Back to Journals » Drug Design, Development and Therapy » Volume 17
Immunoinformatics Prediction and Protective Efficacy of Vaccine Candidate PiuA-PlyD4 Against Streptococcus Pneumoniae
Authors Miao C, Cui Y, Li Y, Qi Q , Shang W, Chen H, Gao Y, Yuan R, Long Q, Wu W, Wang X, Yan Z , Jiang Y
Received 20 September 2023
Accepted for publication 15 December 2023
Published 21 December 2023 Volume 2023:17 Pages 3783—3801
DOI https://doi.org/10.2147/DDDT.S441302
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tuo Deng
Chenglin Miao,1,* Yali Cui,1– 3,* Yingying Li,1 Qianqian Qi,1 Wenling Shang,1 Huilian Chen,2,3 Yujie Gao,2,3 Ruomei Yuan,2,3 Qichen Long,4 Wenjing Wu,1 Xia Wang,1 Ziyi Yan,1 Yongmei Jiang1,5
1Department of Laboratory Medicine, West China Second University Hospital, Sichuan University, Chengdu, Sichuan Province, People’s Republic of China; 2Department of Laboratory Medicine, Meishan Women and Children’s Hospital, Alliance Hospital of West China Second University Hospital, Sichuan University, Meishan, Sichuan Province, People’s Republic of China; 3Department of Laboratory Medicine, West China Second University Hospital (Tianfu), Sichuan University/Sichuan Provincial Children’s Hospital, Meishan, Sichuan Province, People’s Republic of China; 4Department of Laboratory Medicine, The Second Xiangya Hospital, Central South University, Changsha, Hunan Province, People’s Republic of China; 5Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, Chengdu, Sichuan Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ziyi Yan; Yongmei Jiang, Department of Laboratory Medicine, West China Second University Hospital, Sichuan University, Chengdu, Sichuan Province, People’s Republic of China, Tel +86 28 8857050 ; +86 28 85501543, Email [email protected]; [email protected]
Purpose: This study was designed to evaluate the immune protective efficacy of the novel Streptococcus pneumoniae (S. pneumoniae) protein vaccine PiuA-PlyD4 through immunoinformatics prediction and in vitro and in vivo experiments.
Methods: In this study, we conducted immunoinformatics prediction and protection analysis on the fusion protein PiuA-PlyD4. The epitope composition of the vaccine was analyzed based on the prediction of B-cell and helper T-cell epitopes. Meanwhile, the molecular docking of PiuA and TLR2/4 was simulated. After immunizing C57BL/6 mice with the prepared vaccine, the biological safety, immunogenicity and conservation were evaluated. By constructing different infection models and from the aspects of adhesion inhibition and cytokines, the protective effect of the fusion protein vaccine PiuA-PlyD4 on S. pneumoniae infection was explored.
Results: PiuA-PlyD4 has abundant B-cell and helper T-cell epitopes and shows a high antigenicity score and structural stability. Molecular docking analysis suggested the potential interaction between PiuA and TLR2/4. The specific antibody titer of fusion protein antiserum was as high as (7.81± 2.32) × 105. The protective effect of the immunized mice on nasal and lung colonization was significantly better than that of the control group, and the survival rate against S. pneumoniae infection of serotype 3 reached 50%. Cytokine detection showed that the humoral immune response, Th1, Th2 and Th17 cellular immune pathways were all involved in the process.
Conclusion: The study indicates that PiuA-PlyD4, whether the results are predicted by immunoinformatics or experimentally validated in vivo and in vitro, has good immunogenicity and immunoreactivity and can provide effective protection against S. pneumoniae infection. Therefore, it can be considered a promising prophylactic vaccine candidate for S. pneumoniae.
Keywords: Streptococcus pneumoniae, fusion protein vaccine, PiuA-PlyD4, immunoinformatics prediction, protective efficacy
Introduction
Streptococcus pneumoniae (S. pneumoniae) is a pathogen that can infect people of all ages, and the highest incidence is in children under 2 years old, elderly individuals, and immunocompromised individuals. It has a high mortality rate and is responsible for causing severe pneumococcal disease (PD) worldwide. According to the World Health Organization (WHO), approximately 14.5 million people suffer from severe PD each year, leading to 1.6 million deaths and making it a serious public health issue.1,2 Recently, the WHO listed S. pneumoniae as one of the 12 pathogens that require attention and research for developing new antibacterial strategies.3
In recent years, commercially available S. pneumoniae vaccines have been based on the development of pneumococcal polysaccharide vaccines (PPVs) and pneumococcal polysaccharide conjugate vaccines (PCVs) targeting capsule serotypes,4 including PPV23, PCV10, PCV13, PCV15, PCV20, and PCV24. The use of these vaccines has effectively reduced the morbidity and mortality of S. pneumoniae infections, but there is still a drawback of inducing only serotype-specific immune responses.5 PPV23 has protective effects against invasive pneumococcal disease (IPD) and community-acquired pneumonia (CAP) but cannot prevent their occurrence and has no protective effect against colonizing bacteria carried by the population. More significantly, currently available PPVs are poorly immunogenic and have difficulty inducing immune memory in children under 2 years of age due to their T-independent (TI) nature.6,7 PCV13 has a protective effect against IPD and CAP caused by vaccine types (VTs) and significantly reduces S. pneumoniae infections in children. However, it still has problems such as an increase in nonvaccine type (NVT) strains that cause infections and colonize vaccinated individuals.8 Furthermore, increasing serotypes blindly is not feasible from both the complexity and cost aspects in the production of PCVs.9 Therefore, developing new and universal vaccines is an urgent issue that needs to be addressed.
Existing reports have shown that PiuA is present in all known S. pneumoniae strains, is highly conserved, and has a high affinity for iron ions, which is essential for the growth and development of bacteria.10 Furthermore, membrane-exposed PiuA, an essential virulence factor in a mouse infection model, is protective against IPD, making it a candidate vaccine factor and drug target.11,12 Pneumolysin (Ply) is a cholesterol-dependent pore-forming toxin, and its C-terminal D4 domain (AA-360-471) is a functional domain that recognizes mammalian cells, binds to mannose dose-dependently, and activates Toll-like receptor (TLR), with potential adjuvant effects.13 Previous research results have also shown that PlyD4 is an excellent fusion protein component and is expected to become a “good partner” for protein vaccines.14,15
In this study, we selected the full-length PiuA sequence without the signal peptide and transmembrane structure domain and connected it with PlyD4 through a flexible linker to form the fusion protein PiuA-PlyD4 that consists of two antigenic structures, aiming to obtain a fusion protein vaccine with better protective efficacy.14,16 Our study provides an important basis for the development of universal protein vaccines against S. pneumoniae infection.
