Back to Journals » Journal of Pain Research » Volume 17
LncRNA 51325 Alleviates Bone Cancer Induced Hyperalgesia Through Inhibition of Pum2
Authors Wang Y , Xu C, Liu P, He Q, Zhang S, Liu Z, Ni C, Chen L, Zhi T, Xu L , Cheng L, Lin X, Yao M , Ni H
Received 3 November 2023
Accepted for publication 7 January 2024
Published 16 January 2024 Volume 2024:17 Pages 265—284
DOI https://doi.org/10.2147/JPR.S446635
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Wendy Imlach
Yahui Wang,1,2,* Chengfei Xu,3,* Peng Liu,2 Qiuli He,1 Shihua Zhang,1 Zhihao Liu,1 Chaobo Ni,1 Liping Chen,1 Tong Zhi,1 Longsheng Xu,1 Liang Cheng,3 Xuewu Lin,2 Ming Yao,1 Huadong Ni1,4
1Department of Anesthesiology and Pain Research Center, the Affiliated Hospital of Jiaxing University, Jiaxing, 314001, People’s Republic of China; 2Department of Pain Management, the First Affiliated Hospital of Bengbu Medical College, Bengbu City, 233000, People’s Republic of China; 3Department of Anesthesiology, Bengbu Third People’s Hospital, Bengbu City, 233000, People’s Republic of China; 4Institute of Neuroscience, Soochow University, Suzhou, 215123, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Huadong Ni; Ming Yao, Department of Anesthesiology and Pain Research center, the Affiliated Hospital of Jiaxing University, 1882 Zhonghuan South Road, Jiaxing, 314001, People’s Republic of China, Email [email protected]; [email protected]
Background: Bone cancer pain (BCP) represents one of the most challenging comorbidities associated with cancer metastasis. Long non-coding RNAs (lncRNAs) have garnered attention as potential therapeutic agents in managing neuropathic pain. However, their role in the regulation of nociceptive information processing remains poorly understood. In this study, we observed a significant down-regulation of the spinal lncRNA ENSRNOG00000051325 (lncRNA51325) in a rat model of bone cancer pain. Our study sought to elucidate the potential involvement of lncRNA51325 in the development of BCP by modulating the expression of molecules associated with pain modulation.
Methods: We established the BCP model by injecting Walker 256 cells into the tibial plateau of rats. We conducted tests on the pain behaviors and anxiety-like responses of rats through von-Frey test, Gait analysis, and Open Field Test. Spinal lumbar expansion was harvested for molecular biology experiments to explore the relationship between lncRNA51325 and Pumilio RNA binding family member 2 (Pum2).
Results: Notably, the overexpression of lncRNA51325 effectively attenuated mechanical allodynia in rats afflicted with BCP, whereas the knockdown of lncRNA51325 induced pain behaviors and anxiety-like responses in naïve rats. Additionally, we observed a time-dependent increase in the expression of Pum2 in BCP-afflicted rats, and intrathecal injection of Pum2-siRNA alleviated hyperalgesia. Furthermore, our investigations revealed that lncRNA51325 exerts a negative modulatory effect on Pum2 expression. The overexpression of lncRNA51325 significantly suppressed Pum2 expression in BCP rats, while the knockdown of lncRNA51325 led to elevated Pum2 protein levels in the spinal cord of naïve rats. Subsequent treatment with Pum2-siRNA mitigated the downregulation of lncRNA51325-induced mechanical allodynia in naïve rats.
Conclusion: Our findings indicate that lncRNA51325 plays a role in regulating bone cancer pain by inhibiting Pum2 expression, offering a promising avenue for novel treatments targeting nociceptive hypersensitivity induced by bone metastatic cancer.
Keywords: long non-coding RNA, bone cancer pain, Pum2, spinal cord
Introduction
Numerous cancer types,1 including lung, breast, prostate, thyroid, and kidney cancer, have been identified to exhibit bone metastasis in advanced stages.2 This metastatic process can lead to adverse consequences, including fractures,3 pain, hypercalcemia, and compression of the spinal cord and nerves.4 Notably, bone cancer pain (BCP) is a persistent and intricate form of pain that markedly diminishes a patient’s quality of life.5 Thus, the imperative task at hand is the discovery of novel therapeutic modalities and medications aimed at enhancing the quality of life for individuals afflicted by bone cancer pain.
Long non-coding RNAs (lncRNAs) are non-protein-coding transcripts exceeding 200 nucleotides in length.6,7 Accumulating research underscores their pivotal roles in diverse physiological and pathological phenomena,7 including lung metastasis in breast cancer,6 ischemic stroke,8 cartilage differentiation in osteoarthritis,9 peripheral neuropathies,10 and various neuropathic pain conditions.11–13
Pumilio protein, a member of the RNA-binding PUF family, which includes Pum1 and Pum2,14 serves as a vital post-transcriptional regulatory factor with implications for genomic stability.15 It has been established that long non-coding RNAs (lncRNAs) exert their influence on genomic stability by modulating Pumilio activity.16 Notably, PUM genes exhibit ubiquitous expression across various human tissues, with significant variations observed in at least 17 distinct cancer types.17 While some studies have begun to elucidate their roles in tumor progression, the precise mechanism underlying the involvement of Pum2 in bone cancer pain remains unresolved.
In this investigation, a notable reduction in lncRNA51325 expression was discerned in rats afflicted with bone cancer pain. Modulating this gene’s expression subsequently impacted the pre-existing pain-related behaviors in these rats. Subsequently, through database predictions, we uncovered Pum2 as a downstream protein associated with lncRNA51325, a connection hitherto unexplored in the context of pain. Remarkably, heightened Pum2 expression was observed in rats experiencing bone cancer pain. Furthermore, our observations confirmed that diminished lncRNA51325 expression correlates with an elevation in Pum2 levels. These findings illuminate novel prospects for identifying therapeutic targets in the context of cancer-related pain.
Materials and Methods
Experimental Animal
We acquired adult female Sprague-Dawley rats, aged 6–7 weeks and weighing 180–200g, from Zhejiang Vital River Laboratory Animal Technology Co. These rats were housed in specific pathogen-free (SPF) facilities under controlled conditions, with a room temperature of 22 ± 2°C and humidity maintained at 45–55%. Each cage accommodated two to five rats, adhering to a 12:12 light/dark cycle, and had access to food and water ad libitum. Ethical approval for all rat procedures was obtained from the Animal Use and Care Committee for Research and Education of Jiaxing University in Jiaxing, China. These animal experiments strictly adhered to the guidelines outlined in the International Association for the Study of Pain Guide for the Care and Use of Laboratory Animals.
Induction of Bone Cancer Pain
Rats participating in the study were randomly allocated to either a treatment or control group. Initially, these rats were anesthetized with sodium pentobarbital (50mg/kg, i.p.) and placed in a supine position. Subsequently, the left tibial plateau of each rat was prepared by shaving and disinfection using iodophor. A 1 mL syringe needle was employed to create an aperture in the tibial plateau, through which Walker 256 cells were meticulously introduced, following which the opening was sealed with bone wax after a brief two-minute interval. In the sham group, the same surgical procedure was enacted as in the treatment group, with the exception that an equivalent quantity of carcinoma cells was subjected to a 30-minute heat treatment prior to injection. Following suturing of the skin with absorbable sutures, the rats were placed on an animal heating pad until they regained consciousness from the anesthesia.
