long noncoding RNAs (lncRNAs) are noncoding transcripts usually longer than 200 nts that appears to play major roles in diverse biological functions, such as genomic imprinting, cell differentiation, immune response, especially tumor development, etc.5–7 Lots of experiments have shown that lncRNAs was involved in reproductive diseases such as ovarian cancer, cervical carcinoma and other female reproductive disorders. Yang et al found that NORAD/miR-608/STAT3 axis was pivotal in mediating the antineoplastic impacts of PG on ovarian cancer cells.8 Long non-coding RNA RP11-480I12.5 was proved to promote cervical carcinoma progression by regulating the Wnt/β-catenin signaling pathway.9 Besides, Wang et al provided the evidence for the involvement of lncRNAs in polycystic ovary syndrome (PCOS) and premature ovarian insufficiency (POI).10 Many lncRNAs have recently been reported to be crucial for the pathogenesis of endometrial cancer. For example, Xie et al demonstrated that lncRNA CCAT2 was highly expressed in endometrial cancer tissues, and knockdown of lncRNA CCAT2 inhibited endometrial cancer cells growth and metastasis via sponging miR-216b.11 Besides, previous study showed that lncRNA DLEU1 promoted the development of endometrial cancer by sponging miR-490 to regulate SP1 expression.12 Moreover, lncRNA MEG3 was found to inhibit endometrial carcinoma tumorigenesis and progression through PI3K pathway.13 However, the molecular function of lncRNAs in EC remains largely unknown.
MCTP1-AS1 was recently discovered downregulated in endometrial cancer tissues comparing to its paired normal adjacent tissues, which indicated that MCTP1-AS1 might also be an oncogene in endometrial cancer.14 However, few studies have studied MCTP1-AS1 in EC and the exact mechanism have not been clarified. LncRNAs played role in progression of many diseases by binding with different microRNAs (miRNAs) as their targets. In our study, MCTP1-AS1 was proved to be the target of miR-650 and reversely correlated with its expression. Previous study showed that miR-650 was upregulated in endometrial cancer tissues, and regulate tumor progression.15 For example, miR-650 promoted the metastasis and Epithelial-Mesenchymal Transition (EMT) of hepatocellular carcinoma by directly inhibiting LATS2 expression.16 Orlandella et al found that miR-650 promoted the motility of anaplastic thyroid cancer cells by targeting PPP2CA.17
SMAD family plays an important role in mediating TGF-β signaling pathway, which includes three functional classes: common SMADs, receptor-regulated SMADs (RSMADs), and inhibitory SMADs (I-SMADs).18 SMAD7, belonging to I-SMADs, was reported to be an inhibitor of TGF-β signaling, and associated with tumor malignancy.19 Parikh et al indicated that in ovarian cancer, SMAD7 has been proved to regulate TGF-β signaling pathway by affecting the expression and activation of SMAD2/3, thereby affecting EMT.20 In the present study, we demonstrated that lncRNA MCTP1-AS1 regulated the proliferation, migration, invasion and EMT process of EC cells through miR-650/SMAD7 axis, which provided new ideas for the pathogenesis and clinical research of endometrial cancer.
Materials and Methods
Patient Samples
A total of 60 pieces of pathologically proven EC tissues and adjacent normal tissues were collected from Pu Ren Hospital between Apr 2017 and Jan 2018. All patients received no radiotherapy or chemotherapy before surgery. All samples were frozen in liquid nitrogen immediately after resection. This study was approved by the Ethics Committee of Pu Ren hospital. Written informed consent was obtained from every patient.