In addition, the continuous progress of immunoinformatics methods and the wide range of vaccine design tools allowed us to predict the feasibility of the fusion protein PiuA-PlyD4 as a candidate vaccine quickly through immunoinformatics predictions. Therefore, in this study, we analyzed the physicochemical characteristics and structural quality of PiuA-PlyD4 through online modeling and obtained the purified PiuA-PlyD4 fusion protein through a prokaryotic expression system. Then, we immunized C57BL/6 mice to evaluate biosafety immunogenicity and conservation. The protective efficacy of active immunization was studied, and we validated adhesion inhibition and cytokines. Our prediction results and experimental studies provide a basis for the development of S. pneumoniae protein vaccines.
Material and Methods
Immunoinformatics Analysis
Protein Sequence Retrieval and Fusion Protein Design
The nucleotide sequence of PiuA in S. pneumoniae standard strain D39 (NC_008533.2) was obtained from the NCBI (https://www.ncbi.nlm.nih.gov/gene/), which can be translated into protein sequences by Editseq software of the DNAStar package. The presence of transmembrane helical segments in PiuA was tested by TMHMM Server v. 2.0 (https://services.healthtech.dtu.dk/services/TMHMM-2.0/). The SignalP 5.0 server (https://services.healthtech.dtu.dk/services/SignalP-5.0/) was employed to detect the signal peptide region. In addition, the protein sequence of PlyD4 (AA360-471) was fetched from our previous study.14
Evaluation of Physicochemical Parameters and Immunological Characteristics
The ExPASy ProtParam online tool (https://web.expasy.org/protparam/) was applied to determine the molecular weight (MW), theoretical isoelectric point (pI), half-life, instability index, aliphatic index, grand average of hydropathicity (GRAVY), and other physiochemical properties of PiuA-PlyD4.17 The VaxiJen v2.0 server (http://www.ddgpharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was used to evaluate the antigenicity potential of PiuA-PlyD4 with a threshold value of 0.4.18 Additionally, the allergenicity of the designed construct was checked by the AllerTOP (http://www.ddg-pharmfac.net/AllerTOP/) server.19
Tertiary Structure Validation
The 3D structure of PiuA-PlyD4 was modeled using AlphaFold2 (https://cryonet.ai/af2/). Furthermore, the overall structural quality of the generated model of PiuA-PlyD4 was compared through the Z score, ERRAT score (https://saves.mbi.ucla.edu/) and Ramachandran plot analysis. The ProSA server (https://prosa.services.came.sbg.ac.at/prosa.php) validated the reliability of PiuA-PlyD4 based on the predicted Z score. The Ramachandran plot is one of the best quality indicators for experimental structure models,20 which can report the geometry and stereochemistry of the construct using the MolProbity Ramachandran map (http://molprobity.biochem.duke.edu/).
Epitope Prediction
The sequential B-cell epitopes of the fusion protein PiuA-PlyD4 were predicted through the IEDB tool (http://www.iedb.org/).21 The BepiPred-2.0 server at IEDB was used for the prediction of linear B-cell epitopes by the Random Forest algorithm. The residues with scores above the threshold (default value is 0.5) are predicted to be the composition of an epitope. Based on the tertiary structure of PiuA-PlyD4, conformational B-cell epitopes were performed by DiscoTope 1.1 at IEDB (default value is −7.7). For projecting MHCII binding peptides, the NetMHCIIpan 4.0 (https://services.healthtech.dtu.dk/services/NetMHCIIpan-4.0/) online tool was used to predict binding to MHCII molecules using artificial neural networks (ANNs). Both Binding Affinity (BA) and Eluted Ligand mass spectrometry (EL) on NetMHCIIpan 4.0 were used to elucidate appropriate MHCII binding epitopes.22
Molecular Docking Analysis and Visualization
Toll-like receptors (TLRs) play a vital role in the immune response, whether innate or acquired. TLR2 and TLR4 are responsible for recognizing microbial components, and TLR2 is also the receptor for gram-positive bacteria.23,24 Molecular docking between piuA and TLR2/4 was performed by the HDOCK server (http://hdock.phys.hust.edu.cn/), a fully automated protein-protein docking server. This server automatically simulates the interaction through a hybrid strategy of template-based modeling and template-free docking.25 For the docking analysis, the 3D structures of piuA (PDB ID: 4JCC), human TLR2 tetramer (PDB ID: 6NIG) and TLR4 dimer (PDB ID: 3FXI) were retrieved from the RCSB PDB site (https://www.rcsb.org/). Thereafter, the docked complexes were further visualized with PyMOL and ChimeraX software, which provided us with analyses of the protein-ligand interactions by hydrogen bonds. Furthermore, the binding free energy of the complexes obtained using GROMACS 2019.6 and MM-PBSA.26
Bacteria
Escherichia coli (E. coli) DH5α (Invitrogen, CA, USA) was used as the host for plasmid molecules, and E. coli BL21 (Invitrogen, CA, USA) was used as the protein expression vector. The different serotypes of S. pneumoniae strains used in this experiment were all preserved in our previous clinical research (Table 1). The S. pneumoniae strains were inoculated on TSA plates containing 5% sheep blood (blood agar) or Todd-Hewitt medium (THY) and cultured at 37°C with 5% CO2 in an incubator.
 |
Table 1 Information About the Strains Used to Detect the Conservation of PiuA-PlyD4 by Western Blotting |
Mice
Female C57BL/6 mice aged 4 to 6 weeks were purchased from Chengdu Dashuo Experimental Animal Co., Ltd. and raised under specific pathogen-free (SPF) conditions in the Animal Experimental Center at the Second West China Hospital of Sichuan University.
Expression and Identification of the Fusion Protein PiuA-PlyD4
Construction and Expression of Recombinant Plasmid
The recombinant plasmid piuA-plyD4-pET28a was synthesized by Tsingke Biotechnology Co., Ltd. based on the gene sequence of S. pneumoniae standard strain R6 (NC_003098.1) with the addition of BamHI-XhoI restriction sites.
The plasmid was transformed into E. coli DH5α by the heat shock method and amplified. Single colonies growing on the surface of Luria-Bertani (LB) agar plates were picked and cultured for plasmid amplification, and the recombinant plasmids were sent to Tsingke Biotechnology Co., Ltd. for Sanger sequencing identification.
Purification and Identification of PiuA-PlyD4
After breaking open the strains by high pressure, the clarified lysate was purified by sequential Ni2+-NTA chromatography, anion exchange chromatography (Source Q, Cytiva, USA), gel filtration chromatography (Superdex 200 Increase, Cytiva, USA), and polymyxin B resin gravity column (GenScript, NJ, USA) to obtain purified PiuA-PlyD4 fusion protein. The purified protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad, CA, USA). Anti-6×His tag antibody (diluted 1:1000 in 5% BSA solution) was used as the primary antibody, and HRP-labeled goat anti-mouse IgG (diluted 1:5000) was used as the secondary antibody. The PVDF membrane was visualized using an imaging system, and the protein concentration was determined using an ultraviolet-visible spectrophotometer (DeNovix, USA).
