Back to Journals » Stem Cells and Cloning: Advances and Applications » Volume 13
Informatics Approaches for Harmonized Intelligent Integration of Stem Cell Research
Authors Finkelstein J , Parvanova I , Zhang F
Received 5 November 2019
Accepted for publication 11 January 2020
Published 28 January 2020 Volume 2020:13 Pages 1—20
DOI https://doi.org/10.2147/SCCAA.S237361
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Bernard Binetruy
Joseph Finkelstein, 1 Irena Parvanova, 1 Frederick Zhang 2
1Department of Population Health Science and Policy, Icahn School of Medicine at Mount Sinai, New York, NY, USA; 2Center for Bioinformatics and Data Analytics, Columbia University, New York, NY, USA
Correspondence: Joseph Finkelstein
Department of Population Health Science and Policy, Icahn School of Medicine at Mount Sinai, 1425 Madison Ave, Icahn L2-36, New York, NY 10029, USA
Tel +1 212-659-9596
Email [email protected]
Abstract: As biomedical data integration and analytics play an increasing role in the field of stem cell research, it becomes important to develop ways to standardize, aggregate, and share data among researchers. For this reason, many databases have been developed in recent years in an attempt to systematically warehouse data from different stem cell projects and experiments at the same time. However, these databases vary widely in their implementation and structure. The aim of this scoping review is to characterize the main features of available stem cell databases in order to identify specifications useful for implementation in future stem cell databases. We conducted a scoping review of peer-reviewed literature and online resources to identify and review available stem cell databases. To identify the relevant databases, we performed a PubMed search using relevant MeSH terms followed by a web search for databases which may not have an associated journal article. In total, we identified 16 databases to include in this review. The data elements reported in these databases represented a broad spectrum of parameters from basic socio-demographic variables to various cells characteristics, cell surface markers expression, and clinical trial results. Three broad sets of functional features that provide utility for future stem cell research and facilitate bioinformatics workflows were identified. These features consisted of the following: common data elements, data visualization and analysis tools, and biomedical ontologies for data integration. Stem cell bioinformatics is a quickly evolving field that generates a growing number of heterogeneous data sets. Further progress in the stem cell research may be greatly facilitated by development of applications for intelligent stem cell data aggregation, sharing and collaboration process.
Keywords: stem cells, data integration, databases
Introduction
Stem cells are defined as cells with the capacity for self-renewal and development into a specialized cell that composes healthy tissue.1 These cells were first described in 1961, when researchers James Till and Ernest McCulloch discovered the existence of self-renewing cell colonies in mice.2,3 The cells they discovered were later classified as hematopoietic stem cells, the first of many breakthroughs in the field of stem cell research.3 Since then, different types of stem cells have been discovered with the ability to differentiate into many different types of human tissue, including tissues that previously exhibited limited healing capacity such as neurons.4 The discovery of these cells has revolutionized the field of regenerative medicine, with many exciting potential applications for stem cell therapy in a variety of diseases and conditions previously thought to be incurable.
However, the field of stem cell studies is expensive and difficult to access for the majority of researchers. A major reason for this is the controversial nature of stem cell research and the ethical discussions which have ensued. Many of the developed nations in the world, including the United States and several European countries, have restrictive policies regarding stem cell research.5 The United States, in particular, has had an evolving history regarding the accessibility of stem cell research. Under the previous administrations, federal funding for research on new embryonic stem cell lines was halted, leading to a major slowdown in stem cell research in the US. This decision was later reversed under the next administration.6 The result is that the approaches to generate stem cells and use them in research are governed by a set of ethical and regulatory considerations. Under the current conditions, it is both expensive and challenging to create pluripotent stem cell lines for complex disorders.7
Part of the challenge in developing large numbers of stem cell lines is the difficulty in standardizing and optimizing stem cell differentiation protocols. In 2006, Kazutoshi Takahashi and Shinya Yamanaka discovered that pluripotent stem cells could be induced from fibroblasts through the expression of just four transcription factors.8 Since then, a number of different methods have evolved to induce pluripotency in cells. These methods involve alterations at multiple levels of cellular regulation.9,10 This ranges from DNA reprogramming factor delivery using viral or plasmid vectors, to mRNA or miRNA transfection, to direct delivery of proteins or other small molecule compounds.10–13 Although the development of this number of techniques has vastly improved stem cell differentiation efficiency, the eclectic and complicated nature of these techniques makes it difficult for these protocols to be disseminated between researchers. In order for a protocol to be successfully replicated, the researcher needs to have a detailed understanding of the biomarkers being expressed at each stage of differentiation as the gene expression of the final cell produced. For this reason, an important step in the progression of stem cell research is the development of stem cell data aggregates that collect detailed information regarding many stem cell lines into a single, easily accessible database. The potential amount of data being generated in each stem cell line analysis is immense. Providing centralized and easily accessible data repositories is necessary so that the data being generated can be fully utilized in stem cell research using various bioinformatics pipelines.
The creation of efficient and accessible data aggregates for stem cell research faces many challenges. In recent years, biological research has been revolutionized by the rise of the “OMICS”. The OMICS refers to a set of technologies which can provide a comprehensive understanding of the molecules and biomarkers present in a cell.14 This includes fields such as genomics, proteomics, and metabolomics, all of which contribute valuable information regarding the biological activity present in a cell at any given time. Naturally, OMICS techniques have played an integral role in the development of stem cell research. High throughput sequencing techniques have greatly enhanced researchers’ understanding of the gene regulation networks involved in establishing pluripotency in a cell.15 Accordingly, many of the challenges in data aggregation for stem cell research mirror the challenges in data aggregation of OMICS technologies. OMICS technologies generate massive amounts of data that often lack standardization and curation.16 Data aggregates which integrate OMICS datasets need to employ a standardized format for the data they collect and then develop a methodology for curating the data they choose to host. An additional significant challenge facing the integration of OMICS data into research is the need to relate raw data to a biological context.