Cell Culture and Transfection
Endometrial adenocarcinoma cell lines HEC-1B, HEC-1A, Ishikawa and RL-952 and human normal esophageal epithelial cell line hEEC were purchased from the American Type Culture Collection (ATCC). Cells above were maintained in DMEM (Hyclone) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Hyclone) at 37°C in a humidified incubator containing 5% CO2. Stable expression cell lines were established by virus infection. Briefly, 1×106 293FT cells were plated into 6-well plate coated with collagen I (BD Bioscience, #354236), transfection was performed with retroviral constructs together with packaging plasmids, and viruses were collected twice at 24 h intervals. To infect cells, cells were cultured in 1 mL viral supernatant mixed with 1 µL polybrene of 10 µg/mL stock for 6 h followed with regular media. Cells were selected with 2 µg/mL puromycin (Sigma) for 7 days. The overexpression plasmid pcDNA3.1-MCTP1-AS1 and small-interfering RNA (siRNA) for SMAD7 were designed by GenePharma.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from cells or frozen tissues using TRIzol reagent (Invitrogen, #15596026), with its quantity and quality being examined by NanoDrop ND-1000 (Thermo). One microgram of total RNA was converted into cDNA using TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen Biotech, #AT311-02) according to manufacturer’s instruction. The quantitative PCR (qPCR) was performed on 15 ng of cDNA from each sample using SYBR Green Real-time PCR Master Mix (TOYOBO, #QPK-201) based on the recommendations of manufacturer. The qPCR reactions run on the following conditions: initial denaturing at 95°C for 30 sec, followed by 35–40 cycles of 95°C for 5 sec, 60°C for 10 sec and 72°C for 15 sec, melting curves were examined at 37°C for 30 sec before cooling. Each result was from three independent biological replicates for all analyses performed in this work. The qPCR results were analyzed using 2−ΔΔCT method and presented as relative quantity of transcripts with GAPDH as the reference gene. The primers sequences were: MCTP1-AS1: 5ʹ-GGCAGATTCTTAGTTCCG-3ʹ and 5ʹ-ATTCTTGGCTATACTCTACCTC-3ʹ; miR-650: 5ʹ-AGAGGAGGCAGCGCTCT-3ʹ and 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ; SMAD7: 5ʹ-TTCCTCCGCTGAAACAGGG-3ʹ and 5ʹ-CCTCCCAGTATGCCACCAC-3ʹ; GAPDH: 5ʹ-TCTCCGCCCCTTCCGCTGAT-3ʹ and 5ʹ-CCACAGCCTTGGCAGCACCA-3ʹ.
Western Blot Assay
The cell lysates were prepared by using RIPA lysis buffer (Beyotime) and then the protein concentration was measured using the Bicinchoninic acid (BCA) Protein quantification kit (Thermo). 30 μg protein samples were subjected to SDS-PAGE and then transferred onto polyvinylidene fluoride (PVDF) membrane at 100 V for 90 mins. Then, 5% skim milk in PBST was used to block the membranes at room temperature for 1 hr, after which appropriate antibodies were used to incubate with membranes at 4°C overnight. After washing, secondary antibodies were used to incubate with membranes in 5% fat-free milk at room temperature for 1 h. The protein expression was determined by SuperEnhanced chemiluminescence detection reagent (Applygen technologies). GAPDH was used as internal reference. The primary antibodies were used for immunoblotting: E-cadherin (BD Biosciences, 610181), N-cadherin (BD Biosciences, 610920), Vimentin (Cell Signaling Technology, 5741), AGO2 (Millipore, SAB4200085), SMAD7 (R&D Systems, MAB2029), SMAD2 (Cell Signaling Technology, 5339), P-SMAD2 (Cell Signaling Technology, 3108), SMAD3 (Cell Signaling Technology, 9523), P-SMAD3 (Cell Signaling Technology, 9520), TGF-β1 (Invitrogen, MA1-169), GAPDH (Proteintech, 60004-1-Ig). Secondary antibodies were purchased from Thermo Fisher including HRP-linked anti-mouse IgG (Cell Signaling Technology, 7076) and HRP-linked anti-rabbit IgG (Cell Signaling Technology, 7074).
Cell Counting Kit-8 (CCK-8)
Cell proliferation was assessed by Cell Counting Kit-8 assay. Briefly, about 1×103 cells suspended in 100 μL culture medium were seeded into 96-well plates (Corning). After incubation at 37°C in a humidified incubator containing 5% CO2 for 24, 48 and 72 h, respectively, 10 μL of CCK-8 solution (Beyotime) was added to each well. After 2 h of incubation at 37°C, the absorbance at 450 nm was measured using an automatic microplate reader (Infinite M200, TECAN).
