Back to Journals » Journal of Inflammation Research » Volume 16
Integrated DNA Methylation and Transcriptomics Analyses of Lacrimal Glands Identify the Potential Genes Implicated in the Development of Sjögren’s Syndrome-Related Dry Eye
Authors Sun M , Wei Y, Zhang C, Nian H, Du B, Wei R
Received 15 September 2023
Accepted for publication 21 November 2023
Published 29 November 2023 Volume 2023:16 Pages 5697—5714
DOI https://doi.org/10.2147/JIR.S440263
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Mei Sun,* Yankai Wei,* Chengyuan Zhang, Hong Nian, Bei Du, Ruihua Wei
Tianjin Key Laboratory of Retinal Functions and Diseases, Tianjin Branch of National Clinical Research Center for Ocular Disease, Eye Institute and School of Optometry, Tianjin Medical University Eye Hospital, Tianjin, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ruihua Wei, Tianjin Key Laboratory of Retinal Functions and Diseases, Tianjin Branch of National Clinical Research Center for Ocular Disease, Eye Institute and School of Optometry, Tianjin Medical University Eye Hospital, 251 Fukang Road, Nankai District, Tianjin, 300384, People’s Republic of China, Tel/Fax +86 86428862, Email [email protected]
Purpose: Sjögren’s syndrome-related dry eye (SS-related dry eye) is an intractable autoimmune disease characterized by chronic inflammation of lacrimal glands (LGs), where epigenetic factors are proven to play a crucial role in the pathogenesis of this disease. However, the alteration of DNA methylation in LGs and its role in the pathogenesis of SS-related dry eye is still unknown. Here, we performed an integrated analysis of DNA methylation and RNA-Seq data in LGs to identify novel DNA methylation-regulated differentially expressed genes (MeDEGs) in the pathogenesis of SS-related dry eye.
Methods: The DNA methylation and transcription profiles of LGs in NOD mice at different stages of SS-related dry eye (4-, 8-, 12- and 16 weeks old) were generated by reduced representation bisulfite sequencing (RRBS) and RNA-Seq. The differentially methylated genes (DMGs) and differentially expressed genes (DEGs) were analyzed by MethylKit R package and edgeR. Correlation analysis between methylation level and mRNA expression was conducted with R software. The functional correlation of DMGs and DEGs was analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Finally, LG tissues from another litter of NOD mice were collected for methylation-specific polymerase chain reaction (MSP) and quantitative real-time PCR (qRT-PCR) to validate the methylation and expression levels of key genes. CD4+ cell infiltration of LGs was detected by immunofluorescence staining.
Results: Hypermethylation of LGs was identified in NOD mice with the progression of SS-related dry eye and the DMGs were mainly enriched in the GTPases activation and Ras signaling pathway. RNA-seq analysis revealed 1321, 2549, and 3712 DEGs in the 8-, 12- and 16-week-old NOD mice compared with 4-week-old normal control mice. For GO analysis, the DEGs were mainly enriched in T cell immune responses. Further, a total of 140 MeDEGs were obtained by integrated analysis of methylome and transcriptome, which were primarily enriched in T cell activation, proliferation and differentiation. Based on the main GO terms and KEGG pathways of MeDEGs, 8 genes were screened out. The expression levels of these key genes, especially Itgal, Vav1, Irf4 and Icosl, were verified to elevate after the onset of SS-related dry eye in NOD mice and positively correlated with the extent of inflammatory cell infiltration in LGs. Immunofluorescence assay revealed that CD4+ cell infiltration dramatically increased in LGs of SS-related dry eye mice compared with the control mice. And the expression levels of four genes showed significantly positive correlation with the extent of CD4+ cell infiltration in LGs. MSP showed the hypomethylation of the Irf 4 and Itgal promoters in NOD mice with SS-related dry eye compared to control group.
Conclusion: Our study revealed the critical role of epigenetic regulation of T cell immunity-related genes in the progression of SS-related dry eye and reminded us that DNA methylation-regulated genes such as Itgal, Vav1, Irf4 and Icosl may be used as new targets for SS-related dry eye therapy.
Keywords: Sjögren’s syndrome-related dry eye, lacrimal gland, DNA methylation, RNA-seq, T cell-mediated immune response
Introduction
Sjögren’s syndrome (SS) is a chronic autoimmune disease characterized by lymphocytic infiltration of salivary and lacrimal glands (LGs) leading to a clinical picture of dry mouth and dry eye.1 SS-related dry eye is a kind of refractory dry eye disease accompanied by secretory dysfunction of LGs, which may result in severe ocular surface damage and visual impairment.2 At present, the etiology of SS-related dry eye is incompletely understood, which limits the development of diagnosis, management, and effective therapies for SS-related dry eye. Although genetic and environmental factors are thought to contribute to the development of SS-related dry eye,3 increasing evidence has demonstrated that epigenetic modifications, such as DNA methylation, histone modification, non-coding RNA, and m6A methylation, play a key role in the development of various autoimmune eye diseases, including SS-related dry eye.1,4–6
DNA methylation is an important epigenetic modification that can block transcription by recruiting proteins involved in gene repression or by interfering with the binding of sequence-specific transcription factors to their target sequences in candidate genes and lead to changes in gene expression.7 It has been reported that abnormal DNA methylation is critically implicated in the development and progression of autoimmune diseases, including SS.8–10 For example, Yu et al observed that CD4+ T cells of primary SS patients had a significantly hypermethylated FOXP3 promoter compared to healthy controls, and the mRNA and protein levels of FOXP3 were negatively correlated with the mean methylation level in the promoter region.11 Besides, Konsta et al found decreased global DNA methylation in minor salivary glands from SS patients compared with those in non-SS patients. They further demonstrated that the reduction of global DNA methylation was associated with lymphocyte infiltration and the levels of circulating anti-SSB/La Ab, but not with disease activity.12 In a recent paper published in 2023, Chi et al identified two subgroups of SS patients with significantly different DNA methylation levels at the MHC region, based on the genome-wide DNA methylation data from labial salivary gland tissue of SS patients.13 Although the change of DNA methylation in peripheral blood cells and salivary glands of SS patients has been widely studied, little is known about the DNA methylation profile in LGs as well as the correlation between DNA methylation and disease progression of SS-related dry eye.
