Back to Journals » Veterinary Medicine: Research and Reports » Volume 13
Isolation and Molecular Detection of Newcastle Disease Virus from Field Outbreaks in Chickens in Central Ethiopia
Authors Worku T , Dandecha M, Shegu D, Aliy A, Negessu D
Received 13 January 2022
Accepted for publication 5 April 2022
Published 19 April 2022 Volume 2022:13 Pages 65—73
DOI https://doi.org/10.2147/VMRR.S352727
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Young Lyoo
Takele Worku,1 Morka Dandecha,2 Deraje Shegu,1 Abde Aliy,1 Demessa Negessu1
1Department of Virology and Molecular Biology, National Animal Health Diagnostic and Investigation Center, Sebeta, Oromia, Ethiopia; 2Department of Veterinary Laboratory Technology, Ambo University, Ambo, Oromia, Ethiopia
Correspondence: Morka Dandecha, Department of Veterinary Laboratory Technology, Ambo University, Ambo, Oromia, Ethiopia, Tel +251-910309600, Email [email protected]
Background: Newcastle disease is a major viral disease of poultry. The virus is a major problem for chickens in Ethiopia and there is a scarcity of updated information on the virological and molecular status of confirmation of Newcastle disease outbreak cases in the country.
Methods: Newcastle disease outbreaks were investigated from February 2021 to October 2021 in central Ethiopia to isolate and detect the virus by cell culture and reverse transcriptase PCR. A total of 44 pooled tissue specimens were sampled from sick and recently dead chickens showing typical clinical signs of Newcastle disease. Virus isolation were performed using DF-1 cells and detection of the virus was done by real-time PCR.
Results: Out of 44 collected tissue samples, 38.63% (17/44) were positive on DF-1 cells. The result shows 17 of the clinically sick and dead chickens were positive for the virus by reverse transcriptase polymerase chain reaction. Based on the sample type, 54.54% (6/11) of the brain samples, 36.36% (4/11) of the intestines, 54.54% (6/11) of lung and trachea, 9% (1/11) of pooled liver, kidney, heart, and spleen samples were positive. Viruses were isolated in the proportions 37.5% (6/16), 25% (2/8), 50% (2/4), 25% (1/4), 50% (2/4) and 50% (4/8) from Sebeta, Bishoftu, Sululta, Nifas Silk, Kolfe and Yeka, respectively.
Conclusion: This study showed that Newcastle disease is a major viral disease causing death of chickens in the study area. Therefore, any control approach should focus on the appropriate characterization of the virus strain causing the outbreak in the study area.
Keywords: central Ethiopia, chickens, isolation, Newcastle diseases virus, RT-PCR
Introduction
Ethiopia is gifted with numerous livestock populations. The total poultry population in the country is 56.06 million.1 This poultry population contains both exotic and indigenous chickens. They are widely distributed in rural and peri-urban areas where they play important roles in income generation, food production and social interactions.2 The production of these chickens is affected by different obstacles such as disease, management problems and genetics of the chickens. The primary cause of the reduction of production and productivity of the chickens is a viral disease.3 Newcastle disease is the most common viral disease of these birds and is often responsible for various disorders, including gastrointestinal, nervous system, respiratory system and non-gastrointestinal disorders.4,5 Newcastle disease virus (NDV) is an RNA virus with a negative sense and composed of six genes, which are generated through RNA editing.6–8
Newcastle disease virus affects a widespread range of poultry globally.9 It is a major cause of economic harm worldwide.10 In many undeveloped countries, it is widespread and causes great problems in poultry farming.11,12 In Ethiopia, the disease was first reported in 197213 and it can cause up to 80% death in poultry farms. The virus affects the nervous, respiratory and digestive systems.14,15 The clinical signs and severity of NDV can vary depending on the strain of the virus. According to variation in strains, the death of chickens in a flock ranges from 90–100%.14,16,17 There is recurrent occurrence of the disease in commercial poultry farms in different parts of Ethiopia. But, confirmations of outbreaks are uncommon and inadequate data exist on the type of virus responsible for these outbreaks. Generally, information about the isolation and molecular detection of the virus from chickens is insufficient in Ethiopia in general and in the study area in particular. Therefore, the objectives of this study were the isolation and molecular detection of Newcastle disease virus from outbreak cases in the study area.
