Back to Journals » OncoTargets and Therapy » Volume 14
LncRNA FER1L4 Promotes Oral Squamous Cell Carcinoma Progression via Targeting miR-133a-5p/Prx1 Axis
Authors Zhang N , Zeng L , Wang S , Wang R, Yang R, Jin Z, Tao H
Received 17 August 2020
Accepted for publication 29 December 2020
Published 4 February 2021 Volume 2021:14 Pages 795—806
DOI https://doi.org/10.2147/OTT.S277351
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Nan Zhang,1,* Lingfang Zeng,2,* Shouyi Wang,3,* Ronghua Wang,4 Rui Yang,5 Zuolin Jin,6 Hong Tao1
1Department of Stomatology, The First Affiliated Hospital of Xi’an Jiaotong University, Xian, Shanxi, 710061, People’s Republic of China; 2Department of Pediatric Stomatology, Jinan Stomatological Hospital, Jinan, Shandong, 250000, People’s Republic of China; 3Department of Oral and Maxillofacial Surgery, Jinan Stomatological Hospital, Jinan, Shandong, 250000, People’s Republic of China; 4Department of Pediatrics, The First Affiliated Hospital of Xi’an Jiaotong University, Xian, Shanxi, 710061, People’s Republic of China; 5Department of Dental, Xi ‘an Tianrui Institute of Stomatology, Xian, Shanxi, 710061, People’s Republic of China; 6Department of Orthodontics, Oral Hospital of the Fourth Military Medical University, Xian, Shanxi, 710032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zuolin Jin
Department of Orthodontics, Oral Hospital of the Fourth Military Medical University, No. 145, Changle Weste Road, Xian, Shanxi, 710032, People’s Republic of China
Email [email protected]
Hong Tao
Department of Stomatology, The First Affiliated Hospital of Xi’an Jiaotong University, No. 227, Yanta West Road, Xian, Shanxi, 710061, People’s Republic of China
Email [email protected]
Background: Oral squamous cell carcinoma (OSCC) is a common cancer especially young people in the world. The long non-coding RNA Fer-1-like protein 4 (FER1L4) has been reported to be closely associated with the progression of various human cancers. However, the role of FER1L4 in OSCC remains unclear.
Methods: The expression level of FER1L4 in OSCC tissues and cancer cell lines was detected by using quantitative real-time polymerase chain reaction (qRT-PCR). Cell proliferation was evaluated by cell counting kit-8 (CCK-8) assay and EdU staining assay. Cell invasion and migration were evaluated by Transwell assay. Cell apoptosis was detected by flow cytometry. Luciferase reporter assay was performed to determine the targeting relationship between FER1L4, miR-133a-5p and Prx1. The protein expression of Prx1 was detected by Western blot. In addition, a xenograft tumor model in vivo was constructed to confirm the function of FER1L4.
Results: FERIL4 was significantly upregulated in OSCC tissues and cancer cell lines. Moreover, high level of FER1L4 predicted a poor prognosis of OSCC patients. Silencing of FER1L4 not only significantly inhibited cell growth, invasion, migration and induced apoptosis in SCC-9 and HN4 cells in vitro, but also effectively suppressed the tumorigenesis of OSCC cells in vivo. Knockdown of FER1L4 significantly enhanced the expression of miR-133a-5p by sponging it, and then downregulated Prx1 expression.
Conclusion: Our study elucidated a new mechanism of lncRNA FER1L4 that promoting OSCC progression by directly targeting miR-133a-5p/Prx1 axis and provided novel therapeutic targets for OSCC.
Keywords: OSCC, lncRNA FER1L4, miR-133a-5p, Prx1, tumorigenesis
Introduction
Oral squamous cell carcinoma (OSCC) has become a common cancer worldwide due to tumor invasion, orofacial destruction, cervical lymph node metastasis and ultimate blood-borne dissemination.1 It has been reported that approximately 300,000 cases with OSCC are diagnosed every year recently also with an obvious rise in incidence.2 Although numbers of critical progresses have been achieved in recent decades, the overall 5-year survival rate of OSCC patients is still less than 50%.3 Chemotherapy is an efficient treatment for OSCC, while the emergence and development of drug resistance limit the therapeutic effects of chemotherapy drugs and lead to a median overall survival time of OSCC patients with approximately 6 to 9 months.4,5 Therefore, a well understanding of specific molecular mechanisms in OSCC progression contributes to identify and develop new therapeutic strategies for OSCC.
Long chain non-coding RNAs (lncRNAs) are a group of RNAs longer than 200 nucleotides and lack protein coding function, and lncRNAs can regulate the expression of target genes at the post-transcriptional level in eukaryotic cells.6 Recently, lncRNA FER1L4 (Fer-1-like protein 4) has been identified to be closely associated with the development of various human cancers. FER1L4 induces apoptosis and inhibits epithelial-mesenchymal transition (EMT) in osteosarcoma cells through miR-18a-5p-mediated upregulation of SOCS5.7 FER1L4 suppresses the invasion and growth of tumor cells in esophageal squamous cell carcinoma (ESCC), and silencing of FER1L4 promotes the invasion, proliferation and inhibits apoptosis of ESCC cells in vitro.8 FER1L4 also inhibits cell proliferation and cycle by targeting PTEN in endometrial carcinoma.9 FER1L4 overexpression inhibits paclitaxel tolerance of ovarian cancer cells through MAPK signaling pathway.10 In addition, FER1L4 has also been identified to act an important role in renal-cell carcinoma, hepatocellular carcinoma, gastric cancer and lung cancer, and so on.11–14 However, its role in OSCC progression remains unclear.