Biosafety Evaluation
Two weeks after the last immunization (day 42), mice were euthanized by sodium pentobarbital (180 mg/kg), and organs including lungs, brain tissue, heart, liver, spleen, and kidneys were dissected for fixation in 4% paraformaldehyde (Solarbio, Beijing, China). Tissue sections were then prepared and stained with hematoxylin and eosin (H&E) for histopathological examination using light microscopy.
Mouse Immunization and Antiserum Preparation
Six- to eight-week-old female C57BL/6 mice were randomly divided into three groups using a double-blind method (Table 2). Mice were immunized with the PiuA-PlyD4 vaccine adjuvanted with Al(OH)3. One week after each immunization (Figure 1), tail-tip blood was collected from three mice per group. One week after the final immunization, antisera were collected from the orbital blood of all mice.
 |
Table 2 Immunization Schedule for C57BL/6 Mice |
 |
Figure 1 Flowchart of immunization and challenge experiments with the PiuA-PlyD4 protein vaccine. |
Mouse Antiserum Titer Assessment and Cross-Reactivity
Immunogenicity Assessment
Immunogenicity was evaluated using an indirect enzyme-linked immunosorbent assay (ELISA). Mouse sera collected from each group were incubated with PiuA-PlyD4 fusion protein precoated on the microwell plate, and HRP-labeled goat anti-mouse IgG (diluted 1:5000, ZEN BIO, China) was used as the secondary antibody. After color development, the absorbance was measured at 450 nm to determine the specific IgG antibody titers in each sample.
Antibody Subtype Detection
One week after the final immunization (Day 35), immune sera from each immunization group were collected, and an indirect ELISA method was used to detect antibody subtypes. HRP-labeled goat anti-mouse IgG1, IgG2a, IgG2b, and IgG3 (diluted 1:5000, Cell Signaling, USA) were used as secondary antibodies to determine the specific IgG antibody subtype titers in each sample.
Conservation Assessment
The conservative assessment was conducted by selecting clinical strains with high serotype isolation rates in the local area that were identified in previous studies by our research group (Table 1). These included strains 19F, 19A, 3, 6A, 23A, 14, 34, and 15A, with E. coli strains used as negative controls. Bacterial lysates were prepared by heating at 95°C and separated by SDS-PAGE, followed by transfer onto PVDF membranes. Each group of antisera was added (diluted 1:1000), and HRP-labeled goat anti-mouse IgG antibody (diluted 1:5000) was used as the secondary antibody. Western blotting analysis was then used to assess the conservation of PiuA-PlyD4.
Protection Efficacy with Active Immunization
Anti-Colonization Protective Effect
Two weeks after the final immunization (day 42, Table 2), mice were briefly anesthetized with sodium pentobarbital (50 mg/kg), and S. pneumoniae strain WCH2203043016 (Serotype 19F, Table 1) was prepared and suspended in amounts of 30 μL per mouse (2.8×107 CFU/μL). The mice were intranasally infected with the bacterial suspension. Seventy-two hours postinfection, the mice were euthanized by sodium pentobarbital (180 mg/kg), and nasal lavage fluids (NLF) were collected by instilling 500 μL of sterile PBS into the exposed trachea and then recovering it from the nasal cavity. Bacterial counts were determined by plating serially diluted NLF samples for colony-forming units (CFUs). Lung homogenates were also prepared and plated before CFUs were counted.
In addition, lung tissues from the immunized mice were histologically examined by preparing tissue sections and staining them with H&E to evaluate any pathological changes.
Protective Effect Against Lethal Infection
Two weeks after the final immunization (Table 2), S. pneumoniae strain WCH2102011128 (Serotype 3, Table 1) was selected to construct the lethal infection model (3.2×104 CFU/200 μL). The optimal bacterial dose (2.8×107 CFU/μL) was used for intraperitoneal inoculation based on preliminary LD90 experiments. The mice were infected to establish the S. pneumoniae lethal infection model, after which their conditions were observed daily for 21 days, and their survival status was recorded.
Adhesion Inhibition and Cytokines
Adhesion Inhibition Assay with Immune Serum
The A549 cell (Type II epithelial lung carcinoma cells, ATCC) concentration was adjusted to 3×105 cells/mL, and 1 mL was transferred to a 24-well plate. The bacterial suspension of S. pneumoniae strain WCH2203043016 (serotype 19F, Table 1) was adjusted to 5×106 CFU/10 μL. Then, 100 μL of immune serum (diluted 1:10 in 10% FBS) with an antibody titer of 7.5×105 was added to each well. The samples were centrifuged at 1300×g for 5 min and incubated for 1 h at 37 °C and 5% CO2. After dilution, the samples were plated, and CFU values were calculated by bacterial colony counts.
Cytokine Detection
Two weeks after the final immunization (day 42, Table 2), mice were euthanized using sodium pentobarbital (180 mg/kg), and splenocytes were isolated to prepare cell suspensions of 5×106 cells/mL. Then, 1 mL of the suspension was added to each well of a 24-well cell culture plate and cultured at 37 °C with 5% CO2. The PiuA-PlyD4 fusion protein was adjusted to a final concentration of 5 μg/mL and added to the splenocyte suspension. The supernatants were collected at 24, 48, and 72 h. Standard solutions containing ILs, IFN-γ, and TNF were prepared, and cytokine concentrations were measured using a Mouse Th1/Th2/Th17 CBA Kit (BD Biosciences, NJ, USA).
Statistical Analysis
IBM SPSS Statistics software (SPSS, version 22.0, USA) was used to assess the statistical significance of the experimental data. Methods included the chi-square test, Fisher’s exact test, the T-test, one-way ANOVA statistical analysis, and the Mantel-Cox test. The Bonferroni and Tamhane’s T2 methods were used to analyze whether the differences among multiple groups based on one-way ANOVA statistical analysis were statistically significant; the Bonferroni method was used to analyze whether the differences among multiple groups based on the chi-square test were statistically significant. A p value of < 0.05 was considered statistically significant.
Results
Immunoinformatics Analysis
Fusion Protein Design and Characteristic Evaluation
The PiuA with removed transmembrane domain and the signal peptide was connected with PlyD4 through a (GGGGS)2 linker to construct the target protein PiuA-PlyD4.27,28 The physicochemical parameters related to PiuA-PlyD4 were predicted using ExPASy. The results showed that the fusion protein PiuA-PlyD4 was composed of 426 amino acids, with a molecular weight of 46.6 kDa and a theoretical pI of 5.28. Its predicted half-life was 30 h in mammalian reticulocytes, > 10 h in E. coli, and > 20 h in yeast. The GRAVY index was −0.374, with lower scores indicating higher solubility, suggesting that PiuA-PlyD4 is hydrophilic. The instability index of PiuA-PlyD4 was estimated to be 27.68, demonstrating a certain level of stability. The antigenicity of PiuA-PlyD4 was estimated to be 0.684 (threshold: 0.4) by VaxiJen v2.0. Using the AllerTOP server, the allergenicity of PiuA-PlyD4 was assessed; the results predicted that the construct is nonallergenic.