The evolution and proliferation of OMICS technologies has generated data that has become increasingly granular and specific to the platform the data were collected on. This creates difficulty in integrating data across multiple datasets, particularly datasets using different platforms. In addition, it can be difficult to interpret these data in a biologic framework. For these reasons, the importance of using ontologies for biologic data aggregation has increased significantly in recent years. An ontology, broadly speaking, refers to a semantic method to categorize objects using relations and classes.17–19 The concept of using ontologies in biological sciences has been facilitated by the Gene Ontology project to categorize the role and function of genes.18,19 The Gene Ontology project was designed as a method to standardize the classification and annotations of genes across different research projects. Since the implementation of Gene Ontology, other ontologies for applications in biomedical research have been developed. The use of ontologies to standardize datasets and help researchers to find data and interpret the data they generate from a functional perspective is an important development for the advancement of stem cell research.19 For this reason, the integration of stem cell research into current ontologies is another important challenge facing stem cell database development.
Currently, a variety of databases exist which house information on different stem cell lines. However, these databases vary greatly in the data elements they capture, the search features they offer, and a number of other characteristics. Because of this, it is unclear what the best practices are in developing future stem cell databases. The field of stem cell research is rapidly growing, so naturally, the need for high-quality data storage and curation methods for stem cell data is growing alongside it. In this review, we evaluate both academic and commercial stem cell databases currently available. Our aim is to identify which useful features are available in high-quality stem cell databases, so that future stem cell databases can also incorporate these features for maximum utility and accessibility.
Methods
For this paper, we conducted a scoping review of current stem cell data aggregates available. The scoping review methodology was established by Hilary Arksey and Lisa O’Malley20 as a method to quickly and broadly map out the up-to-date knowledge of a certain research area currently available.
Scoping Review Workflow
The scoping review workflow generally follows five major steps:
- Identify a research question
- Identify relevant studies
- Evaluate and select studies to be included
- Chart the data
- Collect, summarize, and report the results
Research Question
Our goal in conducting this review was to identify the characteristics of the currently available platforms for stem cell researchers to aggregate and share data from multiple sources. The research question we aimed to address was “What are the characteristics of stem cell data aggregates and how can future databases improve upon these past iterations?”
Search Strategy
First, we conducted a search on PubMed using the following MeSH term search: “Databases, Factual AND Stem Cells”. Because both stem cell research and data sharing are rapidly evolving fields, we chose 2010 as a publication cutoff year. We chose this year because, during our search, we found that many databases established prior to 2010 had been discontinued and the links contained within their respective papers led to un-hosted domains. These search terms resulted in 491 papers to be considered for inclusion in our final study.
We also conducted a web search for commercial stem cell databases that do not have associated papers indexed in PubMed.
Study Selection
The following inclusion criteria for the PubMed articles were developed during our search process:
- The paper must describe a stem cell experimental database.
- The database must include human stem cell lines.
- The database must include data extracted from stem cell lines which are retrievable.
- The database must aggregate data from multiple datasets, as opposed to hosting only data from “in-house” cell lines.
- Cancer stem cells databases are also included.
- Databases that are no longer available were also included.
- The database or related paper must be available in English.
The following exclusion criteria were then applied after the inclusion criteria:
- Databases where a paper was published but the project itself was not completed.
- Databases which include some stem cell data but stem cells are not the primary focus.
These same criteria were also applied to all databases found from the web search where applicable. After conducting our search and applying these criteria, we found 14 papers to include in our review from PubMed. Our web search found two additional databases to add to our study.
Data Extraction
First, we obtained a general overview of the characteristics of each database as described by the authors, such as the primary goal of the project, the methodology for developing the database, the species included, data elements. Based on the results of the general overview, we developed seven major dimensions to capture the characteristics of each database. The dimensions are as follows:
- Stored data elements: In this category, we described the data elements available for each individual dataset or data point within the database. For example, some databases provided a detailed information for each cell line alongside the raw data, including the tissue of origin, treatments performed, etc., whereas other databases may have only included the basic characteristics of a cell line such as the cell type and species.
- Available data analysis tools, such as co-expression or covariate analysis: In this category, we included any supplemental tools provided in the database to perform analyses, which were not available from the original data (for instance, heatmap generation or cluster analysis).
- Search features: In this category, we described how data could be searched for each database and any additional features implemented to refine or streamline the search process.
- Storage platform and interface: In this category, we described the architecture used to host both the database itself and the user interface.
- Privileges needed for access: For each database, we described the process for researchers to submit data as well as the privileges necessary to access the data, i.e. if the database was freely available, if it required registration, etc.
- Update methodology: In this category, we described the current or planned methodology for periodic database updates.
- Current status: In this category, we described the current status of the database and any updates or significant changes since the original paper was published.
In addition, we identified general features that could improve the utility of all future stem cell databases. The aim of most stem cell databases at their inception was to facilitate and streamline stem cell research. However, these databases greatly varied in the functionalities they implemented to reach this goal. We identified three important, broad database functional features amongst the reviewed databases that could be implemented in future stem cell research data repositories. They are as follows:
- Data Elements: All available data elements were documented including sequencing data regarding specific stem cell lines such as microarray expression or Next Generation Sequencing (NGS) data. The primary aim of many databases is to aggregate large amounts of experimental data from multiple stem cell lines and trials. In the current landscape of stem cell research, traditional gene expression sequencing data and NGS data have emerged as the primary biomarkers for the understanding of stem cell induction and differentiation. Therefore, providing a direct source of information regarding these biomarkers is an important measurement of the utility of the database.