Peroxiredoxin 1 (Prx1) which is a major member of Prxs family and has been demonstrated to participate in a number of biological processes including oxidative stress, cell growth and apoptosis.15 High expression of Prx1 is shown to promote invasion and migration capacity through regulating EMT in OSCC progression.16,17 Wang et al demonstrated that nicotine inhibits cell apoptosis and promotes the growth of oral precancerous lesion cells by directly regulating α7nAChR/Prx1 during carcinogenesis of OSCC.18 MiR-133a-5p has been known as an anti-cancer factor, and can inhibit androgen receptor (AR)-induced proliferation through targeting Fused in prostate cancer.19 In addition, propofol has been revealed that can protect against hepatic I/R injury through targeting miR-133a-5p/MAPK6 axis.20 MiR-133a-5p also can downregulate osteoblast differentiation-associated genes by directly targeting the 3ʹ UTR of RUNX2.21 However, the regulatory networks involved in miR-133a-5p and Prx1 in OSCC are lack.
In this study, we provided a new regulatory pathway in OSCC progression, that was, FER1L4 promoted OSCC progression through directly targeting miR-133a-5p/Prx1 axis. Our results extended the understanding of the roles between lncRNAs and miRNAs in OSCC progression and suggested that FER1L4/miR-133a-5p/Prx1 axis might be potential therapeutic targets for OSCC treatment.
Materials and Methods
Patient Tissues
Total of 45 OSCC patients were recruited in this study and all patients had given their written informed consent. The relevant clinical information of OSCC patients including age, sex, and histopathological characteristics are shown in Supplementary Table 1. All OSCC tissues and matched adjacent normal tissues were collected within 15 min after being removed from the patients by surgical resection and immediately snap-frozen in liquid nitrogen before storage at –80°C. The expression level of target genes between OSCC tissues and matched adjacent normal tissues was evaluated. All OSCC patients were used to evaluate the prognostic value based on the median expression of lncRNA FER1L4. This study was approved by the human Ethic Community of The First Affiliated Hospital of Xi’an Jiaotong University and the research has been carried out in accordance with the World Medical Association Declaration of Helsinki.
Cell Culture
The OSCC cell lines including SCC-9, SCC-25, HN4, Tca-811 and hNOK cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Human normal oral keratinocyte cell line hNOK cells were used as the negative control. For cell lines, we extracted DNA from various cells (SCC 9, SCC 25, HN4, Tca 811 and hNOK) using the Genome extraction kit of Axygen, amplified by the 21-STR amplification scheme. And then we detected STR spot and sex gene Amelogenin on the ABI 7900HT Fast Real-Time PCR System (Supplementary Table 2–6). No multiple alleles were found in these cell lines in this test. All cells were cultured in DMEM medium supplemented with 10% FBS and 100 μg/mL penicillin/streptomycin at 37°C with 5% CO2.
Cell Transfection
For cell transfection, si-FER1L4, miR-133a-5p mimics, miR-133a-5p inhibitor and negative controls (miR-NC and inhibitor NC) were transfected into cells by using Lipofectamine 2000 kit. To construct Prx1 overexpression plasmids, the cDNA sequence of Prx1 was cloned into pcD-ciR vector, which was purchased from Geneseed Biotech Co., Ltd., followed by transfection by Lipofectamine 2000. After transfection for 48 h, cells were collected and used for the subsequent experiments.
qRT-PCR
Total RNA of cultured cells or tissue samples was extracted by using the TRIzol reagent according to the manufacturer’s instructions. The cDNA was synthesized by using the iScript™ cDNA Synthesis Kit. Quantitative Real Time-PCR (qRT-PCR) was performed by using the SYBR Premix Ex Taq II kit and ABI 7900HT Fast Real-Time PCR System. The relative expression levels of target genes were analyzed by 2−ΔΔCt method.22 And GAPDH and U6 were considered as the internal references. The primers used as follows: lncRNA FER1L4: forward primer 5′- CTTAATCACTTGGCGGGAA-3′, reverse primer 5′- CGCTTAGATTCCAAAGG-3′; miR-133a-5p: forward primer: 5′- CTGAATGTGGAGAGAATGT-3′, reverse primer: 5′-GTTCCTCGACATTCGGGCCG-3′; Prx1: forward primer: 5′-CCACAGGATCCGGGAAACT-3′, reverse primer: 5′-GAGGTCATAACAGCGGTGCC-3′; GAPDH: forward primer: 5′-CGAGAGAATCCGCGGACAT-3′, reverse primer: 5′-TTGTGCAATACAGCGTGGAC-3′; U6: forward primer: 5′-CGAGAGAATCCGCGGACAT-3′, reverse primer: 5′-TTGTGCAATACAGCGTGGAC-3′.