Tertiary Structure Validation
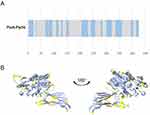
To validate the overall quality of PiuA-PlyD4 after obtaining the model through AlphaFold2, the Z score, the ERRAT score, and the Ramachandran plot were evaluated. The quality factor predicted by the Z score was −9.63 (Figure 2A), suggesting that our construct is reliable.29,30 Moreover, the ERRAT score for PiuA-PlyD4 was 94.4 (Figure 2B), and a higher ERRAT score indicates that the predicted model is more trustworthy.31 MolProbity was used to evaluate the structure quantity and generate the Ramachandran plot (Figure 2C). The Ramachandran plot revealed that 91.7, 95.0, and 0.5% of the residues were in the favored, allowed, and disallowed regions, respectively. Typically, more than 85% of amino acids in favored zones are acceptable,32,33 which further verifies the reliability of our data.
Identification of B‑cell Epitopes
As B cells play an essential role in the adaptive immune system, especially for humoral immunity, the sequential and discontinuous B-cell epitopes of PiuA-PlyD4 were predicted using the IEDB tool, and the prediction results of the sequential (Figure 3A) and discontinuous (Figure 3B) epitopes obtained by the servers were visualized on the 3D structure.
Identification of MHCII Binding Epitopes
According to the common alleles and well-documented alleles (CWD),34 five HLA-DRB1 alleles that are most common in the Chinese population were selected, and MHCII binding epitope prediction was performed using NetMHCIIpan-4.0. The prediction results (Table 3) showed that HLA-DRB1*0701, HLA-DRB1*0803, HLA-DRB1*0901, HLA-DRB1*1202, and HLA-DRB1*1501, which are commonly found in the Chinese population, could all strongly bind to the PiuA-PlyD4 protein sequence, indicating that the fusion protein has good immunoreactivity.
 |
Table 3 MHCII Binding Epitope Prediction |
Molecular Docking Analysis and Visualization
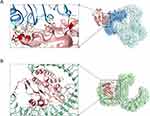
Innate immunity is the first line of defense against pathogen invasion. The TLR family serves as a vital sensor that detects diverse microbial components and elicits innate immune responses. We speculate that PiuA may have potential binding sites with TLR2/4. The processes of protein-ligand docking between TLR2/4 receptors and PiuA were performed by the HDOCK online server. A more negative docking score means a more possible binding model. Meanwhile, when the confidence score is higher than 0.7, the two proteins would be very likely to bind.25,35 Based on these, the models were chosen for PiuA-TLR2 and PiuA-TLR4 complexes with the best docking and confidence scores, respectively; the results showed the potential interaction between PiuA and TLR2/4. The top 10 predictions in HDOCK for PiuA-TLR2/4 were shown in Supplementary Figures 1 and 2. To display and evaluate the interaction of protein-ligand complexes, PyMOL and ChimeraX were used. PiuA-TLR2 formed 11 hydrogen bonds (Figure 4A): Asp267-Ser222, Ser269-Ser222, Leu262-Lys192, Ala263-Lys192, Asp297-Lys192, Asp297-Arg167, Glu276-Thr174, Ala286-Thr149 and Lys290-His147. Additionally, PiuA-TLR4 formed 18 hydrogen bonds (Figure 4B): Ser37-Glu605, Glu136-His529, Lys133-Gln505, Asp132-Arg460, Glu111-Lys477, Glu111-Gln430, Lys129-Glu439, Asp109-Arg382, Ser236-Asn417, Lys207-Ser392, Lys207-Ser394, Gly265-Lys362, Asn268-Tyr296, Ala263-Arg264, Leu262-Arg264 and Lys172-Glu42. Additionally, the average binding free energy of PiuA-TLR2 and PiuA-TLR4 were shown in Supplementary Table 1.
Expression and Identification of the Fusion Protein PiuA-PlyD4
Construction and Expression of Recombinant Plasmid
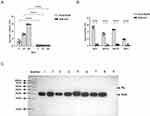
The recombinant plasmid was double-digested with BamHI and XhoI and identified by Sanger sequencing, confirming that the nucleotide sequence was as expected. After inducing expression with IPTG, the results of SDS-PAGE confirmed the presence of the PiuA-PlyD4 fusion protein band at the corresponding position (Figure 5A).
Purification and Identification of PiuA-PlyD4
After purification using the Ni2+-NTA gravity column (Figure 5A), anion exchange chromatography, and gel filtration chromatography, the candidate vaccine PiuA-PlyD4 was obtained with a purity > 90% (Figure 5B and C). After removal of endotoxin and filtration, the final protein concentration was determined to be 2.5 mg/mL, which was confirmed by Western blotting (Figure 5D).
Biosafety Assessment
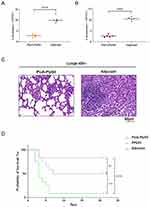
Two weeks after immunization, the heart, lungs, liver, brain tissue, spleen, and kidneys were isolated from C57BL/6 mice in each group, and tissue slices were made and stained with H&E. Compared with the adjuvant control group, no significant pathological damage was observed in the immunized group, indicating that the PiuA-PlyD4 fusion protein has good biosafety and can be used as an immunogen (Figure 6).
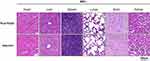 |
Figure 6 Pathological changes in the organs of mice after immunization. From left to right are heart tissue, liver tissue, spleen tissue, lung tissue, brain tissue and kidney tissue respectively. |
Mouse Antiserum Titer Assessment and Cross-Reactivity
Immunogenicity Assessment
After 7 days on each immunization day, serum was collected from mice in each group, and indirect ELISA was performed to measure specific IgG antibody titers (Figure 7A). The titer significantly increased after the third immunization of PiuA-PlyD4, and in the adjuvant control group, there were few specific antibody reactions as follows: (7.81±2.32) ×105 vs (1.40±0.55) (P<0.001).
IgG Antibody Subtype Detection
The specific antibody subtypes (IgG1, IgG2a, IgG2b, and IgG3) in each group were detected by indirect ELISA (Figure 7B). The immunized mice mainly produced IgG1, IgG2a, and IgG2b antibodies, with a certain amount of IgG3. The specific effector titers of the PiuA-PlyD4 and adjuvant groups were as follows: IgG1: (2.68±1.79) ×105 vs (2.20±1.30) (P=0.010), IgG2a: (3.29±2.09) ×103 vs (1.60±0.89) (P=0.008), IgG2b: (4.18±1.06) ×104 vs (2.40±0.89) (P<0.001), IgG3: (75.40±23.27) vs (1.80±1.30) (P<0.001).