Transwell Migration and Invasion Assay
Cell invasion was measured using 24-well Transwell Chamber (8-μm pore membrane, BD Biosciences). 1×105 cells were harvested and planted into the Matrigel pre-coated upper chamber with 100 μL serum-free medium while the bottom chamber was filled with 600 μL completed medium. Inserts were incubated at 37°C, 5% CO2 for 24h. Non-invading cells were removed with a cotton swab soaked in medium. Cells that had moved through the pores (to the lower surface of the filters) were fixed with methanol (Macklin) for 1 h and stained with crystal violet (Solarbio) for 30 min, then counted invasive cells on a microscope at ECLIPSE Ti-U epi-fluorescence microscope (Nikon). Three inserts were counted for each treatment in each experiment and experiments were carried out at least three times.
In vitro Wound-Healing Assay
Cells were removed by trypsinisation (Hyclone), counted and 5×105 cells were added into 6-well plate (Corning). Cells were incubated overnight yielding confluent monolayers for wounding. Wounds were made using a pipette tip and photographs taken by ECLIPSE Ti-U epi-fluorescence microscope (Nikon) immediately (time zero) and 24 h after wounding. Experiments were carried out in triplicate and repeated at least five times.
Luciferase Reporter Assay
The 3ʹ-UTR sequence of miR-650 binding sites within the predicted target sites was amplified by PCR and then cloned into the pGL3 luciferase vector. Ishikawa cells were co-transfected with the pGL3-WT or -MUT or miR-513 mimics/NC using Lipo3000 (Invitrogen). After 48 hrs transfection, the cells were washed twice with PBS twice and digested using lysis buffer. After centrifuge, cells lysates were collected. The luciferase activity was analyzed by Microplate Reader (Infinite M200, TECAN).
RNA Binding Protein Immunoprecipitation (RIP)
RNA immunoprecipitation (RIP) assays were performed to confirm the interaction between MCTP1-AS1 and miR-650. Briefly, RIP was performed using the EZMagna RIP RNA-binding protein immunoprecipitation kit (Millipore) according to the manufacturer’s instructions. Cells in different groups were lysed using RNA lysis buffer containing protease inhibitor and RNase inhibitor. Then, cells lysates were incubated with the RIP buffer containing magnetic beads coated with Ago2 antibodies (Millipore). IgG served as a negative control (input group).
Statistical Analysis
All of the experiments were carried out for at least three times. The results were shown as mean ± standard deviations (SD). Statistical analysis was performed using SPSS 20.0 or GraphPad Prism software. P-values were calculated using two-tailed Student’s t-test from Excel or GraphPad Prism software, and P-values less than 0.05 were considered statistically significant.
Results
MCTP1-AS1 Is Downregulated in EC Tissues and Cell Lines and Correlated with Prognosis in EC Patients
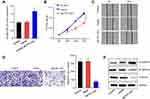
The expression levels of MCTP1-AS1 in EC tissues and adjacent normal tissues were evaluated by qRT-PCR. The results showed the significantly decreased expression of MCTP1-AS1 in tumor tissues compared with adjacent normal tissues (p<0.0001) (Figure 1A). 60 patients of endometrial cancer from Figure 1A were divided into MCTP1-AS1 low expression group (n=30) and MCTP1-AS1 high expression group (n=30) according to the expression level of MCTP1-AS1 in EC tissue. Regardless of patient age, higher MCTP1-AS1 expression was closely related to tumor size, TNM state, differentiation, lymph node metastasis and distant metastasis (p<0.001) (Table 1). Besides, we found that low MCTP1-AS1 expression was associated with significantly shorter overall survival as revealed by Kaplan-Meier analysis (Figure 1B). Moreover, the significant downregulation of MCTP1-AS1 in EC cell lines (especially Ishikawa cell line) was observed compared with normal cell line hEEC (p<0.01) (Figure 1C).