Experimental Group
In this experiment we used a total of 100 rats, which were randomly divided into the following groups. 1) A total of 64 rats were used to verify the effects of lncRNA51325 on the pain behavior of BCP rats. First, we randomly divided 16 rats into two groups: sham group and BCP group (8 rats in each group), to validate the successful establishment of bone cancer pain modeling. To investigate whether overexpression of lncRNA51325 in BCP rats reduces pain, we randomly divided 24 rats into sham group, BCP+lncRNA51325-LV-NC (BCP+LV-NC) group, BCP+lncRNA51325-LV (BCP+LV) group (8 rats in each group). To investigate whether knockdown of lncRNA51325 in Naïve rats could cause pain-like symptoms in rats, we randomly divided 24 rats into Naïve group, Naïve+lncRNA51325-scr-siRNA (Naïve + scr-siRNA) group, Naïve+lncRNA51325-siRNA (Naïve + siRNA) group (8 rats in each group). 2) A total of 36 rats were used to validate the role of Pum2 in BCP rats. We found that the expression of Pum2 was significantly increased in BCP rats by PCR results. Therefore, we designed siRNAs to knock down the expression of Pum2. To verify which siRNA had the best knockdown effect on Pum2, we randomly divided 12 rats into Naïve + Pum2-scr-siRNA group, Naïve + Pum2-siRNA1 group, Naïve + Pum2-siRNA2 group, Naïve + Pum2-siRNA3 group (3 rats in each group). We then chose the siRNA3 with the best knockdown effect for further in vitro experiments, randomizing 24 rats into sham group, BCP+Pum2-scr-siRNA group, and BCP+Pum2-siRNA3 (BCP+Pum2-siRNA) group (8 rats in each group).
In this experiment we used a double-blind approach: behavioral tests on rats were conducted by experimenter who did not know the information about the experimental grouping, and data were collected and analyzed by experimenter with professional statistical background.
Behavioral Assessment
Mechanical pain was assessed using von-Frey filaments (BME-404, Institute of Biological Sciences, Chinese Academy of Medical Sciences) in accordance with established protocols. Each rat underwent a preliminary period of environmental acclimatization within a transparent acrylic chamber measuring 10 cm x 10 cm x 20 cm, lasting 20–30 minutes. Subsequently, mechanical allodynia was evaluated on the left hind paw, specifically the area devoid of a flesh pad, by applying the filaments at a 90-degree angle. These filaments, configured as either “C” or ‘S’ shapes, were maintained in contact for 6–8 seconds. Withdrawal responses of the rat’s paw were observed and documented using the “Up & Down” method. The mechanical pain threshold of the rats was calculated using a sequence of “O” or “X” combinations, with the determination derived from a specific formula.
Gait analysis in rats was conducted employing the Catwalk automatic system. Rats were introduced at the commencement of a sealed glass tunnel, allowing unimpeded movement through its length. A high-speed camera positioned below the tunnel effectively captured the footprint area of the rats as they traversed the apparatus. Subsequently, data analysis was carried out using gait analysis software, specifically CATWALK XT by Aster Wee Information Technology. Three distinct parameters were utilized for the comprehensive evaluation of rat behavior: 1) Maximum contact area, denoting the largest area covered by the left hind paw during the entire locomotion sequence. 2) Maximum contact maximum intensity, representing the peak intensity of the left hind paw at the point of maximum contact area. 3) Mean intensity, signifying the average intensity recorded for the left hind paw throughout the entire movement. To mitigate the influence of extraneous factors, we normalized our analysis by calculating the ratio between the left hind paw and right hind paw, thereby facilitating the assessment of changes in rat gait.
Open Field Test: To assess potential motor impairment or anxiety-like behavior in rats, we placed them within an 80x80x40 cm open field, located in a softly illuminated environment with ambient temperature akin to their natural surroundings. Over a 10-minute duration under low-light conditions, a high-speed camera was positioned above the enclosure to record the rats’ exploratory actions. Data analysis was executed using Jiliang Behavior Analysis Software from Shanghai, with particular emphasis on distance-time measurements regarding the rats’ positions within the central area of the field.
Prior to testing initiation, a two-day pre-training phase was administered to all experimental rats for behavioral assessments. In efforts to ensure result precision, a blinded evaluation approach was employed for all behavioral experiments conducted on the experimental rats. Furthermore, following the completion of each test by an individual rat, meticulous measures were taken to cleanse the experimental environment using alcohol wipes. This process effectively eliminated residual odors and excrement from prior subjects, thus averting any potential influence on subsequent results.
RNA Extraction and Quantitative Real-Time PCR
RNA Extraction: Total RNA was extracted from freshly harvested rat spinal cord tissue using a combination of Trizol, isopropanol, and chloroform. Subsequently, cytoplasmic and nuclear RNA fractions were isolated in accordance with the guidelines provided in the Cytoplasmic & Nuclear RNA Purification Kit (Aidlab, Beijing). RNA concentration was determined using the Thermo Scientific™ NanoDrop™ One microvolume UV-Vis spectrophotometer. Thereafter, 1 μg of RNA underwent reverse transcription into cDNA with the application of 4 μL of 5× PrimeScript RT Master (Takara, Japan). The reverse transcription process involved incubation at 37°C for 15 minutes, followed by enzyme deactivation at 85°C for 5 seconds.
Quantitative Real-time PCR: The quantification of mRNA levels for lncRNA51325, Pum2, and the housekeeping gene GAPDH was performed utilizing the StepOne Real-Time PCR Platform (Applied Biosystems, USA). Each target, lncRNA51325, Pum2, and GAPDH, were separately assessed through polymerase chain reaction (PCR) on the StepOne Real-Time PCR Platform (Applied Biosystems, USA). Reaction conditions encompassed an initial denaturation phase at 50°C and 95°C for 2 minutes, succeeded by 40 amplification cycles (95°C for 15 seconds, 60°C for 1 minute). Relative expression levels for the molecules were subsequently determined employing the 2-ΔΔCt method. Primer sequences were sourced from Sangon Biotech (Shanghai) and are detailed in Table 1.
 |
Table 1 Specific Primer Sequences |
Western Blot
Protein Extraction and Western Blot Analysis: Following isoflurane anesthesia, lumbar spinal cord tissue was swiftly excised, and total proteins were extracted using RIPA lysis buffer supplemented with protease and phosphatase inhibitors. Quantification of the extracted proteins was carried out employing the BCA Protein Assay Kit (Epizyme Biotech, Shanghai). Electrophoresis was conducted using a 7.5% PAGE Gel Fast Preparation Kit (Epizyme Biotech, Shanghai). Subsequent to electrophoresis, proteins were transferred to a 0.45μm PVDF membrane. The membrane was subjected to a 2-hour blocking step with 5% skim milk, followed by thorough washing with PBST to remove residual milk. The primary antibodies (as listed in Table 2) were introduced and incubated overnight on a shaker at 4°C. The subsequent day, after removal of excess primary antibodies, the secondary antibodies (also listed in Table 2) were applied and incubated for 2 hours at room temperature. The protein bands were visualized using Immobilon ECL Ultra Western HRP Substrate (Millipore) and exposed on the Gene Gnome XRQ. Subsequent analysis of the acquired protein bands was performed utilizing ImageJ software to quantify the relative expression levels of the target protein.