The non-obese diabetic (NOD) mice, which spontaneously develop ocular surface dryness and autoimmune dacryoadenitis that resemble human SS-related dry eye, have been widely used in SS dry eye-related studies.14 For example, Shin S et al demonstrated that treatment with mesenchymal stem cells could ameliorate the clinical manifestations of SS-related dry eye in NOD mice by increasing tear secretion, decreasing corneal stain scores, recovering goblet cell counts in the conjunctiva, and attenuating foci infiltrations in the LGs.15 In our previous study, we found that Fenofibrate could alleviate SS–like dacryoadenitis in NOD mice via modulating Th1/Th17 and Treg cell responses.16 Furthermore, it has been reported that the NOD mice have significant genetic homology with humans, which is helpful in studying the pathogenic mechanism of SS-related dry eye.17 Given that male NOD mice primarily developed autoimmune dacryoadenitis18 while female NOD mice developed autoimmune sialadenitis first and were easily affected by diabetes,19,20 we here used male NOD mice for the exploration of SS-related dry eye etiology.
In this paper, we investigated the DNA methylation and transcriptomics alterations in the LGs of male NOD mice from 4 weeks old to 16 weeks old by using RRBS and RNA-seq, and analyzed the correlation between methylation and gene regulation to identify the differential methylation-regulated genes correlated to the pathogenesis of SS-related dry eye.
Materials and Methods
Animals and Management
Male non-obese diabetic mice (NOD/ShiLtJ) were obtained from Gem Pharmatech Co., Ltd (Nanjing, China). Age- and gender-matched normal BALB/c mice, also from Gem Pharmatech Co., Ltd, served as controls.21,22 All mice were bred under specific pathogen-free conditions at the animal center of Tianjin Medical University Eye Hospital. All animal experiments were conducted in accordance with the guidelines of the ARVO Statement on Use of Animals in Ophthalmic and Vision Research and approved by the Laboratory Animal Care and Use Committee of Tianjin Medical University.
The NOD mice were randomly assigned into different groups (4-, 8-, 12- and 16-week-old groups) and those accompanied by any pre-existing eye defects were excluded before the experimentation. Mice in each group were maintained for 4, 8, 12 and 16 weeks with food and water available and ad-libitum, respectively. The number of mice used for RRBS and RNA-seq was 2 in each group. And we used 6–9 NOD mice for each group to investigate the development of SS-related dry eye.
Tear Secretion Test
Tear volume was quantified by phenol red cotton threads (Jingming Co., Ltd, Tianjin, China). Briefly, the threads were held with forceps and inserted into the lower conjunctival fornix close to the lateral canthus for 30 seconds. The levels of basal tear production were expressed as millimeters of the length of thread wetted and turned red by tears. Each eye of the mice was measured and tested three times. The average value was determined as the final length.
Corneal Fluorescein Staining
1 µL of 2% sodium fluorescein was instilled into the lateral conjunctival sac of each eye of mice followed by 3 artificial blinks. After 2 minutes, fluorescein staining of corneal epithelial defects was visualized and photographed by a Phoenix MICRON IV Anterior Segment Slit Lamp system (Phoenix MICRON, New York, USA). The staining of cornea epithelial was scored according to the grading system as described previously.23
Histological Assessment of LGs
For histopathological evaluation of LGs, tissues were fixated in 10% formalin, embedded in paraffin, and sectioned as described previously.16 The LG sections were stained routinely with hematoxylin and eosin (H&E) and then photographed by light microscopy (BX51; Olympus Corporation, Tokyo, Japan). Inflammation was quantified by standard focus-scoring with a focus defined as an aggregate of at least 50 lymphocytes and the focus score defined as the number of foci per 4 mm2 of tissue. Tissue areas were calculated by ImageJ software (National Institutes of Health, Bethesda, MD, USA).24,25
Immunofluorescence Staining
5 μm LG tissue paraffin slides were incubated at 70°C for 1 h and deparaffinized in xylene. After rehydration in a descending series of ethanol, the slides were subjected to citric acid antigen retrieval. Following blocking with 5% goat serum, LG sections were incubated with anti-CD4-rabbit IgG (1:300; Abcam, Boston, MA, USA) at 4°C overnight. Subsequently, tissue sections were incubated with Alexa Fluor 488–conjugated goat antirabbit IgG (1:500; ImmunoWay, Plano, TX, USA) for 2 h at room temperature being protected from light. DAPI (Solarbio) was used to label nuclear DNA. And images were captured using the laser scanning confocal microscope (LSM800, Zeiss, Germany).
Reduced Representation Bisulfite Sequencing (RRBS)
LG tissues were carefully dissected from NOD mice of 4, 8, 12 and 16 weeks of age, respectively. Left LG tissues were subjected to histopathological assessment, and the right LG were immediately immersed into liquid nitrogen, and stored at −80°C for subsequent investigation. Genomic DNA from pooled LG samples was extracted by TIANamp Genomic DNA Kit (TIANGEN) following the manufacturer’s protocols. A total amount of 1 µg genomic DNA spiked with unmethylated lambda DNA was firstly digested by MspI enzyme followed by libraries construction as the Illumina Pair-End protocol and reduced representation bisulfite sequencing (RRBS) described previously.26 Briefly, the purified and fragmented DNA was processed to repair, blunt and phosphorylate ends. After adenylation, cytosine-methylated adaptors were ligated to the fragmented DNA according to the manufacturer’s instructions. The DNA fragments were then purified using MinElute PCR Purification Kit (Qiagen) and treated with ZYMO EZ DNA Methylation-Gold Kit™ to convert unmethylated cytosine into uracil before the PCR amplification was carried out. The insert size of the selected library was analyzed on the Bioanalyzer analysis system (Agilent, Santa Clara, USA), and library concentration was quantified by the real-time quantitative PCR (qRT-PCR), subsequently sequenced on Illumina sequencing platform.
RNA Isolation and RNA-Sequencing
The RNA-sequencing (RNA‑seq) libraries were generated by E-GENE Co., Ltd (Shenzhen, China) following the manufacturer’s protocols. Briefly, the total RNA was extracted from LG tissues using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. After treatment with RNase-free DNase I, 1 µg of total RNA was firstly purified into mRNA containing poly(A) using Oligo(dT) beads. The captured mRNA was then fragmented, reverse transcribed, converted to double-stranded cDNA, and adapter-ligated with unique indices. Finally, PCR was carried out to amplify the adapter-ligated to cDNA and the libraries were validated using Agilent Bioanalyzer 2100 for quality control, followed by qRT-PCR quantification and sequencing on the Illumina sequencing platform.
Data Analysis and Functional Enrichment Analysis
Adapter sequences were trimmed by using cutadapt. And the cleaned reads were mapped back to the genome using BSMAP software version 2.90.27 Only unique mapped reads were used to calculate methylation ratios. Methylation ratios were extracted from BSMAP output (SAM) using a Python script (methratio.py) which is distributed with the BSMAP package. We choose cytosines in a CpG context with sufficient sequencing depth (greater than or equal to 5x coverage) for further analysis. Differentially methylated cytosines (DMCs) and differentially methylated regions (DMRs) were separately detected using MethylKit28 and metilene29 in de-novo mode among CpG sites with at least 5x coverage. Then detected DMRs were filtered according to the standard: (1) P-value must be less than 0.05; (2) methylation level difference must be greater than 0.1; (3) CpG number contained in DMR must be greater than 5; (4) the length of the DMR must be greater than 50bp.