Western Blot
Total protein of cultured cells or tumor tissues was extracted by RIPA Lysis Buffer. Approximately 50 μg protein samples were separated by 12% SDS-PAGE and transferred into PVDF membranes. After blocking with 5% non-fat milk, the membranes were incubated with primary antibodies Prx1 (1:5000, Ab2572, Abcam), and GAPDH (1:1000, Ab8227, Abcam) overnight at 4°C. Subsequently, the membranes were induced with horseradish peroxidase-labeled IgG for 1 h, and the bands of target genes were visualized by using a Bio-Rad imaging system.
Cell Counting-8 (CCK-8) Assay
Cell viability was evaluated by using Cell Counting-8 kit (Dojundo) according to the supplier’s protocols. Briefly, 2 × 103 cells were plated into a 96-well plate and cultured for 24, 48, 72 and 96 h, then 10 μL of CCK-8 reagent was added to each well and the absorbance was detected at a wavelength of 450 nm by a microplate reader. And the difference of cell viability between groups was analyzed at 96 h.
Edu Staining Assay
Cell proliferation was measured by EdU (5-ethynyl-20-deoxyuridine) assay using Cell-Light EdU DNA Cell Proliferation Kit. Briefly, 1×104 cells were plated into 96-well plates and induced for 48 h at 37°C with 5% CO2. 50 mM EdU was added into each well and incubated for another 2 h. Cells were then fixed with 4% paraformaldehyde and stained with Apollo Dye Solution for proliferation detection. Images were captured by using a fluorescence microscope.
Transwell Assay
Cell migration and invasion were determined by using 8.0-μm-pore filters in a Transwell assay as previously described.23 For invasion assay, cells (2×105 cells/well) were diluted in 200 µL serum-free DMEM and inoculated onto the upper wells of a Transwell chamber coated with 20 µL Matrigel (BD Biosciences). DMEM with 10% FBS was added into the lower chambers, which were incubated for 48 h. Then the non-invasive cells were removed by using a cotton swab following overnight incubation. The invasive cells in the lower chambers were fixed and stained with 100% pre-cold methanol and 0.1% crystal violet at room temperature for 30 min. For migration assay, cells were suspended in FBS-free DMEM with 1 µg/mL Mitomycin C and plated in the upper wells of the Transwell chamber and DMEM with 10% FBS was mixed into the lower chambers. Then the following steps were similar to the invasion assay. After 24 h of incubation, cells had invaded and migrated into the lower surface were photographed under a microscope (magnification ×200). Five visual fields of each chamber were randomly chosen.
Flow Cytometry
Approximately 5×105 cells/mL Cells were washed with PBS twice and resuspended in the tube with 195 μL binding buffer (1×). Then, 5 μL Annexin V-FITC solution was added in the tube and incubated for 15 min. After centrifugation, cells were resuspended in solution containing 190 μL binding buffer and stained with 10 μL of propidium iodide (PI). Finally, apoptosis assay was performed with flow cytometry as previously described.24
Luciferase Reporter Assay
The wild-type (WT), mutant (MUT) of FER1L4P and Prx1 were amplified with genomic DNA as the template and cloned into pmirGLO vector. pmirGLO-FER1L4-WT, pmirGLO-FER1L4-MUT, pmirGLO-Prx1-WT or pmirGLO-Prx1-MUT was co-transfected with miR-133a-5p mimics or negative control (miR-NC) into cells by Lipofectamine 2000 kit. After transfection for 48 h, cells were lysed and the relative luciferase activity was measured by using the dual luciferase reporter detection system.
Xenograft Model
A total of 40 male BALB/c nude mice (approximately 19–22 g, 6 weeks old) were obtained from Animal Center of the Chinese Academy of Science (Shanghai, China). All mice were housed and maintained under specific pathogen-free conditions at 18–22°C, with 20% humidity, a 12/12 h light/dark cycle. Approximately 5 × 106 SCC-9 cells transfected with sh-NC, sh-FER1L4, miR-133a-5p inhibitor or co-transfection with sh-FER1L4 and miR-133a-5p inhibitor for 48 h (n = 6), and then separately subcutaneously inoculated into the right flank of the dorsal of each nude mouse to construct the xenograft mice model as previously described.25 The tumor volume was calculated by the formula (length × width2)/2 every week for five weeks. Finally, mice were killed and tumors were obtained and weighed. All animal experiments were approved by the Animal Ethical and Welfare Committee (AEWC) of The First Affiliated Hospital of Xi’an Jiaotong University and in accordance with the Guidelines for Care and Use of Laboratory Animals of “The First Affiliated Hospital of Xi’an Jiaotong University”.
Statistical Analysis
Data were shown as mean ± SD method by GraphPad Prism 5 software. Difference between two groups was determined with two-tailed Student’s t-test and difference among multiple groups was tested by one-way ANOVA. Kaplan-Meier plot was applied to analyze the survival rate of OSCC patients based on the expression median of FER1L4. The correlation between FER1L4 and miR-133a-5p, as well as miR-133a-5p and Prx1 in OSCC tissues was determined by Spearman correlation analysis. Each experiment was repeated three times. With p < 0.05 as the significant threshold.