 |
Table 2 Antibody |
Primary Cell Isolation and Culture
Isolation of Primary Neuronal Cells: Newborn rats, aged 1–2 days, were humanely euthanized by immersion in a 75% alcohol solution, with all ensuing procedures meticulously conducted within a UV-sterilized laminar flow cabinet. The spinal cord was meticulously dissected into small, approximately 1mm^3 fragments, followed by thorough washing with PBS containing dual antibiotics. The tissue fragments were then transferred to a centrifuge tube and subjected to enzymatic digestion in a shaking water bath at 37°C for a duration of 10 minutes. To halt the trypsin reaction, FBS was introduced, and the resultant tissue suspension was subsequently passed through sequential 100μm, 200μm, and 400μm cell strainers. Following this filtration process, the filtrate underwent centrifugation at 300g for 5 minutes to remove the supernatant. The resulting pellet, obtained after resuspension in PBS and subsequent centrifugation at 100g for 5 minutes, represents the isolated rat spinal cord neuronal cells.
Primary Neuronal Cell Culturing: The isolated rat spinal cord neuronal cells were suspended in a fully supplemented culture medium and subsequently seeded into poly-L-lysine-coated dishes. These culture dishes were then incubated in a humidified incubator at 37°C with 5% CO2. After a 24-hour incubation period, cytarabine was introduced into the culture dish to impede the proliferation of non-neuronal cells. Subsequently, at the 48-hour mark, the complete culture medium for rat spinal cord neuronal cells was replaced with fresh medium. As a part of the routine maintenance, half of the culture medium was exchanged every 3 days.
Immunofluorescence
Immunofluorescence Analysis of Tissues: Following the administration of pentobarbital sodium for anesthesia, the spinal cord tissue of the rats underwent immediate perfusion with 0.9% saline and 4% paraformaldehyde. The tissue was subsequently immersed in a centrifuge tube containing 4% paraformaldehyde for a duration of 6–8 hours. Dehydration of the spinal lumbar expansion was achieved by sequential immersion in 15% and 30% sucrose solutions. The processed spinal tissue was embedded in OCT compound and sectioned into slices, each measuring 16μm in thickness, utilizing a cryostat microtome. The sections were then subjected to a series of washes in TBST (Tris-Buffered Saline with Tween) three times, each wash lasting for 10 minutes. Subsequently, a 1-hour blocking phase was instituted, utilizing a solution composed of 7% donkey serum and 0.3% Triton X-100. The primary antibody, as detailed in Table 2, was added and allowed to incubate overnight within a refrigerated environment. The following day, the sections were meticulously washed with TBST on three occasions, and then exposed to the secondary antibody (refer to Table 2) for a duration of 45 minutes, subsequently being subjected to four washes. The resulting images were acquired utilizing a Laser Scanning Confocal Microscopy system (Zeiss), after an incubation period with DAPI.
Cell Immunofluorescence: Commence by positioning sterile cell culture slides within a newly opened 12-well plate. Subsequently, ensure an even distribution of the cell suspension across the 12-well plate, followed by incubation to facilitate cellular adherence. Thereafter, extract the culture medium and conduct two sequential 5-minute sterile PBS rinses of the 12-well plate. Subsequent to this, employ a blocking solution to obstruct cellular activity on the culture slides, proceeding with the remaining steps analogous to those in tissue immunofluorescence. Ultimately, the cells on the culture slides are subjected to DAPI incubation, and the ensuing results are examined microscopically.
Fluorescence in situ Hybridization
RiboBio (Guangzhou) designed a specific probe targeting lncRNA51325, and this probe was employed for Fluorescent In Situ Hybridization (FISH) detection according to the RiboTM FISH Kit protocol. Stringent precautions were observed to prevent light exposure during the co-localization of the lncRNA51325 probe with NeuN, GFAP, and Iba1. Subsequent steps adhered to the immunofluorescence protocol, culminating in the incubation of samples with DAPI. Subsequent image acquisition was performed using a microscope.
Drugs and Administration
RiboBio (Guangzhou) was responsible for the design of siRNA targeting lncRNA51325, and Sangon Biotech (Shanghai) designed the siRNA specific to Pum2, with the respective sequences detailed in Table 3. The lentivirus for lncRNA51325 was sourced from OBiO Tech (Shanghai) and utilized the vector named pSLenti-EF1-EGFP-F2A-Puro-WPRE2-CMV-ENSRNOG00000051325, with an average titer of 2.44E+08.
 |
Table 3 The Target Sequences of siRNA |
Intrathecal Injection
Following pentobarbital sodium anesthesia, rats were positioned in a prone orientation on a warming blanket. A PE-10 catheter was skillfully introduced into the subarachnoid space at the L4-L5 vertebral level, leaving approximately 1 cm of catheter in situ, firmly secured. The opposite end of the catheter was subcutaneously tunneled to the rat’s nuchal region and the catheter aperture was sealed. Subsequently, a 10 μL injection of lidocaine was administered via the catheter on the subsequent day to validate the accurate catheter placement. It is imperative to prescreen rats for pre-existing motor deficits before lidocaine administration and select those demonstrating complete motor function loss in both hind limbs post-lidocaine administration to proceed with subsequent experiments.
Spinal Cord Injection
Following anesthesia, the rats were positioned in a prone orientation on a warming blanket. The vertebral lamina spanning the L1-L2 vertebral bodies was gently excised using a dental drill. Under microscopic guidance, the lumbar enlargement of the spinal cord was meticulously exposed with ophthalmic forceps. Subsequently, a volume of 400 nl of lentivirus was precisely introduced into the dorsal horn of the spinal cord via a 10 μL microsyringe (Gaoge, Shanghai). The microsyringe was controlled at a flow rate of 40 nl/min, and the needle was maintained in situ for a duration of 10 minutes to ensure containment of the lentivirus. Following the completion of the injection, the needle was gently retracted, and the incision was carefully sutured.
Hematoxylin and Eosin Staining
After 24-hour fixation in 4% PFA, the left tibias of rats, encompassing both the sham group and those subjected to 12 days of bone cancer pain modeling, underwent decalcification using 10% EDTA for an additional 24 hours. Following this process, the tibia tissue specimens were paraffin-embedded and subsequently sectioned into 8μm-thick slices. These sections were then subjected to hematoxylin and eosin staining, followed by sealing with neutral resin, culminating in microscopic observation (Olympus BX51, Japan).
CT Three-Dimensional Imaging
To perform CT scans, we procured tibia tissue samples from both the sham group and the left tibia tissue of rats with bone cancer pain following a 12-day modeling period. CT scan parameters were established in alignment with prior studies, and imaging was executed employing the Siemens Syngo MultiModality Workplace (MMWP) post-processing workstation.