Quality control of raw reads was performed using FastQC (version 0.11.5) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and clean reads were obtained following the removal of low‑quality reads with Trimmomatic (version 0.38) software.30 We then use Hisat2 (v2.2.1)31 to perform clean reads mapping to Mus musculus genome reference (mm10). Gene expression profiling was based on the number of mapped reads. Transcripts Per Kilobase of exon model per Million mapped reads values were calculated by StringTie (v2.1.7)32 and were used to estimate the expressed values of genes. The differentially expressed genes (DEGs) among NOD mice groups were obtained using edgeR (v1.34.0)33 with a Padj-value cutoff < 0.05 and an absolute log2(fold-change) of > 1.
Principle component analysis (PCA) was performed to visualize the statistical robustness of expression data by utilizing the R language models (2.18.1). Data were clustered based on hierarchical clustering analysis (HC) using R language factoextra (1.0.7) and each row was normalized using a z-score. The circle plot was based on R language (version 4.1.1). Functional annotations were subsequently conducted to elucidate significant biological implications of differentially methylated-related genes (DMGs) and DEGs in different NOD mice groups according to Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genome (KEGG) pathway analyses, provided by Cluster Profiler (v4.4.2)34 or DAVID Bioinformatics Resources (version 2021).35 Padj-value < 0.05 was defined as significant in all of the enrichment analysis.
Correlation Analysis
DNA methylation is known to have a regulatory effect on gene expression. The DMR annotation results of CGs in the promoter region and gene body region were combined with the DEGs results for VENN analysis and single-sample gene-wide association analysis. Additionally, enrichment analysis of genes related to DMRs was implemented by DAVID Bioinformatics Resources (version 2021) to assess the potential biological functions. GO terms and KEGG pathways with Padj-value <0.05 were considered significantly enriched.
Quantitative Real-Time PCR (qRT-PCR)
The total RNA of LGs was isolated using EZ-press RNA Purification Kit (EZBioscience, Roseville, MN, USA) and reverse-transcribed to synthesize cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocols. qRT-PCR was performed using gene-specific primers and SYBR Green PCR Master Mix (Roche, Mannheim, Germany) on Roche LightCycler 480 II Analyzer. Gene expression relative to housekeeping gene GAPDH was calculated using the comparative threshold cycle (2-ΔΔCt) method. The sequences of qRT-PCR primers are listed in Table 1.
 |
Table 1 mRNA Primer Sequences |
Methylation-Specific PCR (MSP)
The methylation levels in the promotor region of Itgal, Icosl, Vav1 and Irf4 were further examined through MSP. The genomic DNA was extracted from the LGs of NOD mice using the Animal Genomic DNA Quick Extraction Kit (D0065 M, Beyotime, China) and further modified with sodium bisulfite using the EZ DNA Methylation-Gold Kit (Zymo, Foster City, CA) following the manufacturer’s protocols. Primers for methylated and unmethylated promoter sequences were designed using MethPrimer software and listed in Table 2. Subsequently, 2% agarose gels were utilized to analyze the PCR product.
 |
Table 2 Sequences of Primers Used for MSP |
Statistical Analysis
All statistical analyses of the data were performed with GraphPad Prism software (GraphPad Software, San Diego, CA, USA). The normality of continuous variable was evaluated by Shapiro–Wilk test. All data are expressed as mean ± SD. For normally distributed data, differences among groups were analyzed by using one-way ANOVA. Kruskal–Wallis test was used for nonparametric analysis according to the number of groups. The association analysis was evaluated by the Spearman rank correlation test. P-value < 0.05 was considered statistically significant.
Results
The Severity of Dry Eye Showed Progressive Exacerbation with Increasing Age of NOD Mice
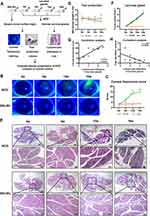
To determine the development of SS-related dry eye in animal models, NOD mice from 4 to 16 weeks of age were selected in our study for the evaluation of corneal fluorescein staining, tear secretion and LGs histology36,37 (Figure 1A). As shown in Figure 1B and C, corneal epithelial defects were detected in NOD mice as early as 8 weeks of age and the corneal staining score showed a significant increase from 12 weeks old compared to those of 4-week-old NOD mice. Tear secretion volume measured by phenol red thread exhibited significant reduction from 12 weeks of age and decreased continually in NOD mice (Figure 1D). In contrast, the corneal fluorescein staining and tear secretion did not show significant alterations in BALB/c mice from 4 to 16 weeks old (Figure 1B-D). Consistent with the clinical results, histological analysis of LGs revealed that lymphocytic infiltration was easily detected as early as 8 weeks old in NOD mice and increased significantly until 16 weeks old, while there was no significant lymphocytic infiltration in the LGs of 4 to 16-week-old BALB/c mice (Figure 1E and F). Furthermore, the correlation analysis revealed that the extent of inflammatory cell infiltration in LGs was positively correlated with the corneal fluorescein score and negatively correlated with tear secretion volume (Figure 1G and H). Altogether, these data suggested that the clinical manifestations and histopathological alterations of SS-related dry eye in NOD mice were observed from 8 weeks and gradually worsened until 16 weeks of age.
DNA Methylation Levels Were Increased in the LGsof NOD Mice During the Development of SS-Related Dry Eye
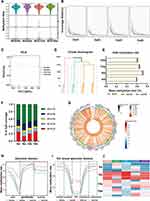
Recently, increasing evidence has demonstrated the contributing role of DNA methylation in the initiation and progression of autoimmune diseases.38 To investigate whether DNA methylation is involved in the development of SS-related dry eye in animal models, we isolated LGs of 4 to 16-week-old NOD mice and profiled DNA methylation across the genome using the RRBS technique. The bisulfite conversion rates of each sample were higher than 98% (Table 3) and the coverage rate of C sites was uniform for each group (Figure 2A and B), which indicated integrity, accuracy, and high uniformity of the sequencing data. Principal component analysis (PCA) and hierarchical clustering (HC) showed distinct DNA methylation profiles in LGs from different ages of NOD mice (Figure 2C and D). By comparing the global methylation rates of all C sites among the four groups, we found that DNA methylation mainly occurred on CG cytosines (mean methylation level, m= 62.8%) and the methylated rates of CG cytosines in mice at 8, 12, and 16 weeks old were higher in comparison to 4-week-old group (Figure 2E-G). Further, we observed that the differential CG sites were mainly located in regions 2-kb upstream of the gene body (promoter region) (Figure 2H). To define the specific position of methylated sites in the promoter, the promoter was further divided into five gene functional regions: CGI-upshelf, CGI-upshore, CG islands (CGI), CGI-downshore, and CGI-downshelf. As depicted in Figure 2I and J, the 8-week-old group showed significantly higher CG methylation levels in the CGI region than the other three groups. These data collectively suggested that the methylation levels of LGs were increased during the development of SS-related dry eye.