Results
LncRNA FER1L4 Was Significantly Upregulated in OSCC Tissues and Predicted a Poor Prognosis of OSCC Patients
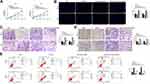
To explore the role of lncRNA FER1L4 in OSCC progression, we first detected the expression of FER1L4 in OSCC tissues and corresponding cancer cell lines. We found that FER1L4 was highly expressed in OSCC tissues compared with matched adjacent normal tissues of OSCC patients (n = 45) (p < 0.01) (Figure 1A). Then, we also evaluated the expression level of FER1L4 between OSCC tissues and matched adjacent normal tissues and the results showed that FER1L4 was significantly elevated in most OSCC tissues compared with that in matched adjacent normal tissues of OSCC patients (n = 45) (p < 0.01) (Figure 1B). To confirm this result, we detected the expression of FER1L4 in OSCC cell lines and found that FER1L4 was also upregulated in various OSCC cell lines including SCC-9, SCC-25, HN4 and Tca-8113 compared with that in the normal cell line hNOK (p < 0.01) (Figure 1C). Moreover, all OSCC patients were divided into two groups based on the expression median of FER1L4 and followed up for 36 months, and survival analysis revealed that OSCC patients in the high FER1L4 level group significantly predicted a poor overall survival compared with that in the low FER1L4 level group (n = 45) (p < 0.01) (Figure 1D). These results suggested a potential oncogenic role of lncRNA FER1L4 in OSCC.
Silencing of FER1L4 Inhibited the Growth of OSCC Cell Lines in vitro
To further examine the oncogenic role of FER1L4, si-FER1L4 (silencing of FER1L4) was transfected into OSCC cell line SCC-9 and HN4 cells to silence the expression of FER1L4. The transfection efficiency was evaluated by qRT-PCR assay and the results showed that si-FER1L4 significantly decreased the expression of FER1L4 in both SCC-9 and HN4 cells (p < 0.001) (Figure 2A). Cell viability of SCC-9 and HN4 cells was obviously inhibited by si-FER1L4 compared with si-NC (p < 0.01) (Figure 2B). Meanwhile, si-FER1L4 significantly decreased the numbers of Edu positive cells in both SCC-9 and HN4 cells compared with si-NC (p < 0.01) (Figure 2C). In addition, si-FER1L4 not only significantly inhibited invasion ability (p < 0.01) (Figure 2D), but also inhibited migration ability (p < 0.01) (Figure 2E) compared with si-NC in both SCC-9 and HN4 cells. As expected, si-FER1L4 remarkedly promoted apoptosis of SCC-9 and HN4 cells compared with si-NC (p < 0.01) (Figure 2F). The results revealed that si-FER1L4 exerted a potent inhibitory role in OSCC progression in vitro.
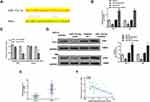
LncRNA FER1L4 Served as a Sponge of miR-133a-5p
We next predicted the target microRNAs (miRNAs) of FER1L4 based on the web tool Starbase, and the results indicated that FER1L4 had a putative binding site with miR-133a-5p (Figure 3B). To determine the specific interaction between LER1L4 and miR-133a-5p, miR-133a-5p mimic or miR-NC was transfected into SCC-9 and HN4 cells and qRT-PCR assay showed that miR-133a-5p mimic significantly increased miR-133a-5p expression compared with miR-NC both in SCC-9 and HN4 cells (p < 0.01) (Figure 3A). The luciferase reporter plasmid FER1L4-WT or FER1L4-MUT containing the putative binding sites with miR-133a-5p was co-transfected with miR-133a-5p mimics or miR-NC into SCC-9 and HN4 cells and luciferase reporter assay was performed. The results revealed that miR-133a-5p mimics significantly decreased the relative luciferase activity of FER1L4-WT compared with miR-NC in both SCC-9 and HN4 cells (p < 0.01), while exhibited no obvious change on FER1L4-MUT (Figure 3C). Further, si-FER1L4 significantly upregulated the expression of miR-133a-5p and pc-FER1L4 (overexpression of FER1L4) inhibited miR-133a-5p expression in both SCC-9 and HN4 cells (p < 0.01) (Figure 3D). Meanwhile, miR-133a-5p was obviously downregulated in various OSCC cell lines compared with that in the normal cell line hNOK (p < 0.01) (Figure 3E). As similar to the results in vitro, miR-133a-5p was also remarkedly downregulated in OSCC tissues compared with that in matched adjacent normal tissues (p < 0.01) (Figure 3F). Meanwhile, a strong negative correlation was observed between the expression of FER1L4 and miR-133a-5p in OSCC tissues (p < 0.001) (Figure 3G). All the results indicated that FER1L4 served as a sponge of miR-133a-5p.