Statistics
In this study, data collection and analysis were performed using GraphPad Prism (8.4.3, United States). Results are presented as mean ± SEM. Two-group comparisons were conducted utilizing t-tests (including nonparametric tests), while One or Two-Way ANOVA with Bonferroni correction was applied for multiple group comparisons. All experimental data were tested for normality before being counted. Statistical significance was defined as a p-value below 0.05.
Results
LncRNA51325 is Down-Regulated in the Spine of BCP Rat
We previously conducted acRIP-seq analysis on dorsal horn spinal cord tissue from both the sham and BCP group rats, identifying 24 lncRNAs with notably distinct acetylation profiles.18 Given the existing body of research highlighting lncRNA involvement in diverse neuropathic pain states, we sought to corroborate the link between lncRNAs and bone cancer pain, commencing with the validation of a successfully established bone cancer pain model.
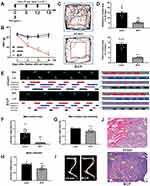
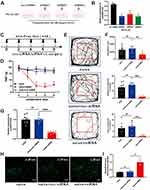
A bone cancer pain model was established utilizing the previously employed methodology of our research team. Induction of bone cancer pain was accomplished in adult female SD rats via the administration of Walker 256 cells. Mechanical pain assessments, specifically Von-Frey tests, were conducted at the time points illustrated in Figure 1A. Notably, BCP rats exhibited a significant reduction in pain threshold from the 6th day through the 18th day in comparison to the sham group (***p<0.001, vs sham group; n=8, two-way ANOVA, Figure 1B). Simultaneously, open field experiments were conducted on both the sham and 12-day-modeled rats, under identical conditions. The results revealed that the BCP group rats spent considerably less time and distance within the central area compared to the sham group (***p<0.001, vs sham group; n=8, Unpaired t test, Figures 1C and D), thus indicating evident manifestations of anxiety-related behaviors among BCP rats.
Furthermore, CatWalk analysis revealed that the BCP group rats displayed a pronounced reduction in maximum paw area, maximum paw pressure at the greatest paw area, and mean paw pressure when contrasted with the sham group (***p<0.001, vs sham group; n=9 or 10, Unpaired t test, Figures 1E–H).
The model’s effectiveness was validated through the utilization of CT three-dimensional imaging and hematoxylin and eosin staining on the tibias of rats belonging to both the sham and BCP groups. These examinations revealed significant bone damage in the tibias of the BCP group rats, whereas no observable changes were noted in the sham group (Figure 1I). Upon staining paraffin sections of bone tissues with hematoxylin and eosin, the sections obtained from the sham group exhibited well-defined bone trabeculae, in contrast to the sections from the BCP group, which displayed numerous tumor cells characterized by prominently darkly stained nuclei within the bone marrow cavity, along with discernible bone resorption pits (Figure 1J). These findings provided confirmation of the successful establishment of the bone cancer pain model.
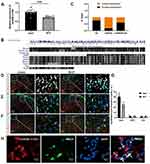
To enhance the robustness of our investigation, we carried out a validation experiment involving an expanded sample size to further elucidate the declining trend of ENSRNOG00000051325 (lncRNA51325). The results showed that the expression level of lncRNA51325 was significantly decreased in rats on day 12 after bone cancer pain modeling compared to the sham group (***p<0.001, vs sham group; n=11 or 12, Unpaired t test, Figure 2A). Additionally, it is worth noting the relatively high degree of biological conservation (Figure 2B) associated with this lncRNA. Given the paucity of prior research on lncRNA51325, we elected to concentrate our efforts on investigating its plausible direct or indirect involvement in bone cancer pain in rats.
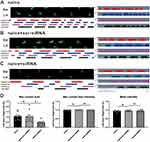
LncRNA51325 Was Primarily Colocalized with the Neuronal Marker NeuN
We conducted quantitative PCR (Q-PCR) on RNA extracted from both the cytoplasm and nucleus of spinal cord tissues from naive rats to ascertain the cellular localization of lncRNA51325. Our findings revealed that lncRNA51325 was distributed in both cellular compartments (n=3, Figure 2C). Moreover, we used fluorescence in situ hybridization (FISH) with nucleic acid probes in both sham and BCP group rats to determine the co-expression of lncRNA51325 with NeuN, GFAP and Iba1. The results demonstrated that lncRNA51325 primarily co-expressed with neurons, and only a small number of lncRNA51325 was co-labeled with astrocytes and microglia (Figures 2D–F). Statistical analysis showed that the percentage of lncRNA51325-positive neurons was markedly reduced in BCP rats when compared with sham rats (***p<0.001, vs sham group; n=3, two-way ANOVA, Figure 2G). To further corroborate this observation, we conducted in vitro FISH experiments with primary neuronal cells isolated from rat spinal cord tissue, which further substantiated the co-expression of lncRNA51325 with neurons (Figure 2H).
Overexpression of lncRNA51325 Alleviates Neuropathic Pain Induced by BCP
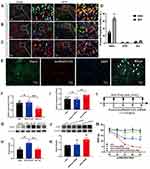
Subsequent to our prior findings revealing reduced lncRNA51325 expression in rats afflicted with bone cancer pain, we sought to affirm its involvement in pain hypersensitivity. A lentivirus (pSLenti-EF1-EGFP-F2A-Puro-WPRE2-CMV-ENSRNOG00000051325, average titer: 2.44E+08) was meticulously constructed for this purpose. Intrathecal injections into the spinal cord, along with von Frey pain assessments, were conducted according to the outlined schematic (Figure 3E) and timeline (Figure 3A). The pain threshold graph delineated that the overexpression of lncRNA51325 in BCP rats significantly alleviated pain hypersensitivity when compared to rats in the LV-NC group (*p<0.05, **p<0.01, ***p<0.001, vs BCP+LV-NC group; n=8, two-way ANOVA, Figure 3B). Simultaneously, an open field test was administered to both the LV-NC and LV-injected bone cancer pain rats. The results disclosed that rats in the LV group exhibited substantially increased central path distance and time in comparison to rats in the LV-NC group (***p<0.001, vs BCP+LV-NC group; n=8, Unpaired t test, Figures 3C and D).
Subsequent to viral injection into the rats, frozen sectioning was conducted, revealing successful viral infiltration within the spinal dorsal horn (Figure 3F). Further substantiating this, we extracted total RNA from the rat spinal cord and performed quantitative PCR (Q-PCR) analysis, which unequivocally demonstrated a significant elevation in lncRNA51325 expression levels within the LV group as opposed to the LV-NC group (***p<0.001, vs BCP+LV-NC group; n=8, Ordinary one-way ANOVA, Figure 3G). Additionally, immunofluorescence staining unveiled that, when compared to the sham group, the model group administered the blank control lentivirus displayed a heightened positive expression rate of c-fos. However, 7 days after the overexpression of lncRNA51325 through intrathecal lentivirus administration, the positive expression rate of c-fos markedly decreased (*p<0.05, **p<0.01, vs BCP+LV-NC group; n=3, Ordinary one-way ANOVA, Figures 3H and I). In parallel, we executed catwalk gait analysis on bone cancer pain rats subjected to spinal cord lentivirus injections and the blank control. The results exhibited that the rats injected with lncRNA51325 lentivirus manifested a significantly augmented maximum paw contact area, maximum paw contact pressure, and average paw contact pressure in comparison to the rats in the blank control group (**p<0.01, ***p<0.001, vs BCP+LV-NC group; n=8, Unpaired t test, Figures 4A–C).