 |
Table 3 Basic sequencing quality control, bisulfite conversion rates, and map rate of RRBS libraries |
Functional Enrichment of the Differentially Methylated Regions-Related Genes (DMGs) Identified in LGs of SS-Related Dry Eye Mice
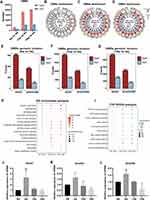
Next, we attempted to explore the association between methylation differences and the pathogenesis of SS-related dry eye by calculating the differentially methylated regions (DMRs) among 4-, 8-, 12-, and 16-week-old NOD mice under CG contexts. Applying the number of differentially methylated CG sites in the region ≥ 5 and the length of the DMR must be greater than 50bp (P-value < 0.05) returned 4132 DMRs (including 2956 hyper- and 1176 hypomethylated), 23,396 DMRs (17,927 hyper- and 5469 hypomethylated) and 26,135 DMRs (20,422 hyper- and 5713 hypomethylated) in 8-, 12-, and 16-week-old groups compared with 4-week-old control group, respectively (Figure 3A). Consistent with this, the circle plot showed that there were more hyper-DMRs than hypo-DMRs along the chromosome. Most DMRs were located on autosomes, while fewer DMRs were located on sex chromosomes (Figure 3B-D). Also, more DMRs were located in exons than promotor regions among different aged mice (Figure 3E-G). To define the function of DMRs-related genes (DMGs), we performed GO enrichment and KEGG pathway analysis. The GO analysis revealed that DMGs were significantly enriched in certain molecular function, including GTPase activator activity, nucleoside-triphosphatase regulator activity and GTPase regulator activity (Figure 3H), which have been considered vital for regulating immune cells migration in SS.39–41 Furthermore, the DMGs were enriched in biological process involved in synapse organization, axonogenesis and cell junction assembly (Figure 3H). PI3K-Akt signaling pathway, MAPK signaling pathway and Ras signaling pathway were also found to be enriched through KEGG analysis (Figure 3I). Each of these pathways has previously been linked to the development of dry eye diseases.42
DNA methylation is predominantly catalyzed through a family of DNA methyltransferases (DMNTs), which consists of three members: DNMT1, DNMT3A and DNMT3B.43–45 Thus, we investigated the mRNA level of three DNMT subtypes (Dnmt1, Dnmt3a, and Dnmt3b) in LGs of different age groups of NOD mice. The results showed that the mRNA level of Dnmt1 and Dnmt3b in LGs was significantly elevated in 8-week-old group compared with 4-week-old group, while there was no significant difference in Dnmt3a expression (Figure 3J-L), suggesting that Dnmt1 and Dnmt3b may play a major role in DNA methylation regulation of LGs.
Transcriptome Analysis of LGs During the Development of SS-Related Dry Eye
As the DNA methylome has been altered during SS-related dry eye progression, it is necessary to study whether the transcriptional profiles are affected. To delineate possible dysregulation of gene transcription during the development of SS-related dry eye, RNA sequencing (RNA-seq) was conducted on LGs from the 4-, 8-, 12-, and 16-week-old NOD mice. High-quality data, characterized by more than 70% mapping rates of clean reads for all samples, were used for further analysis (Figure 4A and Table 4). The PCA and HC results showed a clear separation among the four groups (Figure 4B-C). Transcriptome analysis revealed 1321, 2549 and 3712 differentially expressed genes (DEGs) in the 8-, 12- and 16-week-old groups compared with 4-week-old NOD mice, respectively (Figure 4D). By performing Venn diagram analysis, we further identified 858 overlapping upregulated DEGs in different age groups (Figure 4E). These DEGs were mainly involved in immunological synapse, positive regulation of T cell activation, and MHC class II protein complex binding (Figure 4F-H). Besides, KEGG analysis revealed that the 858 genes were significantly enriched in cell adhesion molecules, Th17 cell differentiation, antigen processing and presentation, and T/B cell receptor signaling pathway (Figure 4I). These results indicated a wide range of immune response-related genes are upregulated in NOD mice during the development of SS-related dry eye.
 |
Table 4 Basic Sequencing Quality Control and Map Rate of RNA-Seq Libraries |
Correlation Analysis between DMRs and genome-Wide Gene Expression
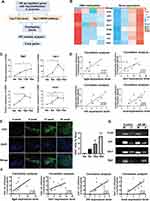
To identify the potential relationship between DNA methylation changes and gene expression in LGs of NOD mice at different disease stages, we performed an integrated analysis of DNA methylation and transcriptomic profiles (Figure 5A). Given that the critical role of DNA methylation within promoter regions in gene transcriptional regulation,46 we focused on DMRs within gene promoter regions for further analysis. As shown in Figure 5B, a total of 140 overexpressed genes with hypomethylated DMRs in the promoters were obtained by the Venn diagram in 8-,12- and 16-week-old NOD mice (2, 46, and 92 genes, respectively) compared to the control group. The results of GO analysis showed that these upregulated genes with hypomethylated DMRs were significantly enriched in positive regulation of T cell proliferation, T cell activation, and T-helper 17 cell lineage commitment (Figure 5C). And the top 5 pathways enriched by KEGG were intestinal immune network for IgA production, T/B cell receptor signaling pathway, Th17 cell differentiation signaling pathway and cell adhesion molecules (Figure 5D).