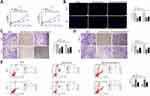
MiR-133a-5p Inhibitor Effectively Reversed Si-FER1L4-Induced Inhibition on Migration and Invasion in vitro
We further explored whether FER1L4 promoted OSCC progression through miR-133a-5p. SCC-9 and HN4 cells were transfected with si-NC, si-FER1L4, miR-133a-5p inhibitor or co-transfected with si-FER1L4 and miR-133a-5p inhibitor. CCK-8 assay indicated that si-FER1L4 inhibited the cell viability (p < 0.01), and miR-133a-5p inhibitor significantly promoted cell viability of SCC-9 and HN4 cells (p < 0.05), while co-transfection of si-FER1L4 with miR-133a-5p inhibitor eliminated the effect of si-FER1L4 induced inhibition on cell proliferation (p < 0.05) (Figure 4A). Meanwhile, si-FER1L4 decreased the number of EdU positive cells in both SCC-9 and HN4 cells (p < 0.01), miR-133a-5p inhibitor increased the number of EdU positive cells (p < 0.01), while co-transfection of si-FER1L4 with miR-133a-5p inhibitor significantly reversed si-FER1L4 induced decrease in the number of EdU positive cells in two cells (p < 0.05) (Figure 4B). MiR-133a-5p inhibitor significantly promoted invasion (p < 0.01) and migration (p < 0.05) capacity in both SCC-9 and HN4 cells, and co-transfection with miR-133a-5p inhibitor and si-FER1L4 could obviously reverse si-FER1L4 induced inhibition on invasion (p < 0.05) and migration capacity in two cells (p < 0.05) (Figure 4C and D). In addition, miR-133a-5p inhibitor inhibited the apoptosis of SCC-9 and HN4 cells (p < 0.05), while co-transfection with si-FER1L4 and miR-133a-5p inhibitor significantly reversed the effect of si-FER1L4 on cell apoptosis (p < 0.05) (Figure 4E). These results indicated that FER1L4 promoted OSCC progression though directly targeting miR-133a-5p.
Prx1 Was a Target of miR-133a-5p
To explore the specific mechanism of FER1L4/miR-133a-5p axis, we predicted the potential targets of miR-133a-5p by using TargetScan. The results showed that Prx1 might be a potential target of miR-133a-5p (Figure 5A). Luciferase reporter assay showed that miR-133a-5p mimic significantly reduced the luciferase activity of Prx1-WT compared with miR-NC, and exhibited no significant change in Prx1-MUT in both Huh7 and HepG2 cells (p < 0.01) (Figure 5B). Meanwhile, miR-133a-5p mimic significantly downregulated the mRNA level of Prx1 compared with miR-NC, and miR-133a-5p inhibitor increased Prx1 expression compared with inhibitor NC in two cells (p < 0.01) (Figure 5C). The protein level of Prx1 was similar to the mRNA trend, miR-133a-5p mimic significantly decreased the protein level of Prx1 compared with miR-NC, and miR-133a-5p inhibitor increased the protein expression of Prx1 compared with inhibitor NC in two cells (p < 0.01) (Figure 5D). Moreover, Prx1 was significantly upregulated in OSCC tissues compared with that in matched adjacent normal tissues (p < 0.01) (Figure 5E). And in OSCC tissues, the expression levels of Prx1 were negatively correlated with miR-133a-5p expression (p < 0.001) (Figure 5F). These results indicated that Prx1 was a direct target of miR-133a-5p.
MiR-133a-5p Mimic Inhibited OSCC Progression Through Prx1
To explore whether miR-133a-5p play its role through inhibiting the expression of Prx1 in OSCC progression, SCC-9 and HN4 cells were transfected with miR-NC, miR-133a-5p mimic, or co-transfected with miR-133a-5p mimic and pc-Prx1 (overexpression of Prx1). qRT-PCR assay showed that miR-133a-5p mimic significantly inhibited the cell proliferation in both SCC-9 and HN4 cells (p < 0.01), while co-transfection with miR-133a-5p and pc-Prx1 obviously reversed miR-133a-5p mimic-induced growth defect (p < 0.05) (Figure 6A). MiR-133a-5p mimic significantly decreased the number of EdU positive cells in both SCC-9 and HN4 cells (p < 0.01), while co-transfection with miR-133a-5p and pc-Prx1 obviously increased the number of EdU positive cells induced by miR-133a-5p mimic (p < 0.05) (Figure 6B). MiR-133a-5p mimic significantly reduced cell invasion and migration capacity in both SCC-9 and HN4 cells (p < 0.01), while co-transfection with miR-133a-5p mimic and pc-Prx1 obviously reversed miR-133a-5p mimic-induced effect on cell invasion (p < 0.05) (Figure 6C) and migration (p < 0.05) (Figure 6D). In addition, flow cytometry assay indicated that miR-133a-5p mimic significantly promoted cell apoptosis in both SCC-9 and HN4 cells (p < 0.01), while co-transfection with miR-133a-5p mimic and pc-Prx1 obviously reversed the effect of miR-133a-5p mimic on cell apoptosis (p < 0.05) (Figure 6E). These results indicated that miR-133a-5p mimic inhibited the growth of OSCC cells through Prx1 in vitro.