Materials and Methods
Study Areas
The study was performed from February 2021 to October 2021 in a selected area of central Ethiopia (Bishoftu, Addis Ababa, Sululta and Sebeta) where NDV outbreaks occurred in commercial poultry farming system, as indicated in Table 1.
 |
Table 1 Detail About the Study Areas |
Study Animals
Chickens of both sexes and all ages managed under commercial poultry farm systems were included. Chickens that had experienced an outbreak of Newcastle disease were used for outbreak investigation.
Study Design
A cross-sectional study design was performed during an active outbreak to isolate and detect NDV from suspected outbreak cases. The study focused on suspected cases of ND. Before the beginning of any outbreaks, proper information channels from concerned bodies regarding the outbreaks were collected through different contact addresses. Depending on the reported outbreak case of ND, a field investigation was conducted at the area of outbreaks, clinical information was recorded and appropriate samples were collected from chickens showing signs suggestive of ND infection.
Sampling
Representative tissue specimens were collected from different organs. About 44 tissues from ill and recently dead chickens showing distinctive clinical signs of ND were sampled. Necropsy examination was performed and affected tissues, i.e. brain, lung and trachea, pooled tissue of liver, spleen, kidney and heart, and intestines were sampled from the same chickens. Collected samples were submitted to the laboratory using an icebox and stored at −80°C for further processing.
Virus Isolation
Tissue specimens were processed by chopping them into small pieces and grinding with sterile sand in mortar and pestle at the virus isolation laboratory. Four tissue specimens, i.e. four brain, four lung and trachea, four pooled tissue of spleen, kidney, liver and heart and four intestines that were sampled from the same outbreak case were pooled to increase the concentration of virus. The suspension of tissue samples (10% (w/v)) were mixed with sterile phosphate buffer saline (PBS) which contains penicillin (100 IU/mL) and streptomycin (1000 μg/mL). The suspension was filled into a sterile Falcon tube and centrifuged at 3000 rpm, +4°C for 20 minutes. The supernatants were collected and filtered with 0.45 µL then 0.1 mL of the samples were inoculated onto confluent DF-1 cells which were cultured on 24-well plates and maintained with DMEM containing 2% calf serum and incubated at 37°C for one week with daily follow-up. The cytopathic effect (CPE) was determined based on a characteristic of NDV on the cell line. Samples that did not show a cytopathic effect were continued up to the third passage. The samples that revealed characteristics of the cytopathic effect were harvested for molecular detection of the virus.
