Back to Journals » International Journal of General Medicine » Volume 14
KLF11 Expression Predicts Poor Prognosis in Glioma Patients
Authors Xi Z , Zhang R, Zhang F, Ma S, Feng T
Received 4 March 2021
Accepted for publication 18 June 2021
Published 28 June 2021 Volume 2021:14 Pages 2923—2929
DOI https://doi.org/10.2147/IJGM.S307784
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Zhuo Xi,1 Rui Zhang,1 Furong Zhang,1 Shuang Ma,2 Tianda Feng1
1Department of Neurosurgery, Shengjing Hospital of China Medical University, Shenyang, Liaoning, People’s Republic of China; 2Department of Neurology, Shengjing Hospital of China Medical University, Shenyang, Liaoning, People’s Republic of China
Correspondence: Tianda Feng
Department of Neurosurgery, Shengjing Hospital of China Medical University, 36 Sanhao St, Heping Qu, Shenyang Shi, Liaoning Sheng, 110001, People’s Republic of China
Email [email protected]
Purpose: Glioma is a primary intracranial malignant tumor with high recurrence and mortality rates. It is very important to study the prognostic factors. KLF11 can function as an oncogene or a tumor suppressor, depending on the tumor and tissue types and the cancer stage. In this study, we aimed to determine whether KLF11 expression is related to the overall survival of glioma patients.
Patients and Methods: We investigated KLF11 expression in 116 glioma patients with different grades using Western blot and immunohistochemistry assay. We analyzed the patients with different glioma grades and KLF11 expression levels by Kaplan–Meier survival curves. Independent prognostic factors for poor overall survival were identified by univariate and multivariate analyses.
Results: There were 37 patients in KLF11 low expression group and 79 patients in high expression group. There was no difference in gender, age, tumor diameter or tumor location between two groups. The patients in KLF11 high expression group had higher ECOG score (P =0.025) and higher WHO grades (P =0.029). Western blot and immunohistochemistry assay showed KLF11 expression was significantly upregulated in glioma groups compared with normal brain tissues group (P < 0.05), and the expression in grades III–IV was significantly higher than those in grades I–II (P < 0.05). Kaplan–Meier survival curve analysis showed high KLF11 expression tended to reduce the overall survival (P < 0.05). After univariate and multivariate analyses, KLF11 expression (P =0.003) and age (P =0.007) were independent prognostic factors for poor survival in glioma patients.
Conclusion: KLF11 expression was increased in glioma tissues, and high KLF11 expression was associated with poor prognosis. KLF11 expression was an independent prognostic factor for poor survival in glioma patients. KLF11 may serve as a novel prognostic marker for gliomas and as a novel treatment target.
Keywords: KLF11, glioma, expression, overall time, prognostic marker
Introduction
Glioma is a kind of malignant tumor that originates from glial cells and accounts for more than 60% of primary intracranial tumors. It is characterized by invasive growth, and there is no clear demarcation between the tumor tissue and healthy brain tissue.1 Current treatment methods mainly include surgery, radiotherapy, chemotherapy, gene therapy, and targeted drug therapy. During surgical treatment, tumor tissues must be removed as much as possible without damaging nerves and/or disrupting their function. However, complete resection is extremely difficult; residual tumor tissue is easily overlooked, and recurrence often occurs, which results in poor prognosis of gliomas. Although conventional radiotherapy and chemotherapy can delay the progression of the disease to a certain extent, the treatment effects are limited due to the existence of the blood–brain barrier and the blood–tumor barrier, and it remains difficult to obtain a satisfactory prognosis.2
Glioma is associated with the inactivation of tumor suppressor genes and the activation of oncogenes. There is an urgent need for more reliable biomarkers for the diagnosis, treatment, and prognosis of glioma patients.