 |
Table 1 Relationship Between Clinico-Pathological Characteristics and MCTP1-AS1 Expression in EC Patients |
MCTP1-AS1 Inhibits Cell Proliferation, Cell Migration, Cell Invasion and EMT Process in EC Cells
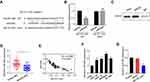
Overexpression of MCTP1-AS1 was achieved by transfection of Ishikawa cells with pcDNA3.1-MCTP1-AS1 and the efficiency of overexpression was verified by qRT-PCR (p<0.01) (Figure 2A). CCK8 assay proved that cell proliferation was inhibited in MCTP1-AS1 overexpression cells compared with control cells (p<0.01) (Figure 2B). In addition, wound-healing assay demonstrated that the migration ability of EC cells was decreased at 24 h upon MCTP1-AS1 overexpression (Figure 2C). Furthermore, the effect of overexpression of MCTP1-AS1 on cell invasion was detected by transwell experiment. The data showed that compared with control cells, cell invasion ability was significantly inhibited in MCTP1-AS1 overexpression cells (p<0.01) (Figure 2D). Moreover, MCTP1-AS1 overexpression significantly promoted the expression of E-cadherin but inhibited N-cadherin and Vimentin compared with control group (Figure 2E). These findings suggested that upregulation of MCTP1-AS1 profoundly inhibited cell proliferation, migration, invasion and EMT process of EC cells.
MCTP1-AS1 Reversely Regulated miR-650 Expression in EC Cells by Directly Targeting
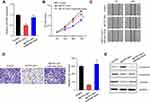
RNA22 (https://cm.jefferson.edu/rna22) was used to predict the potential binding sites between MCTP1-AS1 and miR-650, the result of which was shown in Figure 3A. Next, a dual-luciferase reporter assay was carried out to confirm the binding site between MCTP-AS1 and miR-650. Compared with miR-NC, overexpression of miR-650 inhibited the luciferase activity in 293T cells (p<0.01) while mutation of the predicted miR-650 binding site canceled out the inhibitory effect (Figure 3B). In addition, RIP was performed to verify the direct binding of MCTP1-AS1 and miR-650 (Figure 3C). Besides, miR-650 expression was suppressed in EC tissues in comparison to adjacent non-tumor tissues (p<0.01) (Figure 3D). In addition, an inverse correlation was found between MCTP1-AS1 and miR-650 expression by Pearson correlation analysis (p<0.0001) (Figure 3E). Furthermore, the expression levels of MCTP1-AS1 were increased in EC cells compared with normal cells (p<0.01) (Figure 3F). Meanwhile, we found that miR-650 expression was inhibited in MCTP1-AS1 overexpressed Ishikawa cells (p<0.01) (Figure 3G). Taken together, MCTP1-AS1 reversely regulated miR-650 expression in EC cells.
MCTP1-AS1 Regulated EMT Process via miR-650 in EC Cells
qRT-PCR result showed that miR-650 expression was downregulated in MCTP1-AS1 overexpressed EC cells, while co-transfecting with miR-650 mimic, the miR-650 expression level was partially upregulated (p<0.01) (Figure 4A). CCK8 assay proved that cell proliferation inhibited by MCTP1-AS1 overexpression could be drastically rescued by co-transfecting with miR-650 mimic (p<0.01) (Figure 4B). In addition, wound-healing assay demonstrated that the migration ability of EC cells was increased at 24 h upon MCTP1-AS1 and miR-650 mimic co-transfection compared with MCTP1-AS1 only (Figure 4C). Furthermore, the transwell experiment data showed that miR-650 transfection could significantly alleviate the MCTP1-AS1 inducing cell invasion ability inhibition in EC cells (p<0.01) (Figure 4D). Moreover, miR-650 transfection inhibited the expression level of E-cadherin compared with MCTP1-AS1 overexpression cells, while N-cadherin and Vimentin expression level were increased at the same time (Figure 4E). Therefore, MCTP1-AS1 might inhibit EC cells growth through downregulating the activity of miR-650.