Identification of Key DNA Methylation-Regulated Genes Associated with SS-Related Dry Eye
Based on the literature, GO, KEGG and PPI network analysis, eight upregulated genes with hypomethylation in the promoter region, including Icosl, Batf, Irf4, Itgal, Vav1, Tgfβ1, Jak2 and CD86, were screened (Figures 6A and B, S1A and Tables S1-S3) and validated by qRT-PCR. The data showed that the expression levels of Icosl, Irf4, Vav1 and Jak2 were significantly increased in LGs of 12-week-old NOD mice, and Itgal was significantly upregulated in LGs of 16-week-old mice compared to the control group (Figures 6C and S1B). On the contrary, Batf, CD86 and Tgfβ1 did not have significant alteration among the four groups (Figure S1B). Further correlation analysis revealed that the expression levels of Itgal, Vav1, Irf4, and Icosl were positively correlated with the extent of inflammatory cell infiltration in LGs (Figure 6D). To clarify the relationship between these four genes (Itgal, Vav1, Irf4, and Icosl) and T cell response in this disease context, we performed immunofluorescence assay of LGs and observed that CD4+ cell infiltration dramatically increased in LGs of SS-related dry eye mice compared with the control mice (Figure 6E). Also, correlation analysis revealed that the expression levels of Itgal, Vav1, Irf4, and Icosl showed significantly positive correlation with the extent of CD4+ cell infiltration in LGs (Figure 6F). In addition, using methylation-specific PCR (MSP), we observed hypomethylation of the Irf4 and Itgal promoters in NOD mice with SS-related dry eye compared to those 4-week-old mice without dry eye (Figure 6G). Therefore, these data indicate that Itgal, Vav1, Irf4, and Icosl are closely associated with the development of SS-related dry eye by modulating T cell response, and DNA hypomethylation of the promotor region may be responsible for the upregulation of Itgal and Irf4 in SS-related dry eye.
Discussion
Accumulating evidence has demonstrated that epigenetic factors are critically implicated in the pathogenesis of SS-related dry eye.47 DNA methylation, the main focus of epigenetic modification, has been proven to be altered in peripheral blood cells and salivary glands of SS patients and closely associated with the development of SS.48 Nevertheless, the DNA methylation profiling in LGs and their roles in SS-related dry eye remains unclear. Here, by performing RRBS, we found that the methylation levels were increased in LGs of 8-, 12- and 16-week-old NOD mice when compared with the 4-week-old mice. Further combined analysis of DNA methylome and transcriptome revealed that several genes related to T cell immunity, including Itgal, Icosl, Vav1 and Irf4, were upregulated during the development of SS-related dry eye. Also, hypomethylation of the Irf4 and Itgal promoters were observed in SS-related dry eye mice. Our results thus suggest that altered DNA methylation is closely linked to the occurrence and development of SS-related dry eye, possibly in part because it affects the expression of T cell-related genes in LGs.
It has been reported that CG methylations play a vital role in regulating gene transcription and are critically involved in various autoimmune diseases.49,50 In this study, we found the methylation levels of CG sites were significantly elevated in LGs of NOD mice during the development of SS-related dry eye, suggesting that hypermethylation of genomic DNA in LGs may be closely associated with the pathogenesis of SS-related dry eye. Interestingly, the clinical manifestations of dry eye and inflammatory cell infiltration of LGs in 8-week-old mice were less severe than those in 12- and 16-week-old mice, but the methylome of LGs has changed on a large scale. This might be due to epigenetic changes taking place earlier than pathophysiological changes.51–53 Supporting this, DNA methyltransferases, including Dnmt1 and Dnmt3b, were observed to be upregulated in 8-week-old NOD mice. The causes underlying the alterations in DNA methylation especially DNA methyltransferases await further study. In addition, several previous studies have established the association between DNA methylation changes in salivary glands and peripheral blood cells and SS, revealing that genomic DNA methylation levels were decreased in salivary glands54 and peripheral blood cells of SS patients.55 Differences in species and tissue types may account for the discrepancy in these studies and our findings.
Rab27a, a member of the superfamily of Ras-related small GTPases, has been demonstrated to promote the secretion of cathepsin S in lacrimal gland acinar cells and be closely linked to SS-related dry eye.56,57 Here, our analysis revealed that Ras signaling pathway and its related molecules were enriched in both GO and KEGG analysis, which indicated that these DMGs may contribute to the occurrence and development of SS-related dry eye by affecting Ras pathway and its downstream molecules. In-depth research is warranted to investigate the changes of these DMGs in SS-related dry eye and their effect on the development of SS-related dry eye. T cells are essential for the immune pathogenesis of SS-related dry eye.58 It has been demonstrated that LGs can express major histocompatibility complex class II (MHC II) or human leukocyte antigen-DR and costimulatory molecules, and thereby act as nonprofessional antigen-presenting cells for T cell activation, promoting the development of SS-related dry eye.59 Administration of small molecules inhibitor targeting MHC II could reduce the infiltration of Th1 and Th17 cells in the exocrine glands, and improve SS-like symptoms, such as SS-related dry eye, in NOD mice.60 Consistent with these findings, our study proved that 858 upregulated DEGs in LGs of 8-, 12- and 16-week-old groups were closely related to T cell immune response, such as positive regulation of T cell activation and peptide antigen assembly with MHC class II protein complex,58,61,62 indicating that these upregulated genes may contribute to SS-related dry eye by directly promoting T cell activation or acting on MHC class II molecules in LGs to indirectly drive T cell response.
140 overexpressed genes with hypomethylated DMRs in the promoter based on combined RRBS and RNA-seq data were further studied. These genes were mainly involved in T/B cell receptor signaling pathway and Th17 cell differentiation signaling pathway, which have been identified to play key roles in the pathogenesis of SS-related dry eye.63–65 Among these candidate genes, it was further confirmed that Icosl, Irf4 and Vav1 were upregulated in LGs of 12-week-old NOD mice, and Itgal was upregulated in LGs of 16-week-old mice compared to the control group, suggesting that upregulated Itgal, Icosl, Vav1 and Irf4 might participate in the development of SS-related dry eye. Indeed, we observed that the expression levels of Itgal, Icosl, Vav1 and Irf4 were positively correlated with the severity of CD4+ cell infiltration in LGs. By performing MSP assay, we further found decreased methylation levels in the promotor region of Irf4 and Itgal. This indicated that hypomethylation of promotor region may lead to overexpression of Irf4 and Itgal in LGs of SS-related dry eye mice and contribute to the pathogenesis of SS-related dry eye. Consistent with our observations is a previous study showing that the promoter of ITGAL was hypomethylated in T cells from lupus patients, which led to elevated mRNA expression of ITGAL.66,67 ITGAL, also known as CD11a or LFA-1, has been confirmed to be critical for the occurrence and development of dry eye,68,69 and the LFA-1 antagonist has been approved by the FDA in 2016 as a new drug for the treatment of dry eye.70,71 Targeting LFA-1 in dry eye mouse model could reduce ocular surface epithelial cell damage and increase goblet cell density.72 Therefore, elucidating the upstream molecules of ITGAL can provide new ideas for the treatment of dry eye diseases. Besides, IRF4, VAV1, and ICOSL have been identified as key regulators of T cell response.73–75 Our results provide a clue that Icosl, Irf4, and Vav1 may be new biomarkers for the diagnosis of SS-related dry eye. However, the specific role and mechanism of Icosl, Irf4, and Vav1 in SS-related dry eye await further investigation.