Krüppel-like factors (KLFs) form a highly conserved family of zinc finger transcription factors, and play important roles in the progression of human malignant tumors, such as breast cancer and colon cancer.3 KLF11, a KLF family member, was first found in osteoblast cells. Its expression is highest in the healthy pancreas. It contains three C2H2-type zinc fingers, two acids, and two regions rich in proline residues at the amino and carboxyl terminals.4 Previous studies have shown that KLF11 can act as an oncogene in breast cancer,3 but other studies demonstrated that KLF11 can play the role of a tumor suppressor gene in pancreatic, renal, lung, and gastric cancer.5,6 There have been no reports about KLF11 in gliomas, so it is very interesting and important to clarify the role of KLF11 in glioma tissues. The purpose of this article is to determine whether KLF11 expression in glioma tissues is related to patient survival.
Materials and Methods
Patient Specimens
All patients were diagnosed with glioma in the pathology department in our hospital and surgically treated by three neurosurgeons in the neurosurgical department between 2010 and 2016. We also collected 10 normal brain tissue samples from non-glioma patients to serve as controls in the Western blot analysis and immunohistochemistry (IHC) assay. Our study was approved by the Ethics Committee of Shengjing Hospital of China Medical University (No. 2017PS211K). All patients had provided informed consent for the use of their tissues and data for research, in accordance with the Declaration of Helsinki. Inclusion criteria were the following: age ≥18 years, intracranial primary glioma, and no radiotherapy or chemotherapy before surgery. Patients were excluded if they had other tumors, they died from diseases other than gliomas, follow-up information was incomplete, and/or the date of death was not recorded. Data were collected from hospital medical records, including demographics such as age, gender, and Eastern Cooperative Oncology Group (ECOG) score and clinical information such as WHO grade, max diameter, and location. The survival time was calculated from the surgery date to death, and the death dates were obtained from public records. Data were collected and analyzed by two attending neurosurgeons and one neurologist.
TCGA Database Analysis
The GEPIA website was used to analyze TCGA database to search KLF11 expression in glioblastoma (GBM, grade IV) and the correlation between KLF11 expression and prognosis. We then extend the samples to all grades of glioma (Grade I–IV) in the following analysis.
Western Blot Analysis
Western blot was used to test the level of KLF11 expression in five normal brain tissues, five low-grade glioma tissues (WHO I–II) and five high-grade glioma tissues (WHO III–IV). Equal amount of total tissue lysate protein samples (20 μg) went electrophoresis in a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride (PVDF) membranes. The PVDF membranes were incubated overnight at 4 °C with respective primary antibodies: anti-KLF11 (1:3000, Proteintech Group, Chicago, USA) and anti-GAPDH (1:1000, Santa Cruz Biotechnology). The second day, membranes were incubated with horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit or goat anti-mouse, 1:4000, respectively; Proteintech Group, Chicago, USA) at room temperature for 2 h. Finally, signals were detected by ECL chemiluminescent detection system (Santa Cruz Biotechnology). The relative integrated density values (IDVs) were measured on FluorChem 2.0 software and calculated based on GAPDH as an internal control.
Immunohistochemistry Assay
Paraffin-embedded tissue was cut into 3 μm thick sections and mounted on glass slides. The sections were dewaxed and rehydrated with ethanol solution, and then washed with phosphate-buffered saline (PBS). Antigens were retrieved at 105°C under high pressure in citrate buffer (pH 6.0). After cooling the tissue sections in buffer and washing with PBS, the sections were immersed in 3% hydrogen peroxide at room temperature for 10 minutes and washed with PBS. Subsequently, the slides were incubated with rabbit anti-KLF11 (1:200, Proteintech Group, USA) at 4°C overnight and with a biotin-labeled goat anti-rabbit secondary antibody at room temperature for 15 minutes. Slides were incubated with streptavidin–horseradish peroxidase conjugate complex at room temperature for 10 minutes, and then the slides were washed three times with PBS. All slides were stained with DAB chromogen (ZSGB Beijing, China). Finally, the sections were counterstained with hematoxylin, rehydrated, and fixed through a series of graded ethanol solutions.