MCTP1-AS1 Regulated SMAD7 Expression via miR-650 in EC Cells
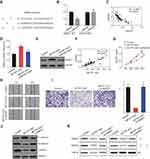
Bioinformatics analysis was performed to predict the potential binding sites between miR-650 and SMAD7 using RNA22 (https://cm.jefferson.edu/rna22) (Figure 5A). Next, the result of dual-luciferase reporter assay showed that compared with miR-NC, overexpression of miR-650 could inhibit the luciferase activity in 293T cells (p<0.001), which was not affected by mutating the predicted binding site between miR-650 and SMAD7 (Figure 5B). In addition, the correlation between miR-650 and SMAD7 was detected by Pearson correlation analysis, the result of which showed that the two molecules had an inverse correlation (p<0.0001) (Figure 5C). Furthermore, we found that mRNA and protein levels of SMAD7 were both dramatically increased in MCTP1-AS1 overexpression cells compared with negative control, while co-transfecting with miR-650, SMAD7 expression level was decreased (p<0.01) (Figure 5D and E). To determine whether MCTP1-AS1 regulated SMAD7 expression in EC cells, correlation between MCTP1-AS1 and SMAD7 was detected. As expected, a positive correlation was found between MCTP1-AS1 and SMAD7 mRNA expressions in EC tissues (p<0.0001) (Figure 5F). CCK8 results showed that cell proliferation was decreased when MCTP1-AS1 was overexpressed, which was reversed by SMAD7 knockdown through siRNA transfecting (p<0.0001) (Figure 5G). Similar results were found in both wound-healing and transwell assay (p<0.01) (Figure 5H and I). Meanwhile, the result of Western blot showed that SMAD7 knockdown could inhibit the expression of E-cadherin but promote the N-cadherin and Vimentin levels compared with MCTP1-AS1 overexpression cells, which proved that MCTP1-AS1 inhibited EMT process of EC cells through SMAD7 (Figure 5J). Besides, as SAMD7 plays an important role in SMAD2/3-TGF-β pathway, we detected the expression level of SMAD2/3, p-SMAD2/3 and TGF-β1 by Western blot. As shown in Figure 5K, overexpression of MCTP1-AS1 induced inhibition of this pathway, while knockdown of SMAD7 attenuated the inhibiting effect. In short, MCTP1-AS1 might regulate cell growth, migration, invasion and EMT via miR-650/SMAD7 axis in EC cells.
Discussion
MCTP1-AS1 expression has been recently discovered to be downregulated in a total of 45 pieces of pathologically proven endometrial cancer tissues comparing to its paired normal adjacent tissues by microarray analysis.14 Similarly, reduced MCTP1-AS1 expressions were found in 60 EC tissues and EC cell lines, especially Ishikawa cells in the current study, which was further proved to be related with poor prognosis in EC patients. However, few studies have studied MCTP1-AS1 in EC or other cancers, its expression and biological role in cancer development is still limited. In this study, our results showed that overexpressing of MCTP1-AS1 significantly inhibited cell growth, migration, invasion and EMT, which indicated that MCTP1-AS1 could potentially serve as a diagnostic biomarker that is beneficial for the diagnosis and therapy of endometrial cancer.
Studies have indicated that miR-650 upregulated in a wide variety of human tumors, such as gastric cancer, hepatocellular carcinoma, colorectal cancer, prostate Cancer and so on.21–24 Notably, previous study showed that miR-650 was upregulated in endometrial cancer tissues.1 Accordingly, we tested the expression level of miR-650 in EC tissues and corresponding adjacent normal tissues. The results showed that miR-650 expression was indeed significantly upregulated in EC tissues compared with adjacent normal tissues. Moreover, MCTP1-AS1 was predicted as a direct target of miR-650 using bioinformatics analysis and was further proved by dual-luciferase reporter assay in our study. Besides, the expression of miR-650 was negatively correlated with the expression of MCTP1-AS1 in EC tissues verified by qRT-PCR. Through co-transfecting with miR-650 mimic, we demonstrated that MCTP1-AS1 regulated EC development via miR-650 in EC cells.
SMAD7, as an inhibitor of TGF-β signaling, was confirmed to have direct binding sites with miR-650 by bioinformatics analysis, which expanded the potential target network of miR-650. Parikh et al indicated that in ovarian cancer, SMAD7 has been proved to regulate TGF-β signaling pathway by affecting the expression and activation of SMAD2/3.20 Kharma et al found that STAT1 drives tumor progression in serous papillary endometrial cancer via regulating SMAD7.25 Nevertheless, in cutaneous melanoma, high expression of SMAD7 was positively associated with several features of tumor aggressiveness.26 Besides, several studies demonstrated SMAD7 played an important role in many inflammation-related diseases.27–29 For example, Xu et al revealed that SMAD7 might play a suppressor role on the development of osteoarthritis.30 The conflicting results of these studies were probably due to its effect under differential conditions, which suggested that MCTP1-AS1 might regulate EC development through SMAD7.