Conservation Assessment
Clinical strains with high isolation rates were selected, including 19F, 19A, 3, 6A, 23A, 14, 34, and 15A. E. coli BL21 was used as the negative control strain. Among them, 19F, 19A, 3, 6A, and 14 are PCV13-covered serotypes, and 23A, 34, and 15A are PCV-uncovered serotypes. After reacting with PiuA-PlyD4-specific antiserum, obvious positive PiuA bands appeared in all S. pneumoniae strains of different serotypes (Figure 7C), indicating that PiuA-PlyD4 fusion protein antiserum can recognize various serotypes of S. pneumoniae and is expected to provide broad-spectrum protection. In addition, the Ply protein showed weak reaction bands or no reaction bands.
Protection Efficacy with Active Immunization
Anti-Colonization Protective Effect
Two weeks after the last immunization, a 19F (ST271) bacterial suspension was prepared at a dose of 2.8×107 CFU/μL to establish colonization infection models. After 72 h of infection, mouse NLF was collected, and lung tissue was separated to prepare homogenate and then diluted to count colonies. The results showed that the number of colonized bacteria in the nasal cavity (Figure 8A) of the PiuA-PlyD4 group and adjuvant control group was (2.74±0.62) ×105 CFU vs (10.34±1.57) ×105 CFU (P<0.001); the number of colonized bacteria in the lungs (Figure 8B) of the PiuA-PlyD4 group and adjuvant control group was (2.57±0.56) ×104 CFU vs (10.47±0.81) ×104 CFU (P<0.001). The number of colonized bacteria in the immunization group was significantly lower than that in the adjuvant control group.
The lung tissue was also collected for histological analysis by H&E staining (Figure 8C). Under an optical microscope, compared to the adjuvant group, mice in the immunization group had milder alveolar tissue damage and exhibited varying degrees of lobar pneumonia symptoms with different degrees of inflammatory cell infiltration and capillary congestion.
Protective Effect Against Lethal Infections
Two weeks after the last immunization, S. pneumoniae serotype 3 was used to prepare bacterial suspensions for the intraperitoneal challenge. The lethal infection models were established based on the optimal challenge dose, and the survival status of the mice was observed every day for 21 consecutive days. The survival time and survival rate of the mice were recorded (Figure 8D). The 21-day survival rates of mice infected with serotype 3 were PiuA-PlyD4: 50.0%; Adjuvant: 8.3%; PPV23: 83.3%. The results showed that PiuA-PlyD4 effectively improved the survival rates and prolonged the survival times of mice infected with serotype 3, which were significantly improved compared with the adjuvant control group (P=0.008); there was no significant difference compared with the PPV23-immunized group (P=0.111).
Adhesion Inhibition and Cytokines
Adhesion Inhibition Assay with the Antiserum
Adhesion inhibition assay using S. pneumoniae and A549 cells with the PiuA-PlyD4 fusion protein antiserum; S. pneumoniae serotype 19F adhered to A549 cells, which was inhibited by PiuA-PlyD4 antiserum, and the adhesion inhibition ability was evaluated by colony counting as follows (Figure 9A): PiuA-PlyD4: (3.73±0.93) ×103 CFU; PPV23: (4.40±0.58) ×103 CFU; adjuvant: (10.81±1.63) ×103 CFU. The inhibitory effect of the PiuA-PlyD4 antiserum on bacterial adhesion was significantly higher than that of the adjuvant group (P<0.001), and there was no significant difference with the PPV23 immune group (P=0.634).
Cytokine Detection
The PiuA-PlyD4 fusion protein effectively stimulated the production of various cytokines in mouse splenocytes. The cytokine concentrations secreted by splenocytes 72 h after stimulation were detected by flow cytometry CBA assay (Figure 9B). The specific concentrations in the PiuA-PlyD4 group and adjuvant group were as follows: IL-2: (14.72±3.16 vs 2.77±1.37) pg/mL (P<0.001), IL-4: (2.98±0.61 vs 2.60±1.16) pg/mL (P=0.526), IL-6: (620.36±185.79 vs 33.37±12.61) pg/mL (P<0.001), IL-10: (996.16±219.05 vs 25.71±6.61) pg/mL (P<0.001), IL-17A: (246.88±72.44 vs 9.32±3.05) pg/mL (P<0.001), TNF-α: (693.07±82.00 vs 17.69±8.00) pg/mL (P<0.001), IFN-γ: (303.74±102.17 vs 11.60±6.41) pg/mL (P<0.001). The levels of secretion of the cytokines IL-2, IL-6, IL-10, IL-17A, TNF-α, and IFN-γ were significantly different between the PiuA-PlyD4 group and the adjuvant group.
Discussion
In addition to the capsule, S. pneumoniae pathogenesis is also closely related to other non-capsule virulence genes. Virulence factors and capsules jointly form a complex, tight and tenacious virulence regulation system in S. pneumoniae, which regulates its pathogenicity. This is also the target that scholars are using to develop vaccines and find new drugs, which may provide a breakthrough for improving the prevention of S. pneumoniae diseases. PiuA is exposed outside the cell membrane of S. pneumoniae and plays an important role during the transfer of bacteria from the nasopharynx to the lung. It is an essential virulence factor in mouse infection models and has the potential to effectively overcome the inherent immunogenicity defects and serotype dependence of pneumococcal polysaccharide vaccines.36
PiuA, as a soluble lipoprotein binding protein of the PiuBCDA ABC transporter, is a tetrameric FeIII-iron carrier binding protein. It promotes exogenous iron absorption by binding tetrameric hydroquinone FeIII, which affects bacterial growth and development and stimulates host immune responses, especially in antibacterial colonization.37,38 Computer modeling studies have shown that multiepitope vaccines composed of high antigenic epitopes of PiuA, along with PspA, CbpA, and PhtD, can provide protection against S. pneumoniae infection and are good early candidate indicators for S. pneumoniae vaccines.39 Ply has strong proinflammatory effects, and the inflammatory response induced by Ply accelerates the clearance of S. pneumoniae from the upper respiratory tract. Its D4 domain at the C-terminus (AA360–471) is a cholesterol-binding site of host cells that exhibits high conservation and immunogenicity. It possesses the TLR4 agonist activity, bacterial virulence and hemolytic activity of full-length Ply. This domain can be used as a good vaccine candidate factor. Our previous studies have also shown that PlyD4, when combined with NanA and Tuf peptide segments to prepare fusion proteins, had certain protective effects when immunized in mice,14 making it a better fusion protein component.13,40
Therefore, in this study, we aimed to increase the exposure of fusion protein epitopes by connecting the full-length sequence of PiuA (excluding the signal peptide and transmembrane structure threshold) and PlyD4 with a flexible linker (GGGGS)2 to prepare the fusion protein PiuA-PlyD4; the aim was to obtain a fusion protein vaccine with better conservation and immune protection effects. (GGGGS)2 can reduce the spatial hindrance of different components of the fusion protein, which is beneficial to the correct folding of each structural domain.41
The Bepipred 2.0 model prediction results of the online IEDB server showed that PiuA-PlyD4 has abundant B-cell epitopes, including both linear and conformational epitopes. In addition, the HLA-DRB1 antigen in the MHC class II gene is important in the immune response and is closely related to CD4+ T-cell immunity. The helper T-cell epitope prediction results of the NetMHCIIpan-4.0 online website showed that in addition to strong binding epitopes, there are also many weak binding epitopes. A large number of epitopes help to stimulate the host to produce higher levels of specific antibodies and better protection, and the above results indicate that the fusion protein PiuA-PlyD4 has good immune reactivity in theory.42
Ply, as the major virulence protein of S. pneumoniae, is known as an agonist of TLR4.13 To identify the potential main residues of the interaction between PiuA, another component of the fusion protein, and TLRs, we performed molecular docking using HDOCK. Following docking, the complexes −209.35 (for PiuA-TLR2) and −254.94 (for PiuA-TLR4) with the best docking scores were selected for analysis. In order to enrich antigenic epitopes, the ligand PiuA is relatively large. Although molecular docking analysis suggests the existence of potential interactions, the binding of the ligand to TLRs still needs further optimization.43,44
Immunoinformatics approaches have been widely used in developing novel vaccines against numerous diseases.45,46 Computing virtual analysis is more efficient in terms of time and cost than conducting extensive screening experimentally.47 Given the good results of immunoinformatics prediction, we performed prokaryotic expression and purification, and detected the impact of PiuA-PlyD4 on pathological tissue sections of mouse organs such as heart, lung, brain, and liver after immunization; we found that PiuA-PlyD4 did not cause damage to the mouse organ tissue, indicating some degree of biosafety and its potential as a candidate protein vaccine for S. pneumoniae.