- Data Analysis Tools: This includes data analysis tools supported by each database. Inclusion of these tools allows for a more streamlined integration of the available data for further analysis. Providing this feature makes stem cell informatics research across multiple datasets more accessible and feasible.
- Ontology Integration: This includes integration with standard ontologies or self-developed nomenclatures to relate raw data to known biological frameworks. The natural overlap of the field of stem cell research with the “omics” (genomics, proteomics, etc.) means that it is often difficult to annotate the experimental data into a standardized and informative format. Mapping the experimental data elements to standard ontology concepts makes stem cell research more accessible to researchers across different disciplines, as well as improves the standardization of the integrated data. For this reason, ontology integration is another important feature to include to improve the utility of stem cell databases.
After this identification, we inspected which of these features each database provided and generated an Euler diagram summarizing the functionality of reviewed databases in Figure 1. All database features were extracted using an iterative manual review. A detailed report of all data elements collected in the course of this review can be found in the Supplementary data.
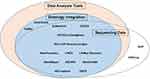 |
Figure 1 A diagram of major utility features in the reviewed databases. |
Results
General Characteristics
In total, we found 14 academic databases from our PubMed search and two databases on the web which did not have an associated paper. These databases varied widely in the data elements, available data analysis tools, search functionality, and update methodology. The most common storage and interface architecture was data storage using a MySQL relational database with a web interface implemented on an Apache server, which was used by five databases. All of the databases were free to access; however, one required registration for full access. Three of the databases no longer had any data available. The oldest active database was the Embryonic Stem Cells Database (ESCD), which was published in 2010. Out of the 16 databases, LifeMap Discovery was the only one that included information about the stem cells applications in clinical trials. A detailed description of the data elements, used in each database can be found in Table 1. A report of the operational characteristics for each database has been outlined in Table 2.
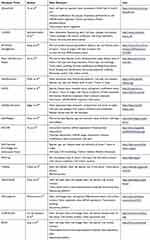 |
Table 1 Data Elements in Stem Cell Databases |
 |
Table 2 Operational Characteristics of Stem Cell Databases |
With regard to major database features, the majority of the academic databases contained sequencing information but did not provide access to the sequencing data. Of all included databases, 14 provided data analysis tools alongside the sequencing information (Table 3). In 14 databases, standard ontology concepts or specifically developed terminologies were integrated with raw data stored in the database and used for further data representation and analysis (Table 4). In total, 12 of the databases we reviewed had all three functional features.
 |
Table 3 Data Analysis Tools Used in Stem Cell Databases |
 |
Table 4 Ontology-Based Applications in Stem Cell Databases |
Overall, all 16 databases included 35 rubrics that characterized these databases (Tables 5 and 6). In Table 6, we listed all rubrics, used to describe stem cell populations, which included characteristics, such as stem cell line names, lineage, cell surface markers, transcription factors, and epigenetic modifications, as well as types of detection assays, data analysis tools, and clinical trials information. Due to the variety of rubrics characterizing stem cells databases, we used minimum information about a cellular assay for regenerative medicine (MIACARM) five-module classification system21 to further group and organize the 35 rubrics under the five MIACARM modules: project, assay, source cell, experimental technology, and data modules (Table 6).
 |
Table 5 Stem Cell Databases are Characterized by Different Number of Rubrics |
 |
Table 6 Representation of Characterization Rubrics in Stem Cell Databases (Plus Sign Indicates Presence in a Database) |
Additionally, in Table 7, we examined in further details the complexity of each of the 35 rubrics with complexity expressed as the breadth of possible options which were used across the reviewed databases for a particular rubric. For instance, for “Cell Lineage”, we listed eight different stem cell types: adult stem/progenitor cells; embryonic progenitor cells; embryonic stem cells (ESC); fetal stem/progenitor cells; induced pluripotent stem cells (iPSC); cell lines; primary cells, and tissue/cells composite. On the other hand, a rubric, such as “Transcription Factors Expression”, features 210 transcription factors just in the CODEX database. Other rubrics, such as Cell “Surface Markers”, include a large number of proteins, which are too diverse to be accurately counted. To complete this portion of the analysis, we represented the frequency of the usage of each rubric for characterizing a stem cell database by calculating the percentage of the appearance of that particular rubric in all 16 databases.
 |
Table 7 Complexity of Rubrics Characterizing Stem Cell Research Databases |
SyStemCell22
SyStemCell is a database that aims to collect stem cell data at seven different levels of regulation: DNA CpG methylation, histone modification, transcript products, microRNA-based regulation, protein products, phosphorylation proteins, and transcription factor regulation. Stem cell data from four species are included: Homo sapiens, Mas musculus, Rattus norwegicus, and Macaca mulatta. Users can search by a specific gene or browse by organism, tissue type, etc. SyStemCell also offers Co-Localization analysis tools to allow researchers to investigate correlations between various genes at their different levels of regulation. Currently, the web domain for SyStemCell is no longer functional and the tool no longer seems to be available.
Codex23
CODEX is a database that aims to collect data from Next Generation Sequencing (NGS) experiments and condense them into an easily searchable, curated database. Formats supported by CODEX include ChIP-Seq, RNA-Seq, and DNase-Seq. The primary purpose of CODEX is to provide NGS data for hematopoietic and embryonic stem cells in a standardized format. All data uploaded to CODEX has gone through a bioinformatics pipeline to be processed into a standardized format and curated for all relevant details. CODEX contains data from both human and mouse samples. Users can browse and search by both cell type or transcription factor. CODEX offers a number of different gene analysis tools, such as correlation analysis and motif analysis. Currently, CODEX is available online and being actively updated.