The cumulative data strongly supports the notion that the upregulation of lncRNA51325 in rats experiencing bone cancer pain can mitigate the manifestation of hypersensitivity in these animals. This implies a potential association between reduced expression of lncRNA51325 and the observed hypersensitivity in these rats.
Knockdown of lncRNA51325 Induced Neuropathic Pain-Like Hypersensitivity in Naïve Rats
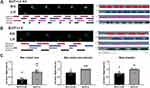
In vitro experiments were conducted using PC12 cells to identify siRNAs capable of effectively downregulating lncRNA51325. Three siRNAs, along with a control siRNA, were designed and transfected into culture dishes containing an equivalent number of PC12 cells (Figure 5A). After 48 hours, total RNA was extracted, and the Q-PCR analysis demonstrated that siRNA1 exhibited the most significant knockdown effect (***p<0.001, vs scr-siRNA group; n=3, Ordinary one-way ANOVA, Figure 5B). Consequently, scr-siRNA and lncRNA51325-siRNA1 (hereafter referred to as siRNA) were chosen for in vivo experiments.
Initially, intrathecal catheters were surgically implanted in naïve rats, and for six consecutive days, scr-siRNA and siRNA were administered intrathecally. Von-Frey pain assessments were performed as outlined in the timeline (Figure 5C). The paw withdrawal threshold (PWT) values of the naïve group rats exhibited no significant disparities when compared to those receiving intrathecal scr-siRNA. Nevertheless, rats receiving intrathecal siRNA experienced a marked reduction in PWT values (**p<0.01, ***p<0.001, vs naïve+scr-siRNA group; n=8, two-way ANOVA, Figure 5D). On the final day, all three groups of rats underwent an open field test under uniform conditions, one hour after the intrathecal administration of scr-siRNA and siRNA. The intrathecal injection of scr-siRNA did not induce any substantial alterations in the total distance traveled, central path percentage, or central time percentage compared to the naïve group. In contrast, rats subjected to intrathecal siRNA exhibited notably lower values in all these measures in comparison to the first two groups (**p<0.01, ***p<0.001, vs naïve+scr-siRNA group; n=8, Ordinary one-way ANOVA, Figures 5E and F). These findings indicate that intrathecal administration of lncRNA51325-siRNA elicited pain-related behavior in rats.
We conducted a gait analysis study employing the Catwalk system to evaluate the gait characteristics of three distinct rat groups. Our findings demonstrated a significant reduction in the maximum paw contact area in the intrathecal siRNA group compared to the other two groups (*p<0.05, vs naïve+scr-siRNA group; n=8, Ordinary one-way ANOVA, Figures 6A-D). However, there were no substantial differences in maximum contact maximum intensity and mean intensity. Visual examination of the paw prints unveiled that both the left and right paw prints of rats in the intrathecal siRNA group exhibited conspicuous reductions in size and depth compared to those of naïve rats. This phenomenon can be attributed to the effective suppression of lncRNA51325 in the dorsal horn of the spinal cord, causing pain-like symptoms in both hind limbs of naïve rats. Importantly, this did not result in a significant alteration in the left hind paw to right hind paw ratio.
Following the behavioral assessments, lumbar spinal cord tissue was collected to extract total RNA for subsequent Q-PCR analysis. The findings revealed a marked reduction in lncRNA51325 expression within the spinal cords of rats treated with intrathecal siRNA, while rats in the scr-siRNA group exhibited no significant variation compared to the naïve group (***p<0.001, vs naïve+scr-siRNA group; n=8, Ordinary one-way ANOVA, Figure 5G). Furthermore, immunofluorescence analyses were performed on the dorsal horns of the spinal cords across the three treatment groups. These analyses unveiled a notably higher positive expression rate of c-fos in the dorsal horns of the spinal cords in rats that received intrathecal siRNA compared to both the naïve group and the scr-siRNA group (*p<0.05, vs naïve+scr-siRNA group; n=3, Ordinary one-way ANOVA, Figures 5H and I).
All the above results collectively indicate that lncRNA51325 is a crucial element in the hyperalgesic behavior of rats with bone cancer pain.
Pum2 is Up-Regulated in the Spinal Cord of BCP Rat
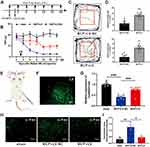
The observed reduction in lncRNA51325 levels in rats with bone cancer pain prompts an exploration of its regulatory mechanisms and its role in pain induction. We employed bioinformatic tools available through the Ensembl website (https://asia.ensembl.org/index.html) and the RBPDB database (RNA-binding protein specificities, http://rbpdb.ccbr.utoronto.ca/) to predict potential downstream molecules with binding affinity to lncRNA51325 (Figure 7A). Subsequent literature analysis led to the selection of Pum2 as a promising candidate due to its high scoring and its likely involvement in pain-related processes. Further inquiry into the gene sequence of Pum2 on the NCBI website (https://www.ncbi.nlm.nih.gov/gene/298874) unveiled multiple sequence segments within Pum2 with the potential to form base-pair interactions with lncRNA51325 (Figure 7C).
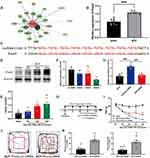 |
Figure 7 Pum2 expression is increased in the spinal cord and contributes to the BCP. (A) Predicted target proteins that may bind to lncRNA51325 can be found at the RBPDB website (the database of RNA-binding protein specificities, http://rbpdb.ccbr.utoronto.ca/). (B) The expression of Pum2 in the spinal cord was detected by Q-PCR after the injection of Walker 256 cell. ***p<0.001, vs sham group; n=8, Unpaired t test. (C)The informatics analysis of lncRNA51325 binding to the bases in Pum2 mRNA. (D and E) The expression of Pum2 in the ipsilateral dorsal horn after the injection of Walker 256 cell. *p<0.05, **p<0.01, vs sham group; n=4, Ordinary one-way ANOVA. (F) The validation of siRNA (Pum2) transfection efficiency in the spinal cord of the naïve rats. **p<0.01 vs Pum2-scr-siRNA group; n=3, Ordinary one-way ANOVA. (G) Timeline of Pum2-siRNA intrathecal injection and von-Frey pain measurement. (H) RT-PCR data showed that the expression of Pum2 in the BCP spinal cord treated with siRNA was significantly lower than in the BCP+Pum2-scr-siRNA group. ***p<0.001, vs BCP+Pum2-scr-siRNA group; n=8, Ordinary one-way ANOVA. (I) siRNA silencing of Pum2 alleviated mechanical hyperalgesia in the BCP rats. ***p<0.001, vs BCP+Pum2-scr-siRNA group; n=8, two-way ANOVA. (J and K) Quantification of behavioral parameters from the BCP+Pum2-scr-siRNA, and BCP+Pum2-siRNA groups in open fields. **p<0.01, ***p<0.001, vs BCP+scr-siRNA group; n=8, Unpaired t test. |
To corroborate the functional relevance of Pum2, we conducted an initial Q-PCR analysis on both the sham group and rats afflicted with bone cancer pain. Notably, Pum2 exhibited a substantial upregulation in rats on day 12 after bone cancer pain modeling (***p<0.001, vs sham group; n=8, Unpaired t test, Figure 7B). This elevation was further validated by Western blot (WB) findings, revealing a time-dependent increment in Pum2 levels in the context of bone cancer pain (*p<0.05, **p<0.01, vs sham group; n=4, Ordinary one-way ANOVA, Figures 7D and E).