KLF11 expression was scored on the basis of the proportion of positive cells and the staining intensity independently by two pathologists who did not know the glioma grade and clinical date. The percentage of KLF11-positive stained cells were scored for 0 (<10%), 1 (≥10% to 50%), 2 (>50% to 90%), and 3 (>90%), and the staining intensity was scored 0 for negative, 1 for weak-positive, 2 for moderate-positive, and 3 for strong-positive. Based on the value of ΣPI, the stained sections were defined as low expression [0 (−) to 1 (+)] or high expression [2 (++) to 3 (+++)].7
Statistical Analyses
Categorical variables are presented as numbers, and continuous variables are presented as mean with standard deviation. Normality of the variables was tested using the Kolmogorov–Smirnov test. One-way ANOVA, the independent sample Student’s t-test, and the chi-squared test were used to evaluate differences. Univariate analyses of possible risk factors for overall survival (OS) were performed using a Cox proportional hazards regression model. The variables that achieved a significance level of P < 0.05 were used for the multivariate analysis using the Cox model to screen for independent prognostic factors. The survival curves were drawn using the Kaplan–Meier method. Values of P < 0.05 were considered statistically significant. Statistical analyses were performed using SPSS version 23.0 software (IBM, Illinois, USA).
Results
Database Analysis Showed High KLF11 Expression in GBMs (Grade IV Glioma) and Poor Overall Survival
Results showed high KLF11 expression in 163 GBMs and 207 normal brain tissues (Figure 1A).8 In an OS analysis with 82 GBM patients (41 with high and 41 with low KLF11 expression), high KLF11 expression was found to be associated with poor OS (hazard ratio [HR] = 1.8, P = 0.029, Figure 1B).
 |
Figure 1 The TCGA database predicted (A) the expression of KLF11 was increased in GBM tissues and (B) high KLF11 expression was associated with poor overall survival of GBM patients. |
Western Blot Analysis
As shown in Figure 2, the expression levels of KLF11 were significantly upregulated in glioma groups compared with normal brain tissues group (P < 0.05). Our date also demonstrated that the KLF11 expression in high-grade glioma tissues (grades III–IV) was significantly higher than those in low-grade glioma tissues (grades I–II) (P < 0.05).
Immunohistochemistry Assay
IHC staining indicated 79 cases had high and 37 cases had low KLF11 expression, and the distribution of KLF11 was throughout the nucleoplasm in glioma cells (Figure 3). Among all the 116 glioma tissues, 23 cases of strong positive staining (+++), 56 cases of moderate positive staining (++), 23 cases of weak positive staining (+), and 14 cases of negative staining (−) of KLF11 were detected. In the correlation analysis, KLF11 staining showed positive correlation with WHO grade (P = 0.046, Table 1).
 |
Table 1 Association of KLF11 Staining in IHC with WHO Grade |
 |
Figure 3 Representative patterns of KLF11 expression was detected by IHC in glioma tissues and normal brain tissues. Abbreviations: NBT, normal brain tissue. |
Association of KLF11 Expression with Clinicopathological Characteristics
In the present study, 116 patients (69 males and 47 females) met the inclusion criteria (Table 2). There were no significant differences in age (P = 0.613), gender (P = 0.104), tumor diameter (P = 0.956), and location (P = 0.879) between the high and low expression groups (Supplementary Figure 1). The proportion of cases with high KLF11 expression was higher among patients with high ECOG scores (≥2) than among patients with low ECOG scores (<2). Among high-grade gliomas (WHO grades III–IV), the proportion of high expression cases (36/45, 80%) was higher than among low-grade gliomas (WHO grades I–II) (43/71, 60.6%) (P = 0.029).
 |
Table 2 Association of KLF11 Expression with Clinicopathological Characteristics |
The Relationship Between KLF11 Expression and Overall Survival
We analyzed patients with different glioma grades and KLF11 expression levels with Kaplan–Meier survival curves. In the low expression group, the mean OS was 48.46 ± 4.94 months, and in the high expression group, the mean OS was 22.31 ± 4.52 months (P = 0.002, Figure 4A). Of 71 patients with low-grade gliomas (WHO grades I–II), 43 cases had high KLF11 expression and 28 cases had low KLF11 expression; the mean OS was 48.46 ± 13.09 months in the low-grade low expression group and 26.22 ± 3.67 months in the low-grade high expression group (P = 0.041, Figure 4B). Of 45 patients with high-grade gliomas (WHO grades III–IV), 36 cases had high KLF11 expression and 9 cases had low KLF11 expression; the mean OS was 42.94 ± 19.49 months in the high-grade low expression group and 17.48 ± 7.12 months in the high-grade high expression group (P = 0.042, Figure 4C).