ESTOOLS Data@Hand24
ESTOOLS Data@Hand is a database that aims to allow users to browse gene expression data from published stem cell research. The database was generated from manually selected experiments from Gene Expression Omnibus (GEO) and ArrayExpress which were then manually curated into a standardized format. The data can be browsed or searched by any annotation dimension. The database offers a set of analysis tools based on the Bioconductor package in R, such as co-expression and clustering tools. The authors also created two meta-datasets, one from combining all Affymetrix datasets and the other from combing all Illumina datasets. Currently, the web domain for ESTOOLS Data@Hand is no longer functional and the database no longer seems to be available.
Stem Cell Discovery Engine25
The Stem Cell Discovery Engine is a database designed to integrate data from both tissue stem cells and cancer stem cells experiments. Data from these experiments are both collected from public studies as well as user-submitted and manually curated for relevant data. Metadata from the experiments are stored in the Investigation/Study/Assay format. Genes selected in the database can be queried against other gene annotation databases for related genes or gene annotation information. The Stem Cell Discovery Engine also offers features to share gene sets in order to facilitate collaboration with other researchers. Currently, the Stem Cell Discovery Engine is active and being updated; however, it has been merged into Harvard’s larger Stem Cell Commons project.
StemFormatics26
StemFormatics is a database of public human stem cell datasets manually collected and curated from public sources such as GEO or ArrayExpress. Microarray datasets are subjected to a quality control bioinformatics pipeline before inclusion into the database. StemFormatics allows users to query by gene name or identifier and view gene expression data from a single gene across multiple datasets simultaneously. Clustering and heatmap generation tools are also available as gene analysis tools. The database also provides a Workbench feature that allows users to create and share gene sets for collaborative analysis. Currently, the StemFormatics database is active and being updated.
LINCS27
LINCS is a database designed to capture data from cell-perturbation and response experiments. This includes a large number of stem cell lines reflecting the number of experiments dedicated to discovering the conditions to differentiate and maintain stem cells. LINCS processes data through its associated LINCS Data Registry, which standardizes and curates the data and metadata. These data include information on cell culturing conditions, reagents and perturbation agents used to induce cell responses, such as small proteins and immunoglobulins, and the resulting cell line biomarkers. The LINCS database includes many of the traditional “browse and search” features shared with the other databases, as well as the unique ability to search for projects involving a specific molecule or similar molecule by uploading the mol file.
LifeMap Discovery28
LifeMap Discovery is a comprehensive stem cell database that aims to share stem cell data with researchers along with potential applications to regenerative medicine. The LifeMap Discovery webpage is unique in providing its own ontology tree of the current understanding of embryonic and stem cells differentiation to facilitate stem cell research. This ontology tree also contains reference data and genes which are relevant to that specific cell or tissue type. In addition to its ontology tree, LifeMap Discovery also contains a database of stem cell experiments, which can be browsed by organism, tissue type, gene, and other ontological characteristics. Users can also browse experiments by their application to stem cell and regenerative medicine therapies. The LifeMap Discovery database is partnered with the GeneQuest data analysis suite, which provides its own set of gene analysis features. Currently, the LifeMap Discovery database is active and still being updated.
StemMapper29
StemMapper is a database of gene expression data for both human and mouse stem cell lines manually collected and curated from GEO. StemMapper contains only data from experiments using certain Affymetrix sequencing platforms. Users can query the database by gene, as well as browse by cell and tissue type. Heatmap generation and Principal Components Analysis are available as built-in data analysis tools. In addition, StemMapper offers a unique feature for users to upload their own gene expression datasets for analysis. The StemMapper database is still available but has not been updated since 2017.
ESCAPE30
ESCAPE is a stem cell database which aims to provide stem cell experiment data at multiple levels of regulation, such as epigenetics, transcriptomics, and proteomics. Using ChIP-seq data, the ESCAPE database localizes transcription factors to their genes, characterizes protein/DNA and other protein interactions, among other biomarkers. The database can be searched by cell type, gene name, or platform, among others, or can be browsed by the level of regulation. ESCAPE offers enrichment analysis and a lineage prediction tool as its data analysis features. Currently, the ESCAPE database is available but is not being updated.
SKiP Stemcell Knowledge and Information Portal
SKiP is a stem cell experiment database developed and operated in Japan. The database contains information on stem cell lines which can be searched by characteristics, such as tissue type, cell type, as well as by their potential applications to disease therapy. Currently, SKiP is still available but no longer actively updated.
hPSCreg
hPSCreg is a stem cell experiment registry based in Europe which contains information on stem cell lines and experiments. The database allows users to submit and edit their own stem cell line data as well as requisition access to other stem cell lines. The database can be searched by free text, country of origin, or associated disease. Currently, hPSCreg is active and still being updated.
CellNet31
CellNet database was an online tool, which allowed microarray data of stem cell populations to be uploaded and evaluated. Currently, the database is no longer available; however, its code could be utilized for the microarray data analysis. The output of the analysis provides information about the cell species, lineage, tissue type, and transcription factors expression.
StemCellNet32
StemCellNet is a database, which includes data about (1) physical protein interactions; (2) transcription regulatory interactions, detected by chromatin immunoprecipitation, combined with microarray and sequencing technologies; and (3) gene interactions, which focuses on the genes that define the “stemnesses” of the stem cell lines. The platform allows for the assessment and visualization of the molecular networks. The tool allows as many as 500 genes to be simultaneously analyzed. The platform is versatile, free to access, and still available.