An important feature of an ideal protein vaccine is its ability to induce host immune responses. The protective effect of specific IgG antibodies on the host depends on multiple pathways, including toxin neutralization, complement activation, antibody-dependent cellular cytotoxicity (ADCC), and participation in opsonophagocytosis, among others. In this study, we used conventional modeling methods,48 mixed PiuA-PlyD4 with Al(OH)3 adjuvant at the same volume, and subcutaneously immunized C57BL/6 mice with three doses. High titer-specific IgG antibodies were produced after immunization, indicating that the fusion protein PiuA-PlyD4 we constructed had good immunogenicity.
High levels of IgG antibodies often indicate better immune protection, enhance complement deposition, induce complement-dependent opsonization, and reduce bacterial colonization to exert anti-infective effects.49 However, the maintenance time of antibodies in the host still needs further investigation, and the specific mechanism is also related to the vaccine form.50 In addition, the clearance and protective effects of fusion proteins against S. pneumoniae infections are related to IgG subtypes and cytokine secretion. Studies have shown that immunization with PiuA-PlyD4 and aluminum adjuvant can induce high levels of IgG1, IgG2a, and IgG2b antibodies in the host, indicating that the fusion protein not only stimulates the host to produce a high level of humoral immune response but also stimulates the Th1 and Th2 immune responses at the same time.
Compared with polysaccharide-based vaccines, one huge advantage of an ideal protein vaccine is that it is not restricted by serotype and can provide protection against infections from different serotypes of S. pneumoniae. In this study, we verified the conservation of PiuA-PlyD4. We found that PiuA-PlyD4-specific antiserum could induce immune reactions with the PiuA protein of different serotypes of S. pneumoniae, producing obvious cross-reactive bands, and indicating that PiuA-PlyD4 may provide protection against infections from different serotypes of S. pneumoniae. The fusion protein antiserum had weaker or no reaction bands with bacterial Ply proteins, which was consistent with the previously reported weak reaction intensity of PlyD4 in the NanAT1-Tuf1-PlyD4 fusion protein.14 The reason for this might be related to the strong positive reaction of the PiuA protein, which affected the exposure and coloration or the formation of absolute dominant epitopes of PiuA in the fusion protein, which masked the reaction of PlyD4-peptide segments.
Nasopharyngeal colonization is a risk factor for invasive infection, and colonized bacteria are considered a reservoir of antibiotic resistance genes and polysaccharide variation, leading to antibiotic resistance and vaccine escape, such as seen in serotypes 15BC and 35B. Therefore, preventing bacterial carriage can prevent the occurrence of invasive diseases.51 If a vaccine could reduce S. pneumoniae colonization in the host’s nasopharynx or other parts of the body, it would help protect against colonization and invasive infections. At the same time, it is also likely to block the spread of antibiotic resistance because shortening the duration of asymptomatic carriage may limit the adaptive advantages of antibiotic-resistant pathogens under the use of antibiotics. Based on the above characteristics, we evaluated the effectiveness of the vaccine from the following two aspects: reducing colonization and increasing survival rate.
In the colonization model, we found that the bacterial load of immunized mice in the nasopharynx and lungs was significantly lower than that of the adjuvant control group, and the inflammatory response in their lungs was milder than that of the adjuvant group. In addition, we constructed lethal infection models using S. pneumoniae with high isolation rates in previous clinical isolates, namely, serotype 3. The results showed that PiuA-PlyD4 can prolong the survival time and improve the survival rate after S. pneumoniae infections. However, the survival rate of mice was lower than that of those given the current commercial polysaccharide vaccine PPV23. However, due to the serotype limitation of PPV23 and its low T-cell-independent immunogenicity, fusion protein vaccines can still be used as promising vaccine candidates for in-depth research.50
Humoral immunity is considered the major response against the extracellular pathogen S. pneumoniae.52 Cytokine detection further showed that PiuA-PlyD4 can stimulate spleen cells in the host to produce high levels of the cytokines IL-2, IL-6, IL-10, IL-17A, TNF-α, and IFN-γ, indicating that the fusion protein immune response not only activates Th1 and Th2 pathways but also stimulates Th17 cell immune pathways. This may be an important reason why the fusion protein can reduce S. pneumoniae colonization, and it is consistent with the previously reported protective effects of Th1 and Th17 responses in clearing nasopharyngeal colonization strains and the protective immune conclusion of Th2-induced humoral immunity on lethal infection models after colonization.53
Based on the prediction of immunoinformatics, this study demonstrated that PiuA-PlyD4 is a promising vaccine candidate protein that enhances the immune response and can stimulate high levels of specific anti-pneumococcal antibodies through a series of in vivo and in vitro experiments. The limitation of this study is the lack of long-term monitoring of the protective utility of the fusion protein PiuA-PlyD4. In the future, we will further explore the immune effectiveness and mechanism, including the types of immune cells that play a protective role and whether PiuA-PlyD4 can induce immune memory to better evaluate the protective effect of fusion protein vaccines.