The link between epigenetic factors and autoimmune eye disease has gotten more and more attention. To date, few studies have been performed on the role of DNA methylation in LGs of SS-related dry eye. This work could constitute a baseline for other future studies focusing on the potential influence of DNA methylation on the pathogenesis of SS-related dry eye. However, the present study has several potential limitations. First, considering that the samples were taken from NOD mice, the results in this study need to be explored in SS-related dry eye patients by further research. And second, the biological mechanisms of the DMR-related genes and signaling pathways in the progression of SS-related dry eye should be confirmed by further mechanistic studies. Finally, given that the LG tissue consists of a variety of cells, using the whole LG tissue rather than distinguishing different types of cells for sequencing may affect data mining to some extent. Future application of single-cell techniques might provide more accurate results and further help to reveal the underlying pathogenesis of SS-related dry eye.
Conclusion
We demonstrated that genomic DNA methylation levels were increased in LGs of NOD mice during the development of SS-related dry eye. Itgal, Icosl, Vav1 and Irf4, which were upregulated in LGs of NOD mice, were closely associated with T cell-mediated immune response and may play vital roles in the pathogenesis of SS-related dry eye. Additionally, DNA methylation levels of the Itgal and Irf4 promotor region were reduced, which may be responsible for the upregulation of Itgal and Irf4 in SS-related dry eye. These findings can help to further understanding of SS-related dry eye etiology and provide novel targets for the diagnosis and treatment of SS-related dry eye.
Data Sharing Statement
Datasets presented in this study are available from the corresponding author on request.
Ethical Approval
The animal study protocol was approved by the Institutional Review Board of the Laboratory Animal Care and Use Committee of Tianjin Medical University Eye Hospital (protocol code and date of approval: TJYY2022122074, 21 December 2022).
Acknowledgments
This work was funded by the National Natural Science Foundation of China (82201157, 82070929) and Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK-037A).
Disclosure
The authors reported no conflicts of interest in this work.
References
1. Imgenberg-Kreuz J, Sandling JK, Nordmark G. Epigenetic alterations in primary Sjögren’s syndrome - an overview. Clin Immunol. 2018;196:12–20. doi:10.1016/j.clim.2018.04.004
2. Choudhry HS, Hosseini S, Choudhry HS, Fatahzadeh M, Khianey R, Dastjerdi MH. Updates in diagnostics, treatments, and correlations between oral and ocular manifestations of Sjogren’s syndrome. Ocul Surf. 2022;26:75–87. doi:10.1016/j.jtos.2022.08.001
3. Foulks GN, Forstot SL, Donshik PC, et al. Clinical guidelines for management of dry eye associated with Sjögren disease. Ocul Surf. 2015;13(2):118–132. doi:10.1016/j.jtos.2014.12.001
4. Lafontaine N, Wilson SG, Walsh JP. DNA Methylation in Autoimmune Thyroid Disease. J Clin Endocrinol Metab. 2023;108(3):604–613. doi:10.1210/clinem/dgac664
5. Qiu Y, Zhu Y, Yu H, Zhou C, Kijlstra A, Yang P. Dynamic DNA Methylation Changes of Tbx21 and Rorc during Experimental Autoimmune Uveitis in Mice. Mediators Inflamm. 2018;2018:9129163. doi:10.1155/2018/9129163
6. Cheng L, Li H, Zhan H, et al. Alterations of m6A RNA methylation regulators contribute to autophagy and immune infiltration in primary Sjögren’s syndrome. Front Immunol. 2022;13:949206. doi:10.3389/fimmu.2022.949206
7. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi:10.1038/npp.2012.112
8. Chuang TJ, Chen FC. DNA methylation is associated with an increased level of conservation at nondegenerate nucleotides in mammals. Mol Biol Evol. 2014;31(2):387–396. doi:10.1093/molbev/mst208
9. Zhao F, Wu W, Wei Q, et al. Exogenous adrenocorticotropic hormone affects genome-wide DNA methylation and transcriptome of corpus luteum in sows. FASEB j. 2019;33(3):3264–3278. doi:10.1096/fj.201801081RRR
10. Du Y, Li J, Wu J, Zeng F, He C. Exploration of the pathogenesis of Sjögren’s syndrome via DNA methylation and transcriptome analyses. Clin Rheumatol. 2022;41(9):2765–2777. doi:10.1007/s10067-022-06200-4
11. Yu X, Liang G, Yin H, et al. DNA hypermethylation leads to lower FOXP3 expression in CD4+ T cells of patients with primary Sjögren’s syndrome. Clin Immunol. 2013;148(2):254–257. doi:10.1016/j.clim.2013.05.005
12. Konsta OD, Le Dantec C, Charras A, et al. Defective DNA methylation in salivary gland epithelial acini from patients with Sjögren’s syndrome is associated with SSB gene expression, anti-SSB/LA detection, and lymphocyte infiltration. J Autoimmun. 2016;68:30–38. doi:10.1016/j.jaut.2015.12.002
13. Chi C, Solomon O, Shiboski C, et al. Identification of Sjögren’s syndrome patient subgroups by clustering of labial salivary gland DNA methylation profiles. PLoS One. 2023;18(3):e0281891. doi:10.1371/journal.pone.0281891
14. Chiorini JA, Cihakova D, Ouellette CE, Caturegli P. Sjögren syndrome: advances in the pathogenesis from animal models. J Autoimmun. 2009;33(3–4):190–196. doi:10.1016/j.jaut.2009.09.009
15. Shin S, Yoon SG, Kim M, et al. The Effect of Mesenchymal Stem Cells on Dry Eye in Sjogren Syndrome Mouse Model. Int J Mol Sci. 2023;24(2):1039. doi:10.3390/ijms24021039
16. Guo X, Dang W, Li N, et al. PPAR-α Agonist Fenofibrate Ameliorates Sjögren Syndrome-Like Dacryoadenitis by Modulating Th1/Th17 and Treg Cell Responses in NOD Mice. Invest Ophthalmol Vis Sci. 2022;63(6):12. doi:10.1167/iovs.63.6.12