RNA Extraction and Reverse Transcription-Polymerase Chain Reaction
Viral RNA extraction was conducted on all positive cell culture samples using Qiagen viral RNA mini kit according to the manufacturer’s instructions. To detect NDV in the isolated samples the specific prime designed M-gene of Newcastle disease virus was used and specific Forward Primer M+4100- 5’-AGT GAT GTG CTC GGA CCT TC-3’, Reverse Primer M-4220- 5ʹCCT GAG GAG AGG CAT TTG CTA-3’, Probe M+4100- 5ʹFAM- TTC TCT AGC AGT GGG ACA GCC -TAMRA −3’ were used to detect M-gene based NDV. Master mix reagents per 25 μL reaction were used from Qiagen one-step RT-PCR kit: 5 μL PCR buffer (5x), 0.5 μL of each primer forward and reverse (20 pmol), 1 μL of probe (6 pmol), 0.8 μL of deoxynucleotide triphosphates, 1.25 μL of 25 mM MgCl2, 0.5 μL of 13.3 u/μL of RNase inhibitor (Promega), 1 μL Qiagen enzyme mix, 6.45 μL Rnase free water and 8 μL of extracted RNA. Real-time PCR was performed using an Applied Biosystems 7500 fast real-time PCR machine. For amplification, reverse transcription at 50°C for 30 min and at 95°C for 15 min was followed by 40 cycles of denaturation at 94°C for 10 s, annealing at 52°C for 30 s and extension at 72°C for 10 s.18 Probe-based fluorescent dye signals were calculated at the extension step of each cycle, and the cycle threshold (Ct) for each sample was observed. The samples that have a Ct value <35 were positive and samples that have >35 Ct value were negative for M genes based on rRT-PCR.19
Results
Clinical Findings
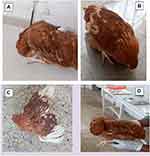
The common clinical signs suggesting Newcastle disease recorded in this study were twisting of the head (Figure 1A), depression (Figure 1B), paralysis of wings (Figure 1C) and paralysis of legs and twisting of the head (Figure 1D). During outbreak investigation of 9 poultry farms a total of 13,000 chickens reared under semi-intensive and intensive poultry farms were examined for Newcastle disease. Out of 13,000 chickens observed 2443 chickens were showing clinical signs and 1233 chickens had died. Overall rates of 18.8%, 9.5% and 50.5% morbidity, mortality and case fatality, respectively were observed in the study area (Table 2).
 |
Table 2 Status of NDV in the Study Area |
Postmortem Findings
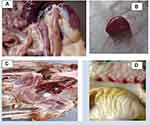
The NDV infected chickens were examined and gross pathological changes were recorded. Postmortem examination of recently dead and humanely killed chickens infected with NDV showed hemorrhagic ulcer in the intestine wall (Figure 2A), enlarged spleen (Figure 2B), degeneration and multifocal necrosis in the liver (Figure 2C) and pin-point hemorrhages in proventriculus (Figure 2D).
Isolation of Newcastle Disease Virus
The present study revealed that among 44 pooled tissue samples of naturally infected chickens, NDV was isolated from 17 (38.63%), as indicated in Table 3. Cytopathic effect was observed in all inoculated samples with clear, small plaques on the DF-1 cell line early from the 3rd day of inoculation. An initial cytopathic effect was observed as small round cells which reflected the light. The foci and syncytia formation occur after a time which causes cell death and detachment from tissue culture plate (Figure 3B arrows).
 |
Table 3 Number of Samples Collected and Cultured Positive Samples from Different NDV Suspected Outbreak Investigations of Chickens |
 |
Figure 3 ND virus grown on DF-1 cells. (A) uninfected monolayer of DF-1 cell and (B) DF-1 cells infected by NDV showing cytopathic effect (arrows). |
In this finding, the virus was isolated from different tissue organs collected from field outbreaks. The descriptions of the isolates by sample type are presented in the Table 4.
 |
Table 4 Newcastle Disease Isolation Rate from Tissue Samples of Chickens |
Reverse Transcriptase Real-Time Polymerase Chain Reaction
A total of 17 isolate samples of RNA were extracted and tested by reverse transcriptase-polymerase chain reaction (RT-PCR) for M gene-based NDV and all of the isolates were positive by RT-PCR. The samples and control RT-PCR amplification curve are indicated in Figure 4B. Ct values ranging from 20.7–34.00 of positive samples, 22.0 ct value of positive control and no ct value for negative control were observed by Applied Biosystems 7500 PCR machine and are indicated in Figure 4B.
 |
Figure 4 Amplification plot result of rRT-PCR. (A) shows rRT-PCR positive samples result with ct value and (B) shows positive and negative controls. |
Discussion
Newcastle disease is a severe viral infection existing worldwide including Ethiopia. The current study was performed for isolation and molecular detection of the virus from active outbreaks.