Conclusion
In conclusion, our study not only successfully verified the feasibility of PiuA-PlyD4 fusion protein as a vaccine candidate by immunoinformatics prediction, but also evaluated its vaccine potential in vivo and in vitro experiments. The fusion protein PiuA-PlyD4 showed desirable biological safety, immunogenicity and conservation. It can stimulate the host to produce high levels of specific IgG antibodies, activates Th1 and Th2 pathways as well as stimulates Th17 cell immune pathways. Therefore, PiuA-PlyD4 can be considered a promising vaccine candidate against S. pneumoniae.
Ethics Approval
The studies involving animal participants were reviewed and approved by the Laboratory Animal Management and Ethics Committee of West China Second University Hospital, Sichuan University (No. 2023070). All animal procedures were conducted in accordance with the AVMA Guidelines for the Euthanasia of Animals (2020).
Acknowledgments
We thank Dr. Dong Deng and Dr. Xiang Wang from West China Second Hospital, Sichuan University for their technical suggestions.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China [No. 82300015], the Sichuan Science and Technology Program [No. 2022YFS0239 and No. 2023YFS0222], the Cadres Healthcare Research Projects in Sichuan Province [No. 2021-1703], the Science and Technology Innovation Project of Maternal and Child Medicine in Sichuan Province [No. 21FX025] and the West China Second University Hospital [No. KL066].
Disclosure
The authors report no conflicts of interest in this work.
References
1. O’Brien KL, Wolfson LJ, Watt JP, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374(9693):893–902. doi:10.1016/S0140-6736(09)61204-6
2. Williams AE, Jose RJ, Brown JS, Chambers RC. Enhanced inflammation in aged mice following infection with Streptococcus pneumoniae is associated with decreased IL-10 and augmented chemokine production. Am J Physiol Lung Cell Mol Physiol. 2015;308(6):L539–49. doi:10.1152/ajplung.00141.2014
3. van de Garde MDB, Knol MJ, Rots NY, van Baarle D, van Els C. Vaccines to protect older adults against pneumococcal disease. Interdiscip Top Gerontol Geriatr. 2020;43:113–130. doi:10.1159/000504490
4. Berical AC, Harris D, Dela Cruz CS, Possick JD. Pneumococcal vaccination strategies. An update and perspective. Ann Am Thorac Soc. 2016;13(6):933–944. doi:10.1513/AnnalsATS.201511-778FR
5. Lagousi T, Basdeki P, De Jonge MI, Spoulou V. Understanding host immune responses to pneumococcal proteins in the upper respiratory tract to develop serotype-independent pneumococcal vaccines. Expert Rev Vaccines. 2020;19(10):959–972. doi:10.1080/14760584.2020.1843433
6. Colombo MJ, Sun G, Alugupalli KR. T-cell-independent immune responses do not require CXC ligand 13-mediated B1 cell migration. Infect Immun. 2010;78(9):3950–3956. doi:10.1128/IAI.00371-10
7. Prinz DM, Smithson SL, Westerink MA. Two different methods result in the selection of peptides that induce a protective antibody response to Neisseria meningitidis serogroup C. J Immunol Methods. 2004;285(1):1–14. doi:10.1016/j.jim.2003.08.005
8. Geno KA, Gilbert GL, Song JY, et al. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev. 2015;28(3):871–899. doi:10.1128/Cmr.00024-15
9. Josefsberg JO, Buckland B. Vaccine process technology. Biotechnol Bioeng. 2012;109(6):1443–1460. doi:10.1002/bit.24493
10. Tai SS. Streptococcus pneumoniae protein vaccine candidates: properties, activities and animal studies. Crit Rev Microbiol. 2006;32(3):139–153. doi:10.1080/10408410600822942
11. Alghofaili F, Najmuldeen H, Kareem BO, et al. Host stress signals stimulate pneumococcal transition from colonization to dissemination into the lungs. mBio. 2021;12(6):e0256921. doi:10.1128/mBio.02569-21
12. Jomaa M, Yuste J, Paton JC, Jones C, Dougan G, Brown JS. Antibodies to the iron uptake ABC transporter lipoproteins PiaA and PiuA promote opsonophagocytosis of Streptococcus pneumoniae. Infect Immun. 2005;73(10):6852–6859. doi:10.1128/IAI.73.10.6852-6859.2005
13. Chiu FF, Leng CH, Ding YJ, et al. Domain 4 of pneumolysin from Streptococcus pneumoniae is a multifunctional domain contributing TLR4 activating and hemolytic activity. Biochem Biophys Res Commun. 2019;517(4):596–602. doi:10.1016/j.bbrc.2019.07.063
14. Cui Y, Miao C, Chen W, et al. Construction and protective efficacy of a novel Streptococcus pneumoniae fusion protein vaccine NanAT1-TufT1-PlyD4. Front Immunol. 2022;13:1043293. doi:10.3389/fimmu.2022.1043293
15. Liu Y, Wang H, Zhang S, et al. Mucosal immunization with recombinant fusion protein DnaJ-DeltaA146Ply enhances cross-protective immunity against Streptococcus pneumoniae infection in mice via interleukin 17A. Infect Immun. 2014;82(4):1666–1675. doi:10.1128/IAI.01391-13
16. Guo X, Sun Q, Xi H, et al. Expression, purification, and characterization of pneumococcal PsaA-PspA fusion protein. Protein Expr Purif. 2021;178:105782. doi:10.1016/j.pep.2020.105782
17. Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788. doi:10.1093/nar/gkg563
18. Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007;8:4. doi:10.1186/1471-2105-8-4
19. Dimitrov I, Flower DR, Doytchinova I. AllerTOP--a server for in silico prediction of allergens. BMC Bioinf. 2013;14(Suppl 6):S4. doi:10.1186/1471-2105-14-S6-S4
20. Sobolev OV, Afonine PV, Moriarty NW, et al. A global Ramachandran score identifies protein structures with unlikely stereochemistry. Structure. 2020;28(11):1249–1258. doi:10.1016/j.str.2020.08.005
21. Ponomarenko J, Bui HH, Li W, et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:514. doi:10.1186/1471-2105-9-514
22. Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48(W1):W449–W454. doi:10.1093/nar/gkaa379
23. Mukherjee S, Huda S, Sinha Babu SP. Toll-like receptor polymorphism in host immune response to infectious diseases: a review. Scand J Immunol. 2019;90(1):e12771. doi:10.1111/sji.12771
24. Mukherjee S, Karmakar S, Babu SP. TLR2 and TLR4 mediated host immune responses in major infectious diseases: a review. Braz J Infect Dis. 2016;20(2):193–204. doi:10.1016/j.bjid.2015.10.011