HSC-Explorer33
HSC-explorer allows access to information on the early differentiation stages of hematopoietic stem cells. It provides information about the species, molecular interactions and signaling process in these cells, as well as the performed assays. The data can be displayed as graphical representation, allowing web-based analysis, where the hematopoietic stem cell repository is publicly available and manually updated.
Cortecon34
CORTECON is a repository of transcriptome analysis of genes in human embryonic stem cells, expressed in the developing human cortex. The transcriptome analysis can be viewed by gene, disease, KEGG pathway, and enriched ontologies. The data can be analyzed using R-software. The database is freely accessible and still updated.
Embryonic Stem Cell Database (ESCD)35
ESCD contains mouse and human embryonic stem cell line information from embryonic stem and carcinoma cells. The included datasets cover perturbation (both knock-down and overexpression) and chromatin immunoprecipitation (ChIP) experiments in stem cell lines. Transcription factor binding data originate from various ChIP experiments (ChIP-PET, ChIP-chip, ChIP-seq). The database enables gene-based queries and supports the search for genes with a specific behavior in selected datasets. Datasets obtained from various array and genome version platforms are linked using Ensembl gene IDs and gene names. The database is publicly accessible and still available.
Discussion
Our review identified 16 databases created since 2010 which aggregate data on stem cell projects. Of those databases, three are no longer available and several others appear to have stopped updating. When we examined the data elements being tracked by each database, we found a great deal of heterogeneity. Some databases warehoused curated and formatted genotype information or next-generation sequencing data, whereas others only provided information regarding the cell line itself. The databases had their own methods of curation and formatting of experimental and clinical data which varied significantly between each database. Many of the databases had similar search functions but differed in the ways they implemented them and in the user-friendliness of their interface. Almost all of the databases were freely available to browse without registration, with the exception of the Stem Cell Discovery Engine, which required registration to access data.
Many of these databases had similar aims: to provide a centralized source of data for stem cell experiments where the data are housed in a standardized, curated, and searchable format. The hope is that in doing so, it becomes easier for researchers to analyze data across multiple data sets. In order to facilitate this goal, we identified three broad features which could be implemented in future stem cell data aggregates to improve their accessibility and utility for researchers. Housing curated sequencing data and providing data analysis tools within the database itself are features which allow the data to be easily integrated into a bioinformatics workflow. Integration of the database with established biomedical ontologies allows the analyzed data to be interpreted in a more standardized format between different researchers, particularly in fields in which our understanding of certain data elements is still rapidly evolving, such as the OMICS. Understanding the database features which improve the stem cell research workflow and collaboration process is important in advancing stem cell research and utilizing the vast amount of aggregated data for further knowledge discovery.
The results of this study reflect a systematic abstraction of all data elements and functional features of reviewed stem cell databases to provide detailed insights for future improvements in accessibility and utility. The current literature examining the characteristics and development of stem cell databases is fairly limited.36,37 A review of stem cell databases was also published in 2015.38 This review provides only summaries of the important features as well as subjective rating score for each database. A more general review of all biological databases was reported in 2015.39 This review provides a broad overview of biological databases covering many different fields in biomedical research and categorizes them based on characteristics such as their scope of coverage and method of curation. However, because the characteristics of databases in different fields of biomedical research serve different purposes, and therefore require different features, the perspectives from this study are difficult to translate to a relatively new and evolving field of stem cell research. Our review differs from previously published work in several aspects. First, a systematic study design based on a scoping review methodology has been employed. Second, we chose to evaluate the individual features of each database, such as their data element characterizations, availability of analytical pipelines, and ontology integration, rather than provide an overall evaluation of each database. This allowed identify valuable and unique features from each database which can be integrated into future databases. Third, we used MIACARM as the current framework for stem cell research data reporting to characterize the scope of the existing databases. Therefore, this review provides new systematic information that potentially can be instrumental in improving the stem cell research analytical databases.
Stem cell studies have become such a highly investigated field in recent years because of their potential for developing new therapies for difficult to treat diseases. For certain conditions, stem cell therapy has already been available for some time. The use of hematopoietic stem cell transplants has been established as an effective therapy for hematologic malignancies such as leukemia and multiple myeloma.40 Because of the success of these treatments as well as the theoretically limitless growth potential of stem cells, stem cells have become the new frontier for the treatment of a variety of chronic conditions.41,42 For example, stem cells are now being investigated for their potential to treat degenerative diseases, such as multiple sclerosis,43 Parkinson’s disease44 or myocardial ischemia.45 Despite the fact that many theoretical models for these treatments have been developed, none have yet been translated into routine clinical care. For this reason, a comprehensive and systematic aggregation of current and future results of preclinical studies and stem cell therapeutic interventions is crucial for advancing this field towards practical implementation. Our review identified a number of stem cell databases that stopped updating their content or were not accessible anymore at all. This finding demonstrated the importance of establishing a sustainable framework for continuing and reliable maintenance of stem cell databases. Thus, in addition to effective informatics solutions to aggregate data across different repositories, investment also needs to be made in sustaining these repositories including financial support for the infrastructure and suitable incentives and career recognition for data curators working to update, clean and maintain these databases and keep them available for open access.
Although this scoping review was intended to be comprehensive, several stem cell databases did not satisfy inclusion/exclusion criteria or were not included due to the limitations of the search criteria. There may be additional stem cell databases which may not necessarily have an associated peer-reviewed article or require different keywords for successful web search. Such repositories of stem cell resources as the European Bank for induced pluripotent Stem Cells (EBiSC)46 and RIKEN BioResource Center47 may serve as important additional resources for stem cell data. Since we characterized all reviewed databases in a granular and systematic way and many features were present in at least a subset of the 16 reviewed databases, an omission of a few databases is unlikely to result in loss of substantive features. Overall, this review resulted in a comprehensive and detailed analysis of stem cell databases currently available.