Subsequently, to ascertain the potential influence of Pum2 on pain hypersensitivity in rats with bone cancer pain, we engineered Pum2 siRNA and conducted in vivo validation. Following six days of uninterrupted administration, our Q-PCR assays unveiled that siRNA3 demonstrated the most substantial knockdown efficacy concerning Pum2 (**p<0.01 vs scr-siRNA group; n=3, Ordinary one-way ANOVA, Figure 7F). Consequently, we elected to employ Pum2-siRNA3, henceforth referred to as Pum2-siRNA, for subsequent in vivo investigations.
Subsequently, we reinitiated bone cancer pain modeling, adhering to a predetermined intrathecal injection regimen and subsequently conducting von-Frey pain assessments (Figure 7G). Upon rigorous statistical scrutiny of the mechanical pain measurement data, we discerned a notable elevation in pain threshold among rats afflicted with bone cancer pain who were intrathecally administered Pum2-siRNA compared to those subjected to Pum2-scr-siRNA treatment (***p<0.001, vs BCP+Pum2-scr-siRNA group; n=8, two-way ANOVA, Figure 7I). This observation strongly suggests that the downregulation of Pum2 in rats enduring bone cancer pain significantly mitigated their pain hypersensitivity. An hour following the ultimate administration, both cohorts of rats underwent an open field test. The outcomes of this assessment corroborated our previous findings (**p<0.01, ***p<0.001, vs BCP+scr-siRNA group; n=8, Unpaired t test, Figures 7J and K). Subsequent to the behavioral examinations, Q-PCR analyses yielded results consistent with our expectations (***p<0.001, vs BCP+scr-siRNA group; n=8, Ordinary one-way ANOVA, Figure 7H). In summation, Pum2 exhibited upregulation in rats afflicted by bone cancer pain, and the attenuation of its expression markedly ameliorated pain-related behaviors in these animals.
LncRNA51325 Regulates Pum2 Expression in BCP
Our earlier findings demonstrated a marked discrepancy in Pum2 expression between the sham group and the BCP group of rats. However, the potential interplay between Pum2 and lncRNA51325 warrants further exploration in our forthcoming investigations. Initially, we engaged in the co-localization of Pum2 with NeuN, GFAP, and Iba1 within the tissue samples, revealing a pronounced co-expression of Pum2 with NeuN, but only a few with GFAP and Iba1 (Figures 8A–C). This observation is notably consistent with the cellular localization pattern of lncRNA51325. The percentage of Pum2-positive neurons was markedly increased in BCP rats when compared to that of sham rats (***p<0.001, vs sham group; n=3, two-way ANOVA, Figure 8D). Subsequently, the results of fluorescence double labeling of Pum2 and lncRNA51325 showed that the expressions of both were co-localized (Figure 8E).
To further substantiate the anticipated interaction between lncRNA51325 and Pum2, we conducted a series of molecular experiments to validate this relationship. Both Q-PCR and WB analyses revealed no noteworthy variation in Pum2 expression levels between the BCP group and the BCP group subjected to lncRNA51325 lentivirus blank control injection. However, the overexpression of lncRNA51325 in rats with bone cancer pain led to a significant reduction in Pum2 expression levels within the tissues (Q-PCR, ***p<0.001, vs BCP+LV-NC group; n=8, Ordinary one-way ANOVA, Figure 8F; WB, **p<0.01, vs BCP+LV-NC group; n=4, Ordinary one-way ANOVA, Figures 8G and H). Conversely, the silencing of lncRNA51325 in naïve rats resulted in a marked increase in Pum2 expression levels (Q-PCR, **p<0.01, vs naïve+scr-siRNA group; n=8, Ordinary one-way ANOVA, Figure 8I; WB, ***p<0.001, vs naïve+scr-siRNA group; n=4, Ordinary one-way ANOVA, Figures 8J and K). Furthermore, we introduced lncRNA51325-siRNA and Pum2-siRNA to rats (Figure 8L). The findings indicated that the neuropathic pain inhibitory effect caused by lncRNA51325 downregulation was significantly counteracted by Pum2 downregulation at postoperative days 4, 6, and 8 in the naïve+Pum2-siRNA+lncRNA51325-siRNA group when compared to the naïve+lncRNA51325-siRNA group (*p<0.05, **p<0.01, ***p<0.001, vs naïve+Pum2-siRNA+lncRNA51325-siRNA group; n=8, two-way ANOVA, Figure 8M). In essence, the attenuation of Pum2 effectively nullified the suppressive influence on neuropathic pain resulting from lncRNA51325 downregulation. These comprehensive experimental outcomes collectively affirm the role of lncRNA51325 in mitigating neuropathic pain and neuroinflammation through the downregulation of Pum2.
Discussion
Through prior sequencing analyses of spinal cord tissues obtained from both the sham group and rats afflicted with bone cancer pain, several long non-coding RNAs (lncRNAs) exhibiting significant expression disparities were unveiled.18 In this investigation, particular emphasis was placed on lncRNA51325, which displayed a marked decline in expression within the model group and exhibited commendable biological conservation. As an initial step, we duly substantiated the noteworthy reduction in lncRNA51325 expression in rats afflicted with bone cancer pain, while concurrently confirming the consequential impact of lncRNA51325 expression variations on the pain-related behaviors in these animals. Furthermore, we identified Pum2 as a pivotal player in the context of cancer-associated pain. Our inquiry also divulged the presence of a negative regulatory association between lncRNA51325 and Pum2. As a culmination of our findings, we assert that lncRNA51325 is intricately linked to the manifestation of bone cancer pain in rats.
Annually, a staggering ten million-plus individuals worldwide contend with a diagnosis of cancer,19 with bone metastasis afflicting over 1.5 million among them.20 This ailment designates the skeletal structure as the third most frequently affected site for cancer metastasis, trailing behind only the lungs and liver. The foremost culprits in propagating cancer-associated bone metastases are breast cancer, lung cancer, and prostate cancer, collectively responsible for 80% of such cases.21 Since the year 2015, there has been a discernible escalation in the global utilization of opioid analgesic medications for pain management.22 However, the prevailing concerns encompassing drug misuse and the concomitant side effects, encompassing the development of drug tolerance, nausea, respiratory depression, and addiction,23 engender substantial impediments to their efficacious deployment in the realm of cancer pain management.