Univariate and Multivariate Analyses of Clinical Characteristics in Relation to Overall Survival
In the univariate analyses of prognostic factors for OS in our cohort, high KLF11 expression (P = 0.002), high-grade gliomas (P = 0.015), age ≥50 years (P = 0.004), and ECOG score ≥2 (P = 0.05) were associated with poorer OS (Table 3). Multivariate analysis indicated that high KLF11 expression (HR = 2.436, 95% confidence interval [CI]: 1.347–4.403, P = 0.003) and age ≥50 (HR = 1.977, 95% CI: 1.205–3.242, P = 0.007) were independent prognostic factors for poor OS in glioma patients (Table 4).
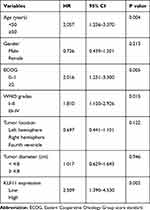 |
Table 3 Univariate Analysis of Clinical Characteristics in Relation to Overall Survival |
 |
Table 4 Multivariate Analysis of Clinical Characteristics in Relation to Overall Survival |
Discussion
Glioma is a worldwide leading cause of cancer mortality, and it is associated with numerous tumor suppressor genes and oncogenes. KLFs are a family of DNA-binding transcriptional regulators which comprises 17 identified members (KLF1–17) with diverse and essential functions in a multitude of cellular processes, ranging from proliferation and apoptosis to differentiation, migration, and pluripotency.9,10 Importantly, some members of the family are ubiquitously expressed, whereas others are tissue specific. In addition, KLF expression patterns can change dramatically during development, such that some members may overlap at some developmental stages but not at others. Although KLF11 expression is decreased in colon, kidney, lung, ovary, and pancreas cancers,11 it is increased in gastric and breast cancers.3,6 In the present study, we collected data from 116 glioma patients to detect KLF11 expression by IHC assay. KLF11 expression was increased in glioma tissues. Interestingly, among high-grade gliomas (WHO grades III–IV), the proportion of tumors with high KLF11 expression (36/45, 80%) was significantly higher than among low-grade gliomas (WHO grades I–II) (43/71, 60.6%). This conclusion was consistent with the predictions on the basis of the TCGA database. Furthermore, in correlation analysis, KLF11 expression was closely correlated with glioma grade and the ECOG score; the percentage of cases with high KLF11 expression was higher among patients with an ECOG score ≥2 than among patients with an ECOG score <2 (P = 0.025), indicating KLF11 might affect clinical behavior of gliomas, especially in patients with high grade gliomas.
We investigated the relationship between KLF11 expression and OS by Kaplan–Meier survival curve analysis. High KLF11 expression tended to reduce the OS in the entire cohort, in the high WHO grade group, and in the low WHO grade group. This was already predicted by the TCGA database. The mean OS was 17 months in the high-grade group, which was consistent with the literature.12–14 Univariate analysis revealed that KLF11 expression, age ≥50 years, ECOG score ≥2, and the WHO grade were prognostic risk factors. Multivariate analysis indicated that only KLF11 expression and age ≥50 years were independent prognostic factors. In our previous study, we also reported that older age was an independent prognostic factor for glioma patients and age ≥50 years predicted poor OS.7 In other prognostic studies, also the glioma grade was demonstrated to be an independent prognostic factor,7,15,16 but in our cohort, glioma grade was a prognostic risk factor but not an independent one.
We wish to mention some limitations to this study. First, this is a study on the relationship between KLF11 expression and clinical prognosis. We did not study the function of KLF11 at the molecular level. Second, the number of patients with high-grade gliomas was slightly low (45 cases). It is well known that higher glioma grades are associated with more obvious malignant biological behavior. In future studies, the number of high-grade gliomas should be increased, especially the number of glioblastoma multiforme cases. To the best of our knowledge, this is the first study that links KLF11 expression with 1) different grades of glioma and 2) prognosis of patients.