Recognizing the importance of establishing an effective ecosystem for data aggregation and sharing, a diverse set of stakeholders representing academia, industry, funding agencies, and scholarly publishers have recently introduced principles for scientific data management and stewardship to improve the findability, accessibility, interoperability, and reuse (FAIR) of digital assets.48 The FAIR guiding principles for research data stewardship could provide a necessary framework for the future development of stem cell databases. Recent publications reported successful approaches for aggregating multiple heterogeneous data streams representing diverse molecular signatures of various cell types in a harmonized and expandable way. For example, the Library of Integrated Network-based Cellular Signatures (LINCS) project, a multi-center NIH-funded program, created a comprehensive library of molecular signatures supporting data integration, modeling and analysis methodologies.49 Broad sharing of genomic- and health-related data requires proper governance and security.50 In the context of stem cell research, data and sample sharing represent a scientific and ethical challenge to ensure appropriate protection of individual interests as well as maintaining public trust. Effective data protection requirements are necessary along with the future data harmonization efforts for building successful stem cell research data sharing.51 Deployment of the common framework for responsible sharing of genomic and health-related data established by the Global Alliance for Genomics and Health (GA4GH) in stem cell databases can facilitate the use of data in compliance with national and international laws and general ethical principles and standards.52
Future informatics approaches for harmonized intelligent integration of stem cell research data are dependent on timely introduction of a spectrum of standards concerning informed consent both from donors and recipients, procurement of biomaterials, manufacturing regulations, cell potency assays, minimally acceptable changes during cell culture, methods of recipient identification for experimental interventions, reporting of preclinical experiments, clinical trial design and reporting, and principles for defining common data elements in stem cell datasets.53 The Guidelines for Stem Cell Research and Clinical Translation established by the International Society for Stem Cell Research (ISSCR) in 2016 provide a comprehensive set of principles for rigor, oversight and transparency in all aspects of stem cell science and practice.54 The International Stem Cell Banking Initiative (ISCBI) promulgates best practices for manufacture, culture, characterization, storage, distribution, and translation of stem cell products.55 For example, ISCBI has taken the lead in defining critical quality attributes for induced pluripotent stem cells (iPSC) and issued recommendations on the minimum dataset required to consider an iPSC line of clinical grade which includes such attributes as identity, microbiological sterility, endotoxin, genetic fidelity & stability, variability, characterization and potency.56 Another example of standardized evaluation of stem cell-based products is the recently proposed multiparametric quality assessment rubric for stem-cell derived cardiac myocytes.57
To catalyze knowledge discovery in stem cell research, the scientific community needs to adopt intelligent technologies that harmonize multiple heterogeneous data streams utilizing internationally accepted standards. Policies that reward approaches supporting interoperability and promoting the expansion of open data commons will facilitate findability, accessibility, interoperability, and reuse of stem cell data. The development of minimum information about stem cell experiments (MIASCE) can further promote intelligent data aggregation and sharing.58 Significant progress has been made in our understanding of effective approaches for supporting shared, annotated stem cell research data.59,60 Strategies to optimize information technology supporting stem cell research and practice are being actively discussed and introduced into practice.61 Recognizing a crucial importance of sustainable information architecture for harmonized and intelligent data aggregation and sharing, the National Institute of Heart and Lung Disease (NHLBI) introduced recently the BioData Catalyst Strategic Framework that provides a forward-looking digital landscape of technologies, science and data.62 The BioData Catalyst is a cloud-based digital platform allowing easily find, access, share, store, cross-link, and compute on large-scale data sets. Innovative programs supporting intelligent stem cell data aggregation and sharing utilizing this platform are under way.63 Further expansion of intelligent bioinformatics solutions64 in stem cell research will provide new opportunities for analysis, evaluation and practical implementation of innovative stem cell technologies.
Acknowledgment
This work was in part funded by NIH grants OT2HL147606 and UL1TR001433.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Biehl JK, Russell B. Introduction to stem cell therapy. J Cardiovasc Nurs. 2009;24(2):98–105. doi:10.1097/JCN.0b013e318197a6a5
2. Weissman I. Stem cell research: paths to cancer therapies and regenerative medicine. JAMA. 2005;294(11):1359–1366. doi:10.1001/jama.294.11.1359
3. Girlovanu M, Susman S, Soritau O, et al. Stem cells - biological update and cell therapy progress. Clujul Med. 2015;88(3):265–271. doi:10.15386/cjmed-483
4. Konagaya S, Iwata H. Microencapsulation of dopamine neurons derived from human induced pluripotent stem cells. Biochim Biophys Acta. 2015;1850(1):22–32. doi:10.1016/j.bbagen.2014.09.025
5. Poulos J. The limited application of stem cells in medicine: a review. Stem Cell Res Ther. 2018;9(1). doi:10.1186/s13287-017-0735-7