The current study revealed that distinctive clinical signs of NDV such as twisting of the head and neck, paralysis of wings and legs, depression, ruffling of feather and gasping were observed in the affected chickens. This result was in agreement with the results of previous studies.20–22 A previous report18 shows enlargement and inflammation of eyes, diarrhea, dizziness and lack of appetite were reported which is a slight variation from this finding. However, the clinical signs of Newcastle disease vary depending on the host organs affected.
From a total of 13,000 chickens observed during outbreaks, 2443 were identified as diseased chickens and 1233 had died. The overall morbidity, mortality and case fatality rates observed in infected chickens in the study area were 18.7%, 9.5% and 50.5%, respectively. According to one study21, 21.21% mortality was reported from chickens exposed to outbreaks of ND. Similarly, Saidu and Abdu23 reported a 97.7% mortality rate which is higher than the present finding. This variation might be associated with the immunity status of the chickens and strain of the virus.
Necropsy was conducted on infected chickens with NDV and gross pathological changes were recorded. Postmortem finding of infected chickens with NDV showed hemorrhagic ulcer in the intestine wall, enlarged spleen, degeneration and multifocal necrosis in the liver and pin-point hemorrhages in proventriculus. These results were in line with some other reports.21,24–26 Hemorrhagic laryngotracheitis, congestion and edema of the lungs have been reported27; the reported lesion is different from the present study, and this variation may be due to different strains of NDV that can affect different organs of the chickens. In this study, the observed lesions were indicative of ND depending on the observed gross pathological lesions. Nevertheless, pathogenicity examinations are obligatory to be conducted to estimate the virulence of the virus.7
The current study showed that from 44 pooled specimens of necropsy examination, NDV was isolated from 38.63% (17/44) samples using DF-1 chicken fibroblast cell line. Isolation of virulent NDV from infected chickens confirms the presence of NDV in the study area. The isolated virus from clinical samples reveals characteristics of NDV cytopathic effect, i.e. rounding of cells, formation of syncytia and cell death. Relatively large number of syncytia was found in the isolates which is related with the virulence of the virus. This finding was in close agreement with previous reports16,27 which found that NDV was isolated from suspected birds and the same CPE characteristics were reported in their findings. In the present study, the virus was isolated from different organs of infected chicken samples, with 54.54% from the brain, 54.54% from the lung and trachea, 36.36% from the intestine and 9% from pooled liver, kidney, spleen and heart. According to one report16, NDV was isolated 100% from the spleen, brain, trachea and colon by using chicken embryo fibroblast cell, which is higher than the present study. This variation may be due to the virus load in those organs during infection.
Reverse transcriptase real-time PCR was used, due to its high sensitivity, high specificity, efficiency and mostly its capacity for detecting the virus. Newcastle disease virus was detected from pooled tissue of 17/44 (38.63%) examined chickens. All 17 isolated viruses were positive by reverse transcriptase real-time PCR. The amplification of matrix gene from isolate samples confirmed the chickens were exposed to Newcastle disease. This finding was in agreement with a report28 which isolated and identified the virus from suspected Newcastle disease in Ethiopia by reverse transcriptase real-time PCR.
Conclusion
The present study revealed that NDV was isolated and detected from active outbreaks in the study areas. It is the primary viral disease in poultry farms in these areas and causes significant economic losses. The current finding also showed that NDV is the most important viral infection causing the death of birds managed in the different production systems in the study areas. Fast identification and isolation of the virus are very important for the prevention and control of the infection. The occurrence of ND in poultry farms of the study area should be considered as the causative agent of poultry death in the study areas. Therefore, further molecular characterization is required to identify the strain of virus circulating in the study area. Awareness training for chicken farmers about the impacts of Newcastle disease infection and regular strategic vaccination are essential.
Statistical Analysis
The data obtained from field and laboratory results were recorded, coded and entered into a Microsoft Excel spreadsheet. Statistical analysis was performed by Statistical Package for Social Sciences (SPSS) version 20. Descriptive statistics including frequencies and percentages were used and results were summarized using tables.