Long non-coding RNAs (lncRNAs), while the scope of their precise biological functions remains a topic of ongoing scholarly discourse,24 undeniably exert a significant influence on various facets of cancer biology.25–28 Notably, aberrations in the functionality of non-coding RNAs have been indisputably associated with the pathogenesis of neuropathic pain.29,30 A myriad of investigations has firmly established the substantial role of lncRNAs in diverse cancer-related processes, encompassing tumor initiation, progression, metastasis, and staging. Intriguingly, their atypical expression profiles or genetic mutations have been intimately linked to the etiology of malignancies.16,31,32 Exemplifying this, lncRNA MEG3 orchestrates the regulation of neurogenic pathological pain through the miR-130a-5p/CXCL12/CXCR4 axis.11 Another instance is lncRNA71132, which modulates bone cancer pain by negatively regulating miR-143-5p, which, in turn, targets the GPR85 protein.12 Furthermore, in the context of rats with paclitaxel-induced peripheral neuropathy, an extensive transcriptome analysis of lncRNAs within the spinal cord has corroborated the pivotal roles lncRNAs play in the underlying mechanisms of neuroinflammation and pain perception.33 This burgeoning body of evidence underscores the potential utility of lncRNAs as both diagnostic biomarkers and promising therapeutic targets within the domain of cancer pain management.27,34 In light of our research group’s comprehensive transcriptome sequencing endeavors, we successfully identified lncRNA51325. This particular lncRNA, notable for its robust evolutionary conservation, had hitherto remained uncharted in the annals of scientific inquiry. Subsequent experimental validation unequivocally affirmed the marked reduction in lncRNA51325 expression within the Bone Cancer Pain (BCP) cohort, a finding congruent with our sequencing data. Consequently, we embarked on an exploration dedicated to deciphering the role of lncRNA51325 in the context of cancer-related pain syndromes.
The Pumilio complex, a multi-subunit repressor,35 represents a pivotal component of post-transcriptional regulation.14 Specifically, it falls within the broader PUF (Pumilio/FBF) family and includes two RNA-binding Pumilio proteins, Pum1 and Pum2, renowned for their influence on the translational modulation of numerous mRNAs across diverse human tissues.17,36 These proteins exhibit the capacity to intricately engage with mRNA networks, thereby exerting nuanced control over protein expression, whether through suppression or activation.35 Nonetheless, the role of Pum2 in the context of cancer development remains a subject of ongoing debate.37–39 Through a review of the literature, no studies have reported the effect of Pum2 on the development of BCP. However, some studies have shown that Pum2 not only regulates body and organ size in mice, but is also strongly associated with cancer. Evidence has illuminated instances where Pumilio proteins, including Pum2, participate in miRNA-mediated regulation of pivotal oncogenes, such as E2F3. Furthermore, the Pum2 protein has been observed to enhance the stemness properties of breast cancer cells, a function attributed to its competitive binding to the NRP-1 3’UTR in concert with miR-376a.38 Recent investigations also propose that the downregulation of SCAMP1-TV2 can attenuate its interaction with Pum2, consequently intensifying the latter’s association with INSM1, thereby constricting the proliferation and metastatic proclivity of breast cancer cells.40 These multifaceted findings underscore the intricate and, at times, paradoxical roles assumed by Pumilio proteins in cellular physiology, underscoring the continued quest to comprehensively elucidate their biological significance.
This study, while informative, bears certain limitations. Notably, it refrained from investigating whether the augmented expression of Pum2 impacts the alterations in pain thresholds among rats afflicted with bone cancer, and it exclusively hinged on Q-PCR and Western Blot analyses to substantiate the interplay between lncRNA51325 and Pum2.
Conclusions
Our research delineates a compelling framework wherein lncRNA51325 within the spinal cord assumes a role in the modulation of pain-like manifestations attendant to bone cancer in rats, a function achieved through the reciprocal regulation of Pum2 expression. This novel revelation opens avenues for potential therapeutic strategies targeting bone cancer pain. Nevertheless, a deeper comprehension of the intricacies governing the regulatory interplay between these entities necessitates further investigative endeavors.
Data Sharing Statement
The data used to support the findings of this study are available from the published literature.
Acknowledgments
This work was supported by the China Postdoctoral Science Foundation (2022M712310), the Natural Science Foundation of Zhejiang Province (LGD22H090002, LTGD24H090002), the Medical and Health General Research Program of Zhejiang Province (2021RC130, 2024KY429), the Science and Technology Project of Jiaxing City (2023AY40022, 2023AY11044), the Bengbu Science and Technology Bureau project (20220115), the Key Speciality of Anhui Province (2022-AH-105), the National Clinical Key Specialty Construction Project-Oncology department (2023-GJZK-001), the Zhejiang Provincial Traditional Chinese Medical Innovation Team of China under Grant No. 2022-19, the Key Medical Subjects Established by Zhejiang Province and Jiaxing City Jointly Pain Medicine and Oncology (2019-ssttyx, 2023-SSGJ-001), the Clinical Key Specialty of Zhejiang Province Anesthesiology (2023-ZJZK001) and the Construction Project of Key Laboratory of Nerve and Pain Medicine in Jiaxing City.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wang K, Gu Y, Liao Y, et al. PD-1 blockade inhibits osteoclast formation and murine bone cancer pain. J Clin Invest. 2020;130(7):3603–3620. doi:10.1172/JCI133334
2. Mercadante S. Malignant bone pain: pathophysiology and treatment. Pain. 1997;69(1–2):1–18. doi:10.1016/s0304-3959(96)03267-8
3. Wu S, Chen X, Huang F, et al. Transcriptomic Analysis of Long Noncoding RNA and mRNA Expression Profiles in the Amygdala of Rats with Bone Cancer Pain-Depression Comorbidity. Life Basel Switz. 2021;11(8):834. doi:10.3390/life11080834
4. Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: a fatal attraction. Nat Rev Cancer. 2011;11(6):411–425. doi:10.1038/nrc3055
5. Zheng XQ, Hao WY, Feng HJ, Wu AM. Neurophysiological mechanisms of cancer-induced bone pain. J Adv Res. 2022;35:117–127. doi:10.1016/j.jare.2021.06.006
6. Kim J, Piao HL, Kim BJ, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50(12):1705–1715. doi:10.1038/s41588-018-0252-3
7. Ratneswaran A, Kapoor M. Osteoarthritis year in review: genetics, genomics, epigenetics. Osteoarthritis Cartilage. 2021;29(2):151–160. doi:10.1016/j.joca.2020.11.003
8. Gao W, Ning Y, Peng Y, Tang X, Zhong S, Zeng H. LncRNA NKILA relieves astrocyte inflammation and neuronal oxidative stress after cerebral ischemia/reperfusion by inhibiting the NF-κB pathway. Mol Immunol. 2021;139:32–41. doi:10.1016/j.molimm.2021.08.002
9. Shi C, Zheng W, Wang J. lncRNA-CRNDE regulates BMSC chondrogenic differentiation and promotes cartilage repair in osteoarthritis through SIRT1/SOX9. Mol Cell Biochem. 2021;476(4):1881–1890. doi:10.1007/s11010-020-04047-4
10. Li Y, Yin C, Liu B, et al. Transcriptome profiling of long noncoding RNAs and mRNAs in spinal cord of a rat model of paclitaxel-induced peripheral neuropathy identifies potential mechanisms mediating neuroinflammation and pain. J Neuroinflammation. 2021;18(1):48. doi:10.1186/s12974-021-02098-y
11. Dong J, Xia R, Zhang Z, Xu C. lncRNA MEG3 aggravated neuropathic pain and astrocyte overaction through mediating miR-130a-5p/CXCL12/CXCR4 axis. Aging. 2021;13(19):23004–23019. doi:10.18632/aging.203592
12. Ni H, Xu M, Kuang J, et al. Upregulation of LncRNA71132 in the spinal cord AU1 regulates hypersensitivity in a rat model of bone cancer pain. 2022.