6. Murugan V. embryonic stem cell research: a decade of debate from bush to Obama. Yale J Biol Med. 2009;82(3):101–103.
7. Panchision DM. Progress and challenges in using human stem cells for biological and therapeutics discovery: neuropsychiatric disorders. Stem Cells. 2016;34(3):523–536. doi:10.1002/stem.2295
8. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi:10.1016/j.cell.2006.07.024
9. Alateeq S, Fortuna P, Wolvetang E. Advances in reprogramming to pluripotency. Curr Stem Cell Res Ther. 2015;10(3):193–207. doi:10.2174/1574888X10666150220154820
10. Revilla A, González C, Iriondo A, et al. Current advances in the generation of human iPS cells: implications in cell-based regenerative medicine. J Tissue Eng Regen Med. 2015;10(11):893–907. doi:10.1002/term.2021
11. Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27(11):2667–2674. doi:10.1002/stem.v27:11
12. Okita K, Matsumura Y, Sato Y, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8(5):409–412. doi:10.1038/nmeth.1591
13. Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi:10.1016/j.stem.2010.08.012
14. Horgan RP, Kenny LC. ‘Omic’ technologies: genomics, transcriptomics, proteomics and metabolomics. Obstet Gynaecol. 2011;13(3):189–195. doi:10.1576/toag.13.3.189.27672
15. Bian Q, Cahan P. Computational tools for stem cell biology. Trends Biotechnol. 2016;34(12):993–1009. doi:10.1016/j.tibtech.2016.05.010
16. Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18(1). doi:10.1186/s13059-017-1215-1
17. Hoehndorf R, Schofield PN, Gkoutos GV. The role of ontologies in biological and biomedical research: a functional perspective. Brief Bioinform. 2015;16(6):1069–1080. doi:10.1093/bib/bbv011
18. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi:10.1038/75556
19. Rubin DL, Shah NH, Noy NF. Biomedical ontologies: a functional perspective. Brief Bioinform. 2007;9(1):75–90. doi:10.1093/bib/bbm059
20. Arksey H, O’Malley L. Scoping studies: towards a methodological framework. Int. J. Socl Rese Methodol. 2005;8:19–32. doi:10.1080/1364557032000119616
21. Sakurai K, Kurtz A, Stacey G, et al. First proposal of minimum information about a cellular assay for regenerative medicine. Stem Cells Transl Med. 2016;5(10):1345–1361. doi:10.5966/sctm.2015-0393
22. Yu J, Xing X, Zeng L, et al. SyStemCell: a database populated with multiple levels of experimental data from stem cell differentiation research. PLoS One. 2012;7(7).
23. Sánchez-Castillo M, Ruau D, Wilkinson AC, et al. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 2014;43(D1).
24. Kong L, Aho K-L, Granberg K, et al. ESTOOLS Data@Hand: human stem cell gene expression resource. Nat Methods. 2013;10(9):814–815. doi:10.1038/nmeth.2576
25. Sui SJH, Begley K, Reilly D, et al. The stem cell discovery engine: an integrated repository and analysis system for cancer stem cell comparisons. Nucleic Acids Res. 2011;40(D1).
26. Wells CA, Mosbergen R, Korn O, et al. Stemformatics: visualisation and sharing of stem cell gene expression. Stem Cell Res. 2013;10(3):387–395. doi:10.1016/j.scr.2012.12.003
27. Koleti A, Terryn R, Stathias V, et al. Data portal for the Library of Integrated Network-based Cellular Signatures (LINCS) program: integrated access to diverse large-scale cellular perturbation response data. Nucleic Acids Res. 2017;46(D1).
28. Edgar R, Mazor Y, Rinon A, et al. LifeMap discovery™: the embryonic development, stem cells, and regenerative medicine research portal. PLoS One. 2013;8(7):e66629. doi:10.1371/journal.pone.0066629
29. Pinto JP, Machado RSR, Magno R, et al. StemMapper: a curated gene expression database for stem cell lineage analysis. Nucleic Acids Res. 2017;46(D1).
30. Xu H, Baroukh C, Dannenfelser R, et al. ESCAPE: database for integrating high-content published data collected from human and mouse embryonic stem cells. Database. 2013;2013.
31. Cahan P, Li H, Morris SA, et al. CellNet: network biology applied to stem cell engineering. Cell. 2014;158(4):903–915. doi:10.1016/j.cell.2014.07.020
32. Pinto JP, Reddy Kalathur RK, Machado RS, et al. StemCellNet: an interactive platform for network-oriented investigations in stem cell biology. Nucleic Acids Res. 2014;42(Web Server issue):W154–W160. doi:10.1093/nar/gku455
33. Montrone C, Kokkallaris KD, Loeffler D, et al. HSC-explorer: a curated database for hematopoietic stem cells. PLoSOne. 2013;8(7):e70348. doi:10.1371/journal.pone.0070348
34. van de Leemput J, Boles NC, Kiehl TR, et al. CORTECON: a temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron. 2014;83(1). doi:10.1016/j.neuron.2014.05.033.
35. Jung M, Peterson H, Chavez L, et al. A data integration approach to mapping OCT4 gene regulatory networks operative in embryonic stem cells and embryonal carcinoma cells. PLoSOne. 2010;5(5):e10709. doi:10.1371/journal.pone.0010709
36. Stemcell Knowledge & Information Portal. SKiP stem cell knowledge and information portal. Available from: https://skip.stemcellinformatics.org/en/link/. Accessed May 18, 2019.
37. Kurtz A, Stachelscheid H, Seltmann S, Müller R, Dewender J, Reimann S Human pluripotent stem cell registry. Human Pluripotent Stem Cell Registry. Available from: https://hpscreg.eu/.