Ethics Approval
An ethical clearance certificate for this research was obtained from National animal health diagnostic and investigation center (Reference ARSERC/EC010/2020).
Acknowledgments
The authors would like to thank the national animal health diagnostic and investigation center for full laboratory access and opportunity during the laboratory work.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. CSA. Central Statistical Agency Agricultural Sample Survey. CSA; 2018.
2. Ebsa YA, Harpal S, Negia GG. Challenges and chicken production status of poultry producers in Bishoftu, Ethiopia. Poult Sci. 2019;98(11):5452–5455. doi:10.3382/ps/pez343
3. Cattoli G, Fusaro A, Monne I, et al. Emergence of a new genetic lineage of Newcastle disease virus in West and Central Africa–implications for diagnosis and control. Vet Microbiol. 2010;142(3–4):168–176. doi:10.1016/j.vetmic.2009.09.063
4. Bello MB, Yusoff K, Ideris A, Hair-Bejo M, Peeters BPH, Omar AR. Diagnostic and vaccination approaches for Newcastle disease virus in poultry: the current and emerging perspectives. Biomed Res Int. 2018;2018:7278459. doi:10.1155/2018/7278459
5. Mayo MA. A summary of taxonomic changes recently approved by ICTV. Arch Virol. 2002;147(8):1655–1663. doi:10.1007/s007050200039
6. Xiao S, Nayak B, Samuel A, et al. Generation by reverse genetics of an effective, stable, live-attenuated newcastle disease virus vaccine based on a currently circulating, highly virulent Indonesian strain. PLoS One. 2012;7(12):e52751. doi:10.1371/journal.pone.0052751
7. OIE. Manual of diagnostic tests and vaccines for terrestrial animals mammals, birds and bees. Newcastle Dis. 2012;1:555–574.
8. Jin J, Zhao J, Ren Y, Zhong Q, Zhang G. Contribution of HN protein length diversity to Newcastle disease virus virulence, replication and biological activities. Sci Rep. 2016;6(1):36890. doi:10.1038/srep36890
9. Madadgar O, Karimi V, Nazaktabar A, et al. A study of Newcastle disease virus obtained from exotic caged birds in Tehran between 2009 and 2010. Avian Pathol. 2013;42(1):27–31. doi:10.1080/03079457.2012.752791
10. Alexander DJ. Ecology and epidemiology of Newcastle disease. In: Avian Influenza and Newcastle Disease. Milano: Springer; 2009:19–26.
11. Mohamed MH, Kumar S, Paldurai A, Samal SK. Sequence analysis of fusion protein gene of Newcastle disease virus isolated from outbreaks in Egypt during 2006. Virol J. 2011;8(1):237. doi:10.1186/1743-422X-8-237
12. Rezaeianzadeh G, Dadras H, Safar A, Ali M, Nazemshirazi MH. Serological and molecular study of Newcastle disease virus circulating in village chickens of Fars province. J Vet Med Anim Health. 2011;3(8):105–111.
13. Aschalew ZS, Bewket R. New Castle Disease in Ethiopian Animal Health Year Book. Animal and Plant Health Regulatory Directorate; 2011:22–23.
14. Nanthakumar T, Kataria RS, Tiwari AK, Butchaiah G, Kataria JM. Pathotyping of Newcastle disease viruses by RT-PCR and restriction enzyme analysis. Vet Res Commun. 2000;24(4):275–286. doi:10.1023/A:1006403017578
15. Tiwari AK, Kataria RS, Nanthakumar T, Dash BB, Desai G. Differential detection of Newcastle disease virus strains by degenerate primers based RT-PCR. Comp Immunol Microbiol Infect Dis. 2004;27(3):163–169. doi:10.1016/j.cimid.2003.09.002
16. Haque MH, Hossain MT, Islam MT, Zinnah MA, Khan MSR, Islam MA. Isolation and detection of Newcastle disease virus from field outbreaks in broiler and layer chickens by reverse transcription–polymerase chain reaction. J Vet Med. 2010;8(2):87–92.