In our study, we found SMAD7 was negatively correlated to the miR-650 expression, and positively correlated to the MCTP1-AS1 expression in EC tissues. MCTP1-AS1 overexpression inhibited EC cells growth, migration, invasion and EMT, while knockdown of SMAD7 expression by transfecting siRNA-SMAD7 rescued the inhibition effect induced by MCTP1-AS1 in EC cells. To detect the role of MCTP1-AS1 in regulating SMAD2/3-TGF-β signaling, SMAD2/3, p-SMAD2/3 and TGF-β1 expressions in EC cells were determined using Western blot. As expected, decreased SMAD2/3, p-SMAD2/3 and TGF-β1 expressions were found in the MCTP1-AS1 overexpression group compared with control group. While co-transfecting with siRNA-SMAD7, the expressions of SMAD2/3, p-SMAD2/3 and TGF-β1 were increased than MCTP1-AS1 overexpression group.
Taken together, our data indicate that the MCTP1-AS1/miR-650/SMAD7 axis functions as an important player in EC cells growth, migration, invasion and EMT, which may have diagnostic and therapeutic potential in EC.
Conclusion
MCTP1-AS1 and SMAD7 were downregulated in EC cells, while miR-650 was significantly upregulated in the EC cells. MCTP1-AS1 regulates the cell proliferation, migration, invasion and EMT in EC cells through regulation of miR-650/SMAD7 axis. Therefore, this study uncovered the expression and biological role of MCTP1-AS1 in EC, which could act as a potential therapeutic target in the future.
Data Sharing Statement
All data and materials supporting the conclusions were included in this paper. More details are available on request.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. McAlpine JN, Temkin SM, Mackay HJ. Endometrial cancer: not your grandmother’s cancer. Cancer. 2016;122:2787–2798. doi:10.1002/cncr.30094
2. Chang Z, Talukdar S, Mullany SA, Winterhoff B. Molecular characterization of endometrial cancer and therapeutic implications. Curr Opin Obstet Gynecol. 2019;31:24–30. doi:10.1097/GCO.0000000000000508
3. Lee YC, Lheureux S, Oza AM. Treatment strategies for endometrial cancer: current practice and perspective. Curr Opin Obstet Gynecol. 2017;29:47–58. doi:10.1097/GCO.0000000000000338
4. Braun MM, Overbeek-wager EA, Grumbo RJ. Diagnosis and management of endometrial cancer. Am Fam Physician. 2016;93:468–474.
5. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi:10.1158/0008-5472.CAN-16-2634
6. Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74. doi:10.1007/978-981-10-5203-3_2
7. Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA interactions. Methods Mol Biol. 2016;1402:271–286. doi:10.1007/978-1-4939-3378-5_21
8. Yang X, Yan Y, Chen Y, Li J, Yang J. Involvement of NORAD/miR-608/STAT3 axis in carcinostasis effects of physcion 8-O-beta-glucopyranoside on ovarian cancer cells. Artif Cells Nanomed Biotechnol. 2019;47:2855–2865. doi:10.1080/21691401.2019.1637884
9. Zhang L, Li Y, Sona L. Long non-coding RNA RP11-480I12.5 promotes cervical carcinoma progression by regulating the Wnt/beta-catenin signaling pathway. Oncol Lett. 2020;19:469–475. doi:10.3892/ol.2019.11120
10. Wang XY, Qin YY. Long non-coding RNAs in biology and female reproductive disorders. Front Biosci. 2019;24:750–764. doi:10.2741/4748
11. Xie P, Cao H, Li Y, Wang J, Cui Z. Knockdown of lncRNA CCAT2 inhibits endometrial cancer cells growth and metastasis via sponging miR-216b. Cancer Biomark. 2017;21:123–133. doi:10.3233/CBM-170388
12. Shao W, Li Y, Chen F, et al. Long non-coding RNA DLEU1 contributes to the development of endometrial cancer by sponging miR-490 to regulate SP1 expression. Pharmazie. 2018;73:379–385. doi:10.1691/ph.2018.8352
13. Sun KX, Wu DD, Chen S, Zhao Y, Zong ZH. LncRNA MEG3 inhibit endometrial carcinoma tumorigenesis and progression through PI3K pathway. Apoptosis. 2017;22:1543–1552. doi:10.1007/s10495-017-1426-7
14. Yang L, Zhang J, Jiang A, et al. Expression profile of long non-coding RNAs is altered in endometrial cancer. Int J Clin Exp Med. 2015;8:5010–5021.