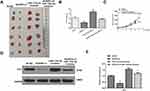
LncRNA FER1L4 Promoted OSCC Progression Through Targeting miR-133a-5p/Prx1 Axis in vivo
To further determine the role of FER1L4/miR-133a-5p/Prx1 axis in vivo, SCC-9 cells were transfected with sh-NC, sh-FER1L4, miR-133a-5p inhibitor or co-transfected with sh-FER1L4 and miR-133a-5p inhibitor, and then separately subcutaneously inoculated into nude mice for the construction of the xenograft tumor model in vivo. The results indicated that sh-FER1L4 significantly decreased the tumor weight and tumor volume compared with sh-NC group, miR-133a-5p inhibitor significantly increased tumor weight and tumor volume, while co-transfection with sh-FER1L4 and miR-1331-5p inhibitor obviously reversed the inhibitory effect of sh-FER1L4 (Figure 7A–C). In addition, we detected the expression of Prx1 in the tumor tissues isolated from different groups of mice, and the results indicated that sh-FER1L4 significantly downregulated the expression of Prx1 compared with sh-NC group (p < 0.01), and miR-133a-5p inhibitor significantly upregulated the protein level of Prx1 (p < 0.001), while co-transfection with sh-FER1L4 and miR-133a-5p inhibitor obviously reversed the inhibitory effect of sh-FER1L4 on the Prx1 expression (p < 0.05) (Figure 7D and E). These results indicated that FER1L4 promoted OSCC progression through miR-133a-5p mediated upregulation of Prx1.
Discussion
Increasing evidences indicated that the abnormal expressed lncRNAs widely participate in various biological processes in OSCC progression.26 LncRNA CASC9 has been identified to promote tumor progression through inhibiting autophagy-mediated cell apoptosis mediated by AKT/mTOR pathway.27 LncRNA AC007271.3 promotes cell proliferation, migration, invasion and inhibits cell apoptosis through regulating Wnt/β-catenin signaling pathway.23 Silencing of lncRNA LEF1-AS1 suppresses the progression of OSCC through targeting the Hippo signaling pathway.28 LncRNA-p23154 has been reported that can promote the invasion-metastasis potential of OSCC cells by regulating Glut1-mediated glycolysis.29 lncRNA PLAC2 promotes cell proliferation and invasion through activating Wnt/β-catenin pathway in OSCC progression.30 LncRNA LUCAT1 promotes growth, invasion and migration of OSCC cells by targeting PCNA.31 Although lncRNA FER1L4 has been identified to play crucial roles in various human cancers, its specific functions in OSCC have not been studied. In this study, we demonstrated for the first time that FER1L4 was significantly upregulated in OSCC tissues and OSCC cell lines. Meanwhile, silencing of FER1L4 remarkedly inhibited proliferation, invasion, migration, but promoted apoptosis of OSCC cells in vitro. These results revealed that lncRNA FER1L4 functioned as an oncogenic role in OSCC and suggested that FER1L4 might be a novel biomarker for the diagnosis and treatment of OSCC.
MiR-133a-5p has been identified to play important roles in human diseases. For instance, Chen et al demonstrated that a novel circular RNA, circFGFR2, can significantly promote skeletal muscle proliferation and differentiation by inhibiting miR-133a-5p.32 CircP4HB promotes the aggressiveness and metastasis non-small cell lung carcinoma (NSCLC) through targeting miR-133a-5p.33 Moreover, lncRNAs always act as a competing endogenous RNA (ceRNA) of miRNAs in OSCC progression. For example, lncRNA UCA1 promotes proliferation and cisplatin resistance of OSCC by suppressing the expression of miR-184.34 LncRNA H1 promotes cell proliferation and invasion by acting as a ceRNA of miR-138 and increasing the release of EZH2 in OSCC.35 LncRNA HOXA11-AS can promote cell proliferation and cisplatin resistance of OSCC through the inhibition of miR-214-3p.36 LncRNA LINC01234 facilitates cell growth and invasiveness of OSCC through targeting miR-637/NUPR1 axis.37 LncRNA OIP5-AS1 promotes the progression of OSCC through regulating miR-338-3p/NRP1 axis.38 To explore whether FER1L4 play its roles through miRNAs in OSCC, we predicted the potential targets of FER1L4 and found that miR-133a-5p of FER1L4. Luciferase reporter assay determined that FER1L4 served as a sponge of miR-133a-5p. Moreover, the function assay was performed and found that co-transfection with si-FER1L4 and miR-133a-5p inhibitor significantly reversed the inhibitory effect of si-FER1L4 on cell proliferation, invasion, migration and apoptosis in vitro, as well as the tumorigenesis of OSCC cells in vivo. These results all demonstrated that FER1L4 promoted OSCC progression though directly targeting miR-133a-5p. Our study extended a new regulatory mechanism of lncRNAs and miRNAs involved in OSCC progression.
Next, we predicted the targets of miR-133a-5p by TargetScan and GENECARD database, and identified 633 potential targets (Supplementary Table 7). Increasing evidences demonstrated that Prx1 is significantly associated with the progression of OSCC. For example, Niu et al demonstrated that Prx1 might play an oncogenic role in tobacco-related OSCC through activating the NF-κB pathway in OSCC cell line SCC15, and silencing of Prx1 obviously suppressed the nicotine-induced EMT, cell invasion and migration of SCC15 cells.16 However, the regulatory network involved in Prx1 is lack. In this study, Prx1 was identified to be a direct target of miR-133a-5p, and luciferase reporter assay confirmed their relationship. Moreover, co-transfection with miR-133a-5p mimic and pc-Prx1 obviously reversed miR-133a-5p mimic-induced effect on cell growth, proliferation, invasion, migration and apoptosis. To further confirm the role of FER1L4/miR-133a-5p/Prx1 axis in OSCC, a xenograft tumor model in vivo was established and confirmed that silencing of FER1L4 attenuated the tumorigenesis of OSCC cells by regulating miR-133a-5p/Prx1 axis.