17. Li X, Qiu Y, Yu A, et al. Degenerate primers based RT-PCR for rapid detection and differentiation of airborne chicken Newcastle disease virus in chicken houses. J Virol Methods. 2009;158(1–2):1–5. doi:10.1016/j.jviromet.2009.01.011
18. Wise MG, Suarez DL, Seal BS, et al. Development of a real-time reverse-transcription PCR for detection of newcastle disease virus RNA in clinical samples. J Clini Microbiol. 2004;42(1):329–338. doi:10.1128/JCM.42.1.329-338.2004
19. OIE. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals: Mammals, Birds and Bees Paris, France. OIE; 2013:2–14.
20. Alexander D. Newcastle disease, other avian paramyxoviruses, and pneumovirus infections. Dis Poultry. 2003;11:89–107.
21. Bereket M, Beilul G, Fitsum N, Yodahi PA, Yohana S. Outbreak investigation of Newcastle disease virus from vaccinated chickens in Eritrea. Afr J Biotech. 2017;16(32):1717–1723. doi:10.5897/AJB2017.15899
22. Khorajiya JH, Pandey S, Ghodasara PD, et al. Patho-epidemiological study on Genotype-XIII Newcastle disease virus infection in commercial vaccinated layer farms. Vet World. 2015;8(3):372–381. doi:10.14202/vetworld.2015.372-381
23. Sa’idu LA, Abdu PA. Outbreak of Viscerotropic Velogenic form of Newcastle disease in vaccinated six weeks old pullets. J Vet Sci. 2008;7(1):37–40.
24. Murree B, Nizamani ZA, Leghari IH, et al. Pathology and transmission of experimental velogenic viscerotropic newcastle disease in wild pigeons, broiler and aseel chickens. Sci Int. 2016;28(4):3965–3971.
25. Ashraf A, Shah MS, Habib M, et al. Isolation, identification and molecular characterization of highly pathogenic Newcastle disease virus from field outbreaks. Braz Arch Biol Technol. 2016;59. doi:10.1590/1678-4324-2016160301
26. Uddin MA, Islam K, Sultana S, et al. Seroprevalence of antibodies against Newcastle disease in layer chicken at Cox’s Bazar, Bangladesh. Res J Vet Pract. 2014;2:36–39. doi:10.14737/journal.rjvp/2014/2.2.36.39
27. Dodovski A, Krstevski K, Dzadzovski IA, Naletoski I. Molecular detection and characterization of velogenic Newcastle disease virus in common starlings in Macedonia. Vet Arh. 2015;85(6):635–645.
28. Belayheh G, Kyule MN, Melese BA, Fufa D. Isolation and identification of Newcastle disease virus from outbreak cases and apparently healthy local chickens in South West Shewa, Ethiopia. Int J Microb Res. 2016;6(1):5–8.
29. Bemnet G, Ameha Y, Alemayehu Z, Jemanesh KA, Tekalign T. (Trinidad) - Fertilizer N effects on yield and grain quality of durum wheat. Trop Agric. 2003;80(2):1–6.
30. Tesfaye A. Steady-State Ground Water Flow and Contaminant Transport Modelling of Akaki Well Field and Its Surrounding Catchment. M.Sc thesis submitted to the International Institute for geo-information science and earth observation; 2009.
31. Dawit L, Addis MA, Gari G. Distribution, seasonality and abundance of Sotoxys flies in selected districts of central Ethiopia. World Appl Sci. 2012;19(7):998–1002.
32. Berhanu KT, Nengcheng C, Xiang ZA, Dev N. Urbanization in small cities and their significant implications on landscape structures: the case in Ethiopia. Sustainability. 2020;12:1235. doi:10.3390/su12031235
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.