13. Tang Y, Liu C, Zhu T, et al. Transcriptome Profiles of IncRNA and mRNA Highlight the Role of Ferroptosis in Chronic Neuropathic Pain With Memory Impairment. Front Cell Dev Biol. 2022;10:843297. doi:10.3389/fcell.2022.843297
14. Goldstrohm AC, Tanaka Hall TM, McKenney KM. Post-transcriptional Regulatory Functions of Mammalian Pumilio Proteins. Trends Genet. 2018;34(12):972–990. doi:10.1016/j.tig.2018.09.006
15. PUMILIO proteins promote colorectal cancer growth via suppressing p21 - PubMed. Available from: https://pubmed.ncbi.nlm.nih.gov/35338151/.
16. Lee S, Kopp F, Chang TC, et al. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164(1–2):69–80. doi:10.1016/j.cell.2015.12.017
17. Smialek MJ, Ilaslan E, Sajek MP, Jaruzelska J. Role of PUM RNA-Binding Proteins in Cancer. Cancers. 2021;13(1):129. doi:10.3390/cancers13010129
18. Xu L, Zheng S, Liu B, et al. Epitranscriptomic profiling of N4-acetylcytidine-related RNA acetylation in the spinal dorsal horn of rat with cancer-induced bone pain. Mol Pain. 2023;19:17448069231178487. doi:10.1177/17448069231178487
19. Snijders RAH, Brom L, Theunissen M. Update on Prevalence of Pain in Patients with Cancer 2022: a Systematic Literature Review and Meta-Analysis. Cancers. 2023;15:591.
20. Clézardin P, Coleman R, Puppo M, et al. Bone metastasis: mechanisms, therapies, and biomarkers. Physiol Rev. 2021;101(3):797–855. doi:10.1152/physrev.00012.2019
21. Zajączkowska R, Kocot-Kępska M, Leppert W, Wordliczek J. Bone Pain in Cancer Patients: mechanisms and Current Treatment. Int J Mol Sci. 2019;20(23):6047. doi:10.3390/ijms20236047
22. J C, W L, Kkc M, et al. Global, regional, and national trends in opioid analgesic consumption from 2015 to 2019: a longitudinal study. Lancet Public Health. 2022;7(4). doi:10.1016/S2468-2667(22)00013-5
23. Toblin RL, Mack KA, Perveen G, Paulozzi LJ. A population-based survey of chronic pain and its treatment with prescription drugs. Pain. 2011;152(6):1249–1255. doi:10.1016/j.pain.2010.12.036
24. Esposito R, Bosch N, Lanzós A, Polidori T, Pulido-Quetglas C, Johnson R. Hacking the Cancer Genome: profiling Therapeutically Actionable Long Non-coding RNAs Using CRISPR-Cas9 Screening. Cancer Cell. 2019;35(4):545–557. doi:10.1016/j.ccell.2019.01.019
25. Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun. 2021;41(2):109–120. doi:10.1002/cac2.12108
26. Shen L, Wu Y, Li A, et al. LncRNA TTN‑AS1 promotes endometrial cancer by sponging miR‑376a‑3p. Oncol Rep. 2020;44(4):1343–1354. doi:10.3892/or.2020.7691
27. Bhan A, Soleimani M, Mandal SS. Long non-coding RNA (LncRNA) and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
28. Du S, Wu S, Feng X, et al. A nerve injury-specific long noncoding RNA promotes neuropathic pain by increasing Ccl2 expression. J Clin Invest. 2022;132(13):e153563. doi:10.1172/JCI153563
29. Chu Q, Gu X, Zheng Q, et al. Long noncoding RNA SNHG4: a novel target in human diseases. Cancer Cell Int. 2021;21(1):583. doi:10.1186/s12935-021-02292-1
30. Hu JZ, Rong ZJ, Li M, et al. Silencing of lncRNA PKIA-AS1 Attenuates Spinal Nerve Ligation-Induced Neuropathic Pain Through Epigenetic Downregulation of CDK6 Expression. Front Cell Neurosci. 2019;13:50. doi:10.3389/fncel.2019.00050
31. Sun RM, Wei J, Wang SS, Xu GY, Jiang GQ. Upregulation of lncRNA-NONRATT021203.2 in the dorsal root ganglion contributes to cancer-induced pain via CXCL9 in rats. Biochem Biophys Res Commun. 2020;524(4):983–989. doi:10.1016/j.bbrc.2020.01.163
32. Wei J, Dou Q, Ba F, Xu GY, Jiang GQ. Identification of lncRNA and mRNA expression profiles in dorsal root ganglion in rats with cancer-induced bone pain. Biochem Biophys Res Commun. 2021;572:98–104. doi:10.1016/j.bbrc.2021.07.063
33. Shen L, Wu Y, Li A. LncRNA TTN‑AS1 promotes endometrial cancer by sponging miR‑376a‑3p. Oncol Rep. 2020;44(4):1343.
34. Zhang C, Gao R, Zhou R, et al. The emerging power and promise of non-coding RNAs in chronic pain. Front Mol Neurosci. 2022;15:1037929. doi:10.3389/fnmol.2022.1037929
35. Miles WO, Tschöp K, Herr A, Ji JY, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26(4):356–368. doi:10.1101/gad.182568.111
36. Lin K, Qiang W, Zhu M, et al. Mammalian Pum1 and Pum2 Control Body Size via Translational Regulation of the Cell Cycle Inhibitor Cdkn1b. Cell Rep. 2019;26(9):2434–2450.e6. doi:10.1016/j.celrep.2019.01.111
37. Wang Y, Sun W, Yang J, et al. PUM2 Promotes Glioblastoma Cell Proliferation and Migration via Repressing BTG1 Expression. Cell Struct Funct. 2019;44(1):29–39. doi:10.1247/csf.18030
38. Zhang L, Chen Y, Li C, et al. RNA binding protein PUM2 promotes the stemness of breast cancer cells via competitively binding to neuropilin-1 (NRP-1) mRNA with miR-376a. Biomed Pharmacother Biomedecine Pharmacother. 2019;114:108772. doi:10.1016/j.biopha.2019.108772
39. Hu R, Zhu X, Chen C, Xu R, Li Y, Xu W. RNA-binding protein PUM2 suppresses osteosarcoma progression via partly and competitively binding to STARD13 3’UTR with miRNAs. Cell Prolif. 2018;51(6):e12508. doi:10.1111/cpr.12508
40. Tao W, Ma J, Zheng J, et al. Silencing SCAMP1-TV2 Inhibited the Malignant Biological Behaviors of Breast Cancer Cells by Interaction With PUM2 to Facilitate INSM1 mRNA Degradation. Front Oncol. 2020;10:613. doi:10.3389/fonc.2020.00613
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.