15. Ratner ES, Tuck D, Richter C, et al. MicroRNA signatures differentiate uterine cancer tumor subtypes. Gynecol Oncol. 2010;118:251–257. doi:10.1016/j.ygyno.2010.05.010
16. Han LL, Yin XR, Zhang SQ. miR-650 promotes the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by directly inhibiting LATS2 expression. Cell Physiol Biochem. 2018;51:1179–1192. doi:10.1159/000495495
17. Orlandella FM, Mariniello RM, Iervolino PLC, et al. miR-650 promotes motility of anaplastic thyroid cancer cells by targeting PPP2CA. Endocrine. 2019;65:582–594. doi:10.1007/s12020-019-01910-3
18. Reguly T, Wrana JL. In or out? The dynamics of Smad nucleocytoplasmic shuttling. Trends Cell Biol. 2003;13:216–220. doi:10.1016/S0962-8924(03)00075-8
19. Javelaud D, Mauviel A. Mammalian transforming growth factor-betas: Smad signaling and physio-pathological roles. Int J Biochem Cell Biol. 2004;36:1161–1165. doi:10.1016/S1357-2725(03)00255-3
20. Parikh A, Lee C, Joseph P, et al. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition. Nat Commun. 2014;5:2977. doi:10.1038/ncomms3977
21. Zhang X, Zhu W, Zhang J, et al. MicroRNA-650 targets ING4 to promote gastric cancer tumorigenicity. Biochem Biophys Res Commun. 2010;395:275–280. doi:10.1016/j.bbrc.2010.04.005
22. Zeng ZL, Li FJ, Gao F, Sun DS, Yao L. Upregulation of miR-650 is correlated with down-regulation of ING4 and progression of hepatocellular carcinoma. J Surg Oncol. 2013;107:105–110. doi:10.1002/jso.23210
23. You Q, Li H, Liu Y, et al. MicroRNA-650 targets inhibitor of growth 4 to promote colorectal cancer progression via mitogen activated protein kinase signaling. Oncol Lett. 2018;16:2326–2334. doi:10.3892/ol.2018.8910
24. Zuo Z-H, Yu YP, Ding Y, et al. Oncogenic activity of miR-650 in prostate cancer is mediated by suppression of CSR1 expression. Am J Pathol. 2015;185:1991–1999. doi:10.1016/j.ajpath.2015.03.015
25. Kharma B, Baba T, Matsumura N, et al. STAT1 drives tumor progression in serous papillary endometrial cancer. Cancer Res. 2014;74:6519–6530. doi:10.1158/0008-5472.CAN-14-0847
26. Kaczorowski M, Biecek P, Donizy P, et al. SMAD7 is a novel independent predictor of survival in patients with cutaneous melanoma. Transl Res. 2019;204:72–81. doi:10.1016/j.trsl.2018.09.002
27. Yan X, Liao H, Cheng M, et al. Smad7 protein interacts with receptor-regulated Smads (R-Smads) to inhibit transforming growth factor-beta (TGF-beta)/Smad signaling. J Biol Chem. 2016;291:382–392. doi:10.1074/jbc.M115.694281
28. Monteleone G, Kumberova A, Croft NM, et al. Blocking Smad7 restores TGF-beta1 signaling in chronic inflammatory bowel disease. J Clin Invest. 2001;108:601–609. doi:10.1172/JCI12821
29. Lan HY. Smad7 as a therapeutic agent for chronic kidney diseases. Front Biosci. 2008;13:4984–4992. doi:10.2741/3057
30. Xu J, Xu Y. The lncRNA MEG3 downregulation leads to osteoarthritis progression via miR-16/SMAD7 axis. Cell Biosci. 2017;7:69. doi:10.1186/s13578-017-0195-x
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.