17. Masli S, Dartt DA. Mouse Models of Sjögren’s Syndrome with Ocular Surface Disease. Int J Mol Sci. 2020;21(23):9112. doi:10.3390/ijms21239112
18. Schenke-Layland K, Xie J, Angelis E, et al. Increased degradation of extracellular matrix structures of lacrimal glands implicated in the pathogenesis of Sjögren’s syndrome. Matrix Biol. 2008;27(1):53–66. doi:10.1016/j.matbio.2007.07.005
19. Robinson CP, Cornelius J, Bounous DI, Yamamoto H, Humphreys-Beher MG, Peck AB. Infiltrating lymphocyte populations and cytokine production in the salivary and lacrimal glands of autoimmune NOD mice. Adv Exp Med Biol. 1998;438:493–497. doi:10.1007/978-1-4615-5359-5_68
20. Yamano S, Atkinson JC, Baum BJ, Fox PC. Salivary gland cytokine expression in NOD and normal BALB/c mice. Clin Immunol. 1999;92(3):265–275. doi:10.1006/clim.1999.4759
21. Guo H, Ju Y, Choi M, et al. Supra-lacrimal protein-based carriers for cyclosporine A reduce Th17-mediated autoimmunity in murine model of Sjögren’s syndrome. Biomaterials. 2022;283:121441. doi:10.1016/j.biomaterials.2022.121441
22. Ohno Y, Satoh K, Shitara A, Into T, Kashimata M. Arginase 1 is involved in lacrimal hyposecretion in male NOD mice, a model of Sjögren’s syndrome, regardless of dacryoadenitis status. J Physiol. 2020;598(21):4907–4925. doi:10.1113/jp280090
23. Xiao X, Luo P, Zhao H, et al. Amniotic membrane extract ameliorates benzalkonium chloride-induced dry eye in a murine model. Exp Eye Res. 2013;115:31–40. doi:10.1016/j.exer.2013.06.005
24. Debreceni IL, Chimenti MS, Serreze DV, Geurts AM, Chen YG, Lieberman SM. Toll-Like Receptor 7 Is Required for Lacrimal Gland Autoimmunity and Type 1 Diabetes Development in Male Nonobese Diabetic Mice. Int J Mol Sci. 2020;21(24):9478. doi:10.3390/ijms21249478
25. Allred MG, Chimenti MS, Ciecko AE, Chen YG, Lieberman SM. Characterization of Type I Interferon-Associated Chemokines and Cytokines in Lacrimal Glands of Nonobese Diabetic Mice. Int J Mol Sci. 2021;22(7):67. doi:10.3390/ijms22073767
26. Meng M, Li X, Ge H, et al. Noninvasive prenatal testing for autosomal recessive conditions by maternal plasma sequencing in a case of congenital deafness. Genet Med. 2014;16(12):972–976. doi:10.1038/gim.2014.51
27. Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinf. 2009;10:232. doi:10.1186/1471-2105-10-232
28. Akalin A, Kormaksson M, Li S, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. doi:10.1186/gb-2012-13-10-r87
29. Jühling F, Kretzmer H, Bernhart SH, Otto C, Stadler PF, Hoffmann S. metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2016;26(2):256–262. doi:10.1101/gr.196394.115
30. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi:10.1093/bioinformatics/btu170
31. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi:10.1038/s41587-019-0201-4
32. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi:10.1038/nprot.2016.095
33. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8
34. Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2(3):100141. doi:10.1016/j.xinn.2021.100141
35. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi:10.1093/nar/gkn923
36. Takahashi M, Ishimaru N, Yanagi K, Haneji N, Saito I, Hayashi Y. High incidence of autoimmune dacryoadenitis in male non-obese diabetic (NOD) mice depending on sex steroid. Clin Exp Immunol. 1997;109(3):555–561. doi:10.1046/j.1365-2249.1997.4691368.x
37. Hunger RE, Carnaud C, Vogt I, Mueller C. Male gonadal environment paradoxically promotes dacryoadenitis in nonobese diabetic mice. J Clin Invest. 1998;101(6):1300–1309. doi:10.1172/jci1230
38. Imgenberg-Kreuz J, Almlöf JC, Leonard D, et al. Shared and Unique Patterns of DNA Methylation in Systemic Lupus Erythematosus and Primary Sjögren’s Syndrome. Front Immunol. 2019;10:1686. doi:10.3389/fimmu.2019.01686
39. Biro M, Munoz MA, Weninger W. Targeting Rho-GTPases in immune cell migration and inflammation. Br J Pharmacol. 2014;171(24):5491–5506. doi:10.1111/bph.12658
40. Peck AB, Nguyen CQ, Ambrus JL. Upregulated Chemokine and Rho-GTPase Genes Define Immune Cell Emigration into Salivary Glands of Sjögren’s Syndrome-Susceptible C57BL/6.NOD-Aec1Aec2 Mice. Int J Mol Sci. 2021;22(13):7176. doi:10.3390/ijms22137176
41. Peck AB, Nguyen CQ, Ambrus JL Jr. A MZB Cell Activation Profile Present in the Lacrimal Glands of Sjögren’s Syndrome-Susceptible C57BL/6.NOD-Aec1Aec2 Mice Defined by Global RNA Transcriptomic Analyses. Int J Mol Sci. 2022;23(11):6160. doi:10.3390/ijms23116106
42. Han Y, Guo S, Li Y, et al. Berberine ameliorate inflammation and apoptosis via modulating PI3K/AKT/NFκB and MAPK pathway on dry eye. Phytomedicine. 2023;121:155081. doi:10.1016/j.phymed.2023.155081
43. Chen YC, Lin IC, Su MC, et al. Autophagy impairment in patients with obstructive sleep apnea modulates intermittent hypoxia-induced oxidative stress and cell apoptosis via hypermethylation of the ATG5 gene promoter region. Eur J Med Res. 2023;28(1):82. doi:10.1186/s40001-023-01051-4
44. Gasque Schoof CR, Izzotti A, Jasiulionis MG, Vasques Ldos R. The Roles of miR-26, miR-29, and miR-203 in the Silencing of the Epigenetic Machinery during Melanocyte Transformation. Biomed Res Int. 2015;2015:634749. doi:10.1155/2015/634749
45. Wang X, Bhandari RK. DNA methylation dynamics during epigenetic reprogramming of medaka embryo. Epigenetics. 2019;14(6):611–622. doi:10.1080/15592294.2019.1605816
46. Zhang Z, Wang J, Shi F, et al. Genome-wide alternation and effect of DNA methylation in the impairments of steroidogenesis and spermatogenesis after PM(2.5) exposure. Environ Int. 2022;169:107544. doi:10.1016/j.envint.2022.107544