However, overexpression of Prx1 should be designed to determine the role of FER1L4/miR-133a-5p in OSCC with the xenograft model. In addition, the effect of PER1L4 on histopathological features such as intratumor inflammation and edema during OSCC should be detailly studied in the future.
Conclusion
In summary, we demonstrated that lncRNA FER1L4 promoted OSCC progression through directly targeting miR-133a-5p/Prx1 axis both in vitro and in vivo. Our results revealed a new mechanism of lncRNA in OSCC progression and provided a novel therapeutic target for OSCC treatment.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jiang S, Dong Y. Human papillomavirus and oral squamous cell carcinoma: a review of HPV-positive oral squamous cell carcinoma and possible strategies for future. Curr Probl Cancer. 2017;41:323–327. doi:10.1016/j.currproblcancer.2017.02.006
2. Thomson PJ. Perspectives on oral squamous cell carcinoma prevention-proliferation, position, progression and prediction. J Oral Pathol Med. 2018;47:803–807. doi:10.1111/jop.12733
3. Kessler P, Grabenbauer G, Leher A, Bloch-Birkholz A, Vairaktaris E, Neukam FW. Neoadjuvant and adjuvant therapy in patients with oral squamous cell carcinoma long-term survival in a prospective, non-randomized study. Br J Oral Maxillofac Surg. 2008;46:1–5. doi:10.1016/j.bjoms.2007.08.006
4. Vermorken JB, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–1127. doi:10.1056/NEJMoa0802656
5. Perez-Sayans M, Somoza-Martin JM, Barros-Angueira F, Diz PG, Rey JM, Garcia-Garcia A. Multidrug resistance in oral squamous cell carcinoma: the role of vacuolar ATPases. Cancer Lett. 2010;295:135–143. doi:10.1016/j.canlet.2010.03.019
6. Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi:10.1038/ng.3192
7. Ye F, Tian L, Zhou Q, Feng D. LncRNA FER1L4 induces apoptosis and suppresses EMT and the activation of PI3K/AKT pathway in osteosarcoma cells via inhibiting miR-18a-5p to promote SOCS5. Gene. 2019;721:144093. doi:10.1016/j.gene.2019.144093
8. Ma W, Zhang CQ, Li HL, et al. LncRNA FER1L4 suppressed cancer cell growth and invasion in esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2018;22:2638–2645. doi:10.26355/eurrev_201805_14958
9. Qiao Q, Li H. LncRNA FER1L4 suppresses cancer cell proliferation and cycle by regulating PTEN expression in endometrial carcinoma. Biochem Biophys Res Commun. 2016;478:507–512. doi:10.1016/j.bbrc.2016.06.160
10. Liu S, Zou B, Tian T, et al. Overexpression of the lncRNA FER1L4 inhibits paclitaxel tolerance of ovarian cancer cells via the regulation of the MAPK signaling pathway. J Cell Biochem. 2018.
11. Wang X, Dong K, Jin Q, Ma Y, Yin S, Wang S. Upregulation of lncRNA FER1L4 suppresses the proliferation and migration of the hepatocellular carcinoma via regulating PI3K/AKT signal pathway. J Cell Biochem. 2019;120:6781–6788. doi:10.1002/jcb.27980
12. Cox A, Tolkach Y, Kristiansen G, Ritter M, Ellinger J. The lncRNA Fer1L4 is an adverse prognostic parameter in clear-cell renal-cell carcinoma. Clin Transl Oncol. 2020;22:1524–1531. doi:10.1007/s12094-020-02291-0
13. Gao X, Wang N, Wu S, Cui H, An X, Yang Y. Long noncoding RNA FER1L4 inhibits cell proliferation and metastasis through regulation of the PI3K/AKT signaling pathway in lung cancer cells. Mol Med Rep. 2019;20:182–190. doi:10.3892/mmr.2019.10219
14. Liu Z, Shao Y, Tan L, Shi H, Chen S, Guo J. Clinical significance of the low expression of FER1L4 in gastric cancer patients. Tumour Biol. 2014;35:9613–9617. doi:10.1007/s13277-014-2259-4
15. Kim YJ, Lee WS, Ip C, Chae HZ, Park EM, Park YM. Prx1 suppresses radiation-induced c-Jun NH2-terminal kinase signaling in lung cancer cells through interaction with the glutathione S-transferase Pi/c-Jun NH2-terminal kinase complex. Cancer Res. 2006;66:7136–7142. doi:10.1158/0008-5472.CAN-05-4446
16. Niu W, Zhang M, Chen H, et al. Peroxiredoxin 1 promotes invasion and migration by regulating epithelial-to-mesenchymal transition during oral carcinogenesis. Oncotarget. 2016;7:47042–47051. doi:10.18632/oncotarget.9705
17. Zhang M, Hou M, Ge L, et al. Induction of peroxiredoxin 1 by hypoxia regulates heme oxygenase-1 via NF-kappaB in oral cancer. PLoS One. 2014;9:e105994. doi:10.1371/journal.pone.0105994