25. Yan Y, Tao H, He J, Huang SY. The HDOCK server for integrated protein-protein docking. Nat Protoc. 2020;15(5):1829–1852. doi:10.1038/s41596-020-0312-x
26. Kumari R, Kumar R. Open source drug discovery consortium, Lynn A. g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54(7):1951–1962. doi:10.1021/ci500020m
27. Mahmud S, Rafi MO, Paul GK, et al. Designing a multi-epitope vaccine candidate to combat MERS-CoV by employing an immunoinformatics approach. Sci Rep. 2021;11(1):15431. doi:10.1038/s41598-021-92176-1
28. Shafaghi M, Bahadori Z, Madanchi H, Ranjbar MM, Shabani AA, Mousavi SF. Immunoinformatics-aided design of a new multi-epitope vaccine adjuvanted with domain 4 of pneumolysin against Streptococcus pneumoniae strains. BMC Bioinf. 2023;24(1):67. doi:10.1186/s12859-023-05175-6
29. Gurung AB, Bhattacharjee A. Impact of a non-synonymous Q281R polymorphism on structure of human Lipoprotein-Associated Phospholipase A(2) (Lp-PLA(2)). J Cell Biochem. 2018;119(8):7009–7021. doi:10.1002/jcb.26909
30. Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–10. doi:10.1093/nar/gkm290
31. Messaoudi A, Belguith H, Ben Hamida J. Homology modeling and virtual screening approaches to identify potent inhibitors of VEB-1 beta-lactamase. Theor Biol Med Model. 2013;10:22. doi:10.1186/1742-4682-10-22
32. Yang Z, Bogdan P, Nazarian S. An in silico deep learning approach to multi-epitope vaccine design: a SARS-CoV-2 case study. Sci Rep. 2021;11(1):3238. doi:10.1038/s41598-021-81749-9
33. Yilmaz Colak C. Computational design of a multi-epitope vaccine against clostridium chauvoei: an immunoinformatics approach. Int J Pept Res Ther. 2021;27(4):2639–2649. doi:10.1007/s10989-021-10279-9
34. He Y, Li J, Mao W, et al. HLA common and well-documented alleles in China. HLA. 2018;92(4):199–205. doi:10.1111/tan.13358
35. Yan Y, Wen Z, Wang X, Huang SY. Addressing recent docking challenges: a hybrid strategy to integrate template-based and free protein-protein docking. Proteins. 2017;85(3):497–512. doi:10.1002/prot.25234
36. Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 2018;16(6):355–367. doi:10.1038/s41579-018-0001-8
37. Whalan RH, Funnell SG, Bowler LD, Hudson MJ, Robinson A, Dowson CG. PiuA and PiaA, iron uptake lipoproteins of Streptococcus pneumoniae, elicit serotype independent antibody responses following human pneumococcal septicaemia. FEMS Immunol Med Microbiol. 2005;43(1):73–80. doi:10.1016/j.femsim.2004.07.010
38. Zhang Y, Edmonds KA, Raines DJ, et al. The pneumococcal iron uptake protein a (PiuA) specifically recognizes Tetradentate Fe(III)bis- and mono-catechol complexes. J Mol Biol. 2020;432(19):5390–5410. doi:10.1016/j.jmb.2020.08.005
39. Dorosti H, Eslami M, Negahdaripour M, et al. Vaccinomics approach for developing multi-epitope peptide pneumococcal vaccine. J Biomol Struct Dyn. 2019;37(13):3524–3535. doi:10.1080/07391102.2018.1519460
40. Matthias KA, Roche AM, Standish AJ, Shchepetov M, Weiser JN. Neutrophil-toxin interactions promote antigen delivery and mucosal clearance of Streptococcus pneumoniae. J Immunol. 2008;180(9):6246–6254. doi:10.4049/jimmunol.180.9.6246
41. Chen X, Zaro JL, Shen WC. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev. 2013;65(10):1357–1369. doi:10.1016/j.addr.2012.09.039
42. van de Garde MDB, van Westen E, Poelen MCM, Rots NY, van Els C. Prediction and validation of immunogenic domains of pneumococcal proteins recognized by human CD4(+) T cells. Infect Immun. 2019;87(6):e00098–19. doi:10.1128/IAI.00098-19
43. Das NC, Sen Gupta PS, Biswal S, Patra R, Rana MK, Mukherjee S. In-silico evidences on filarial cystatin as a putative ligand of human TLR4. J Biomol Struct Dyn. 2022;40(19):8808–8824. doi:10.1080/07391102.2021.1918252
44. Mukherjee S, Mukherjee S, Maiti TK, Bhattacharya S, Sinha Babu SP. A novel ligand of toll-like receptor 4 from the sheath of Wuchereria bancrofti microfilaria induces proinflammatory response in macrophages. J Infect Dis. 2017;215(6):954–965. doi:10.1093/infdis/jix067
45. Choudhury A, Sen Gupta PS, Panda SK, Rana MK, Mukherjee S. Designing AbhiSCoVac - a single potential vaccine for all ‘Corona culprits’: immunoinformatics and immune simulation approaches. J Mol Liq. 2022;351:118633. doi:10.1016/j.molliq.2022.118633
46. Das NC, Gupta PSS, Panda SK, Rana MK, Mukherjee S. Reverse vaccinology assisted design of a novel multi-epitope vaccine to target Wuchereria bancrofti cystatin: an immunoinformatics approach. Int Immunopharmacol. 2023;115:109639. doi:10.1016/j.intimp.2022.109639
47. Usmani SS, Kumar R, Bhalla S, Kumar V, Raghava GPS. In silico tools and databases for designing peptide-based vaccine and drugs. Adv Protein Chem Struct Biol. 2018;112:221–263. doi:10.1016/bs.apcsb.2018.01.006
48. Yu J, Li B, Chen X, et al. Comparison of immunogenicity and protection of two pneumococcal protein vaccines based on PsaA and PspA. Infect Immun. 2018;86(6):e00916–17. doi:10.1128/IAI.00916-17
49. Gil E, Noursadeghi M, Brown JS. Streptococcus pneumoniae interactions with the complement system. Front Cell Infect Microbiol. 2022;12:929483. doi:10.3389/fcimb.2022.929483
50. Chen X, Li B, Yu J, et al. Comparison of four adjuvants revealed the strongest protection against lethal pneumococcal challenge following immunization with PsaA-PspA fusion protein and AS02 as adjuvant. Med Microbiol Immunol. 2019;208(2):215–226. doi:10.1007/s00430-019-00579-9
51. Bogaert D, De Groot R, Hermans PW. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4(3):144–154. doi:10.1016/S1473-3099(04)00938-7
52. Casadevall A. Antibody-based vaccine strategies against intracellular pathogens. Curr Opin Immunol. 2018;53:74–80. doi:10.1016/j.coi.2018.04.011
53. Platt HL, Cardona JF, Haranaka M, et al. A Phase 3 trial of safety, tolerability, and immunogenicity of V114, 15-valent pneumococcal conjugate vaccine, compared with 13-valent pneumococcal conjugate vaccine in adults 50 years of age and older (PNEU-AGE). Vaccine. 2022;40(1):162–172. doi:10.1016/j.vaccine.2021.08.049
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.