47. Thorlacius GE, Björk A, Wahren-Herlenius M. Genetics and epigenetics of primary Sjögren syndrome: implications for future therapies. Nat Rev Rheumatol. 2023;19(5):288–306. doi:10.1038/s41584-023-00932-6
48. Li P, Han M, Zhao X, Ren G, Mei S, Zhong C. Abnormal Epigenetic Regulations in the Immunocytes of Sjögren’s Syndrome Patients and Therapeutic Potentials. Cells. 2022;11(11):767. doi:10.3390/cells11111767
49. Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–285. doi:10.1038/nrg2072
50. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi:10.1038/nrg3230
51. de Oliveira DT, de Paiva NCN, Carneiro CM, Guerra-Sá R. Dynamic changes in hepatic DNA methylation during the development of nonalcoholic fatty liver disease induced by a high-sugar diet. J Physiol Biochem. 2022;78(4):763–775. doi:10.1007/s13105-022-00900-w
52. Komatsu Y, Waku T, Iwasaki N, Ono W, Yamaguchi C, Yanagisawa J. Global analysis of DNA methylation in early-stage liver fibrosis. BMC Med Genomics. 2012;5:5. doi:10.1186/1755-8794-5-5
53. Rui J, Deng S, Lebastchi J, Clark PL, Usmani-Brown S, Herold KC. Methylation of insulin DNA in response to proinflammatory cytokines during the progression of autoimmune diabetes in NOD mice. Diabetologia. 2016;59(5):1021–1029. doi:10.1007/s00125-016-3897-4
54. Cole MB, Quach H, Quach D, et al. Epigenetic Signatures of Salivary Gland Inflammation in Sjögren’s Syndrome. Arthritis Rheumatol. 2016;68(12):2936–2944. doi:10.1002/art.39792
55. Imgenberg-Kreuz J, Sandling JK, Almlöf JC, et al. Genome-wide DNA methylation analysis in multiple tissues in primary Sjögren’s syndrome reveals regulatory effects at interferon-induced genes. Ann Rheum Dis. 2016;75(11):2029–2036. doi:10.1136/annrheumdis-2015-208659
56. Meng Z, Edman MC, Hsueh PY, et al. Imbalanced Rab3D versus Rab27 increases cathepsin S secretion from lacrimal acini in a mouse model of Sjögren’s Syndrome. Am J Physiol Cell Physiol. 2016;310(11):C942–54. doi:10.1152/ajpcell.00275.2015
57. Fu R, Edman MC, Hamm-Alvarez SF. Rab27a Contributes to Cathepsin S Secretion in Lacrimal Gland Acinar Cells. Int J Mol Sci. 2021;22(4):1630. doi:10.3390/ijms22041630
58. Ogawa Y, Shimizu E, Tsubota K. Interferons and Dry Eye in Sjögren’s Syndrome. Int J Mol Sci. 2018;19(11):3548. doi:10.3390/ijms19113548
59. Ogawa Y, Takeuchi T, Tsubota K. Autoimmune Epithelitis and Chronic Inflammation in Sjögren’s Syndrome-Related Dry Eye Disease. Int J Mol Sci. 2021;22(21):11820. doi:10.3390/ijms222111820
60. Gupta S, Li D, Ostrov DA, Nguyen CQ. Blocking IAg(7) class II major histocompatibility complex by drug-like small molecules alleviated Sjögren’s syndrome in NOD mice. Life Sci. 2022;288:120182. doi:10.1016/j.lfs.2021.120182
61. Abdulahad WH, Kroese FG, Vissink A, Bootsma H. Immune regulation and B-cell depletion therapy in patients with primary Sjögren’s syndrome. J Autoimmun. 2012;39(1–2):103–111. doi:10.1016/j.jaut.2012.01.009
62. Karabiyik A, Peck AB, Nguyen CQ. The important role of T cells and receptor expression in Sjögren’s syndrome. Scand J Immunol. 2013;78(2):157–166. doi:10.1111/sji.12079
63. Emamian ES, Leon JM, Lessard CJ, et al. Peripheral blood gene expression profiling in Sjögren’s syndrome. Genes Immun. 2009;10(4):285–296. doi:10.1038/gene.2009.20
64. Lin X, Rui K, Deng J, et al. Th17 cells play a critical role in the development of experimental Sjögren’s syndrome. Ann Rheum Dis. 2015;74(6):1302–1310. doi:10.1136/annrheumdis-2013-204584
65. Xiao F, Rui K, Han M, et al. Artesunate suppresses Th17 response via inhibiting IRF4-mediated glycolysis and ameliorates Sjogren’s syndrome. Signal Transduct Target Ther. 2022;7(1):274. doi:10.1038/s41392-022-01103-x
66. Zhang Y, Zhao M, Sawalha AH, Richardson B, Lu Q. Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. J Autoimmun. 2013;41:92–99. doi:10.1016/j.jaut.2013.01.005
67. Luo Y, Li Y, Su Y, et al. Abnormal DNA methylation in T cells from patients with subacute cutaneous lupus erythematosus. Br J Dermatol. 2008;159(4):827–833. doi:10.1111/j.1365-2133.2008.08758.x
68. Takahashi M, Mimura Y, Hayashi Y. Role of the ICAM-1/LFA-1 pathway during the development of autoimmune dacryoadenitis in an animal model for Sjögren’s syndrome. Pathobiology. 1996;64(5):269–274. doi:10.1159/000164058
69. Gao J, Morgan G, Tieu D, et al. ICAM-1 expression predisposes ocular tissues to immune-based inflammation in dry eye patients and Sjögrens syndrome-like MRL/lpr mice. Exp Eye Res. 2004;78(4):823–835. doi:10.1016/j.exer.2003.10.024
70. Paton DM. Lifitegrast: first LFA-1/ICAM-1 antagonist for treatment of dry eye disease. Drugs Today. 2016;52(9):485–493. doi:10.1358/dot.2016.52.9.2542066
71. Holland EJ, Luchs J, Karpecki PM, et al. Lifitegrast for the Treatment of Dry Eye Disease: results of a Phase III, Randomized, Double-Masked, Placebo-Controlled Trial (OPUS-3). Ophthalmology. 2017;124(1):53–60. doi:10.1016/j.ophtha.2016.09.025
72. Du G, Du W, An Y, et al. Design, synthesis, and LFA-1/ICAM-1 antagonist activity evaluation of Lifitegrast analogues. Med Chem Res. 2022;31(4):555–579. doi:10.1007/s00044-022-02851-9
73. Xu WD, Pan HF, Ye DQ, Xu Y. Targeting IRF4 in autoimmune diseases. Autoimmun Rev. 2012;11(12):918–924. doi:10.1016/j.autrev.2012.08.011
74. Gong YZ, Nititham J, Taylor K, et al. Differentiation of follicular helper T cells by salivary gland epithelial cells in primary Sjögren’s syndrome. J Autoimmun. 2014;51:57–66. doi:10.1016/j.jaut.2013.11.003
75. Tybulewicz VL. Vav-family proteins in T-cell signalling. Curr Opin Immunol. 2005;17(3):267–274. doi:10.1016/j.coi.2005.04.003
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.