Conclusions
KLF11 expression was increased in gliomas. High KLF11 expression was significantly associated with poorer prognosis and short-time survival, especially in high-grade gliomas. High KLF11 expression and older age were independent prognostic factors for glioma patients. KLF11 may serve as a novel prognostic marker for gliomas and as a novel treatment target.
Acknowledgments
This research was funded by the Natural Science Foundation of China (No. 81802500), Natural Science Foundation of Liaoning Province (No. 20180550817) and 345 Talent Project of Shengjing Hospital.
Disclosure
The authors declared no potential conflicts of interest to this article.
References
1. Ostrom QT, Gittleman H, Stetson L, Virk S, Barnholtz-Sloan JS. Epidemiology of intracranial gliomas. Prog Neurol Surg. 2018;30:1–11.
2. Pellerino A, Franchino F, Soffietti R, Ruda R. Overview on current treatment standards in high-grade gliomas. Q J Nucl Med Mol Imaging. 2018;62(3):225–238. doi:10.23736/S1824-4785.18.03096-0
3. Cheng L, Shi L, Dai H. Bioinformatics analysis of potential prognostic biomarkers among Kruppel-like transcription Factors (KLFs) in breast cancer. Cancer Biomark. 2019;26(4):411–420. doi:10.3233/CBM-190199
4. Cook T, Gebelein B, Mesa K, Mladek A, Urrutia R. Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J Biol Chem. 1998;273(40):25929–25936. doi:10.1074/jbc.273.40.25929
5. Wang G, Li X, Tian W, et al. Promoter DNA methylation is associated with KLF11 expression in epithelial ovarian cancer. Genes Chromosomes Cancer. 2015;54(7):453–462. doi:10.1002/gcc.22257
6. Ji Q, Li Y, Zhao Q, et al. KLF11 promotes gastric cancer invasion and migration by increasing Twist1 expression. Neoplasma. 2019;66(1):92–100. doi:10.4149/neo_2018_180325N201
7. Xi Z, Xue Y, Zheng J, Liu X, Ma J, Liu Y. WTAP expression predicts poor prognosis in malignant glioma patients. J Mol Neurosci. 2016;60(2):131–136. doi:10.1007/s12031-016-0788-6
8. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
9. Limame R, Op de Beeck K, Lardon F, De Wever O, Pauwels P. Kruppel-like factors in cancer progression: three fingers on the steering wheel. Oncotarget. 2014;5(1):29–48.
10. Zhang Y, Hao J, Zheng Y, et al. Role of Kruppel-like factors in cancer stem cells. J Physiol Biochem. 2015;71(1):155–164. doi:10.1007/s13105-015-0381-4
11. Son B, Jeon J, Lee S, et al. Radiotherapy in combination with hyperthermia suppresses lung cancer progression via increased NR4A3 and KLF11 expression. Int J Radiat Biol. 2019;95(12):1696–1707. doi:10.1080/09553002.2019.1665213
12. Tamura R, Miyoshi H, Yoshida K, Okano H, Toda M. Recent progress in the research of suicide gene therapy for malignant glioma. Neurosurg Rev. 2019.
13. Fecci PE, Sampson JH. The current state of immunotherapy for gliomas: an eye toward the future. J Neurosurg. 2019;131(3):657–666. doi:10.3171/2019.5.JNS181762
14. Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14(2):284–297. doi:10.1007/s13311-017-0519-x
15. Music D, Dahlrot RH, Hermansen SK, et al. Expression and prognostic value of the WEE1 kinase in gliomas. J Neurooncol. 2016;127(2):381–389. doi:10.1007/s11060-015-2050-4
16. Zeng J, Zhang XK, Chen HD, Zhong ZH, Wu QL, Lin SX. Expression of programmed cell death-ligand 1 and its correlation with clinical outcomes in gliomas. Oncotarget. 2016;7(8):8944–8955. doi:10.18632/oncotarget.6884
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