38. Wei TI, Peng X, Ye L, Wang J, Song F, Bai Z. Web resources for stem cell research. Genomics Proteomics Bioinformatics. 2015;13(1):40–45. doi:10.1016/j.gpb.2015.01.001
39. Zou D, Ma L, Yu J, Zhang Z. Biological databases for human research. Genomics Proteomics Bioinformatics. 2015;13(1):55–63. doi:10.1016/j.gpb.2015.01.006
40. Henig I, Zuckerman T. Hematopoietic stem cell transplantation—50 years of evolution and future perspectives. Rambam Maimonides Med J. 2014;5(4). doi:10.5041/RMMJ.10162
41. Watt F, Driskell RR. The therapeutic potential of stem cells. Philos Trans R Soc Lond B Biol Sci. 2010;365(1537):155–163. doi:10.1098/rstb.2009.0149
42. Preethy S, John S, Ganesh JS, et al. Age-old wisdom concerning cell-based therapies with added knowledge in the stem cell era: our perspectives. Stem Cells Cloning. 2013;6:13–18. doi:10.2147/SCCAA.S41798
43. Shroff G. A review on stem cell therapy for multiple sclerosis: special focus on human embryonic stem cells. Stem Cells Cloning. 2018;11:1–11. doi:10.2147/SCCAA.S135415
44. El-Sadik AO. Potential sources of stem cells as a regenerative therapy for Parkinson’s disease. Stem Cells Cloning. 2010;3:183–191. doi:10.2147/SCCAA.S14626
45. Lu J, Pompili VJ, Das H. Hematopoietic stem cells: ex-vivo expansion and therapeutic potential for myocardial ischemia. Stem Cells Cloning. 2010;3:57–68. doi:10.2147/sccaa.s6908
46. De Sousa PA, Steeg R, Wachter E, et al. Rapid establishment of the European Bank for induced Pluripotent Stem Cells (EBiSC) - the hot start experience. Stem Cell Res. 2017;20:105–114. doi:10.1016/j.scr.2017.03.002
47. Yokoyama KK, Murata T, Pan J, et al. Genetic materials at the gene engineering division, RIKEN bioresource center. Exp Anim. 2010;59(2):115–124. doi:10.1538/expanim.59.115
48. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;15(3):160018. doi:10.1038/sdata.2016.18
49. Stathias V, Koleti A, Vidović D, et al. Sustainable data and metadata management at the BD2K-LINCS data coordination and integration center. Sci Data. 2018;5:180117. doi:10.1038/sdata.2018.117
50. Knoppers BM, Isasi R, Benvenisty N, et al. Publishing SNP genotypes of human embryonic stem cell lines: policy statement of the international stem cell forum ethics working party. Stem Cell Rev Rep. 2011;7(3):482–484. doi:10.1007/s12015-010-9226-2
51. Morrison M, Bell J, George C, et al. The European general data protection regulation: challenges and considerations for iPSC researchers and biobanks. Regen Med. 2017;12(6):693–703. doi:10.2217/rme-2017-0068
52. Bredenoord AL, Mostert M, Isasi R, Knoppers BM. Data sharing in stem cell translational science: policy statement by the international stem cell forum ethics working party. Regen Med. 2015;10(7):857–861. doi:10.2217/rme.15.42
53. Frati P, Scopetti M, Santurro A, Gatto V, Fineschi V. Stem cell research and clinical translation: a roadmap about good clinical practice and patient care. Stem Cells Int. 2017;2017:5080259. doi:10.1155/2017/5080259
54. Daley GQ, Hyun I, Apperley JF, et al. Setting global standards for stem cell research and clinical translation: the 2016 ISSCR guidelines. Stem Cell Rep. 2016;6(6):787–797. doi:10.1016/j.stemcr.2016.05.001
55. Crook JM, Stacey GN. Setting quality standards for stem cell banking, research and translation: the international stem cell banking initiative. In: Ilic D, editor. Stem Cell Banking. Stem Cell Biology and Regenerative Medicine. New York, NY: Springer; 2014.
56. Sullivan S, Stacey GN, Akazawa C, et al. Quality control guidelines for clinical-grade human induced pluripotent stem cell lines. Regen Med. 2018;13(7):859–866. doi:10.2217/rme-2018-0095
57. Sheehy SP, Pasqualini F, Grosberg A, Park SJ, Aratyn-Schaus Y, Parker KK. Quality metrics for stem cell-derived cardiac myocytes. Stem Cell Rep. 2014;2(3):282–294. doi:10.1016/j.stemcr.2014.01.015
58. Litterman NK, Ekins S. Databases and collaboration require standards for human stem cell research. Drug Discov Today. 2015;20(2):247–254. doi:10.1016/j.drudis.2014.10.006
59. Sansone SA, Rocca-Serra P, Field D, et al. Toward interoperable bioscience data. Nat Genet. 2012;44(2):121–126. doi:10.1038/ng.1054
60. Ho Sui SJ, Begley K, Reilly D, et al. The stem cell discovery engine: an integrated repository and analysis system for cancer stem cell comparisons. Nucleic Acids Res. 2012;40(Databaseissue):D984–91. doi:10.1093/nar/gkr1051
61. Jones RB, Martinez C, Majhail NS, et al. Stem cell transplantation and informatics: current considerations. Biol Blood Marrow Transplant. 2018;24(4):659–665. doi:10.1016/j.bbmt.2017.12.792
62. BioData Catalyst. Available from: https://www.nhlbidatastage.org/.
63. Regenerative medicine innovation catalyst. Available from: http://rmidatahub.org/.
64. Finkelstein J, Zhang F, Levitin SA, Cappelli D. Using big data to promote precision oral health in the context of a learning healthcare system. J Public Health Dent. 2020;1–16. doi:10.1111/jphd.12354
 © 2020 The Author(s). This work is published by Dove Medical Press Limited, and licensed under a Creative Commons Attribution License.
The full terms of the License are available at http://creativecommons.org/licenses/by/4.0/.
The license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
© 2020 The Author(s). This work is published by Dove Medical Press Limited, and licensed under a Creative Commons Attribution License.
The full terms of the License are available at http://creativecommons.org/licenses/by/4.0/.
The license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