18. Wang C, Niu W, Chen H, et al. Nicotine suppresses apoptosis by regulating alpha7nAChR/Prx1 axis in oral precancerous lesions. Oncotarget. 2017;8:75065–75075. doi:10.18632/oncotarget.20506
19. Zheng L, Kang Y, Zhang L, Zou W. MiR-133a-5p inhibits androgen receptor (AR)-induced proliferation in prostate cancer cells via targeting FUsed in Sarcoma (FUS) and AR. Cancer Biol Ther. 2020;21:34–42. doi:10.1080/15384047.2019.1665393
20. Hao W, Zhao ZH, Meng QT, Tie ME, Lei SQ, Xia ZY. Propofol protects against hepatic ischemia/reperfusion injury via miR-133a-5p regulating the expression of MAPK6. Cell Biol Int. 2017;41:495–504. doi:10.1002/cbin.10745
21. Zhang W, Wu Y, Shiozaki Y, et al. miRNA-133a-5p inhibits the expression of osteoblast differentiation-associated markers by targeting the 3ʹ UTR of RUNX2. DNA Cell Biol. 2018;37:199–209. doi:10.1089/dna.2017.3936
22. Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol. 2006;15:56–61. doi:10.1097/00019606-200603000-00009
23. Shao TR, Zheng ZN, Chen YC, et al. LncRNA AC007271.3 promotes cell proliferation, invasion, migration and inhibits cell apoptosis of OSCC via the Wnt/beta-catenin signaling pathway. Life Sci. 2019;239:117087. doi:10.1016/j.lfs.2019.117087
24. Zhang X, Feng H, Li Z, Guo J, Li M. Aspirin is involved in the cell cycle arrest, apoptosis, cell migration, and invasion of oral squamous cell carcinoma. Int J Mol Sci. 2018;19.
25. Feng X, Luo Q, Zhang H, et al. The role of NLRP3 inflammasome in 5-fluorouracil resistance of oral squamous cell carcinoma. J Exp Clin Cancer Res. 2017;36:81. doi:10.1186/s13046-017-0553-x
26. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi:10.1038/onc.2017.184
27. Yang Y, Chen D, Liu H, Yang K. Increased expression of lncRNA CASC9 promotes tumor progression by suppressing autophagy-mediated cell apoptosis via the AKT/mTOR pathway in oral squamous cell carcinoma. Cell Death Dis. 2019;10:41. doi:10.1038/s41419-018-1280-8
28. Zhang C, Bao C, Zhang X, Lin X, Pan D, Chen Y. Knockdown of lncRNA LEF1-AS1 inhibited the progression of oral squamous cell carcinoma (OSCC) via Hippo signaling pathway. Cancer Biol Ther. 2019;20:1213–1222. doi:10.1080/15384047.2019.1599671
29. Wang Y, Zhang X, Wang Z, et al. LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating Glut1-mediated glycolysis. Cancer Lett. 2018;434:172–183. doi:10.1016/j.canlet.2018.07.016
30. Chen F, Qi S, Zhang X, Wu J, Yang X, Wang R. lncRNA PLAC2 activated by H3K27 acetylation promotes cell proliferation and invasion via the activation of Wnt/beta-catenin pathway in oral squamous cell carcinoma. Int J Oncol. 2019;54:1183–1194. doi:10.3892/ijo.2019.4707
31. Kong Y, Feng Y, Xiao YY, et al. LncRNA LUCAT1 promotes growth, migration, and invasion of oral squamous cell carcinoma by upregulating PCNA. Eur Rev Med Pharmacol Sci. 2019;23:4770–4776. doi:10.26355/eurrev_201906_18059
32. Chen X, Ouyang H, Wang Z, Chen B, Nie Q. A novel circular RNA generated by FGFR2 gene promotes myoblast proliferation and differentiation by sponging miR-133a-5p and miR-29b-1-5p. Cells. 2018;7:199. doi:10.3390/cells7110199
33. Wang T, Wang X, Du Q, et al. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun. 2019;513:904–911. doi:10.1016/j.bbrc.2019.04.108
34. Fang Z, Zhao J, Xie W, Sun Q, Wang H, Qiao B. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by suppressing miR-184 expression. Cancer Med. 2017;6:2897–2908. doi:10.1002/cam4.1253
35. Hong Y, He H, Sui W, Zhang J, Zhang S, Yang D. Long non-coding RNA H1 promotes cell proliferation and invasion by acting as a ceRNA of miR138 and releasing EZH2 in oral squamous cell carcinoma. Int J Oncol. 2018;52:901–912. doi:10.3892/ijo.2018.4247
36. Wang X, Li H, Shi J. LncRNA HOXA11-AS promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by suppression of miR-214-3p expression. Biomed Res Int. 2019;2019:8645153.
37. Huang W, Cao J, Peng X. LINC01234 facilitates growth and invasiveness of oral squamous cell carcinoma through regulating the miR-637/NUPR1 axis. Biomed Pharmacother. 2019;120:109507. doi:10.1016/j.biopha.2019.109507
38. Li M, Ning J, Li Z, et al. Long noncoding RNA OIP5-AS1 promotes the progression of oral squamous cell carcinoma via regulating miR-338-3p/NRP1 axis. Biomed Pharmacother. 2019;118:109259. doi:10.1016/j.biopha.2019.109259
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.