Back to Journals » Infection and Drug Resistance » Volume 16
Identification of Two Novel Carbapenemase-Encoding Hybrid Plasmids Harboring blaNDM-5 and blaKPC-2 in a Clinical ST11-KL47 Klebsiella pneumoniae
Authors Xiao W, Wang X , Qu Y, Sun M, Chang Y, Li W, Shen Y , Shi X, Jing M, Xu Q
Received 16 February 2023
Accepted for publication 16 June 2023
Published 24 June 2023 Volume 2023:16 Pages 4073—4081
DOI https://doi.org/10.2147/IDR.S408824
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Weiqiang Xiao,1,* Xiaokun Wang,1,* Yuanye Qu,1 Mingyue Sun,1 Yanmin Chang,1 Wenjiao Li,1 Yong Shen,1,2 Xiufang Shi,3 Min Jing,1 Qingxia Xu1,4
1Department of Clinical Laboratory, Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou, People’s Republic of China; 2Institute of Biophysics, Chinese Academy of Sciences, Foshan, People’s Republic of China; 3School of Pharmaceutical Sciences, Zhengzhou University, Zhengzhou, People’s Republic of China; 4Zhengzhou Key Laboratory for Diagnosis of Digestive System Tumor Markers, Zhengzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qingxia Xu; Xiufang Shi, Email [email protected]; [email protected]
Background: Emergence of blaKPC and blaNDM co-harboring Klebsiella pneumoniae has escalated the threat of Carbapenem-resistant Klebsiella pneumoniae (CRKP) to healthcare. It remains unknown the prevalence and molecular characteristics of CRKP co-producing KPC and NDMs carbapenemases in Henan.
Methods and Results: Twenty-seven CRKP strains isolated from different times were selected randomly in affiliated cancer hospital of Zhengzhou University from January 2019 to January 2021, among which one KPC-2 and NDM-5 positive CRKP named K9 was isolated from an abdominal pus sample of a 63-year-old male patient with leukemia. Sequencing of K9 determined that K9 belonged to ST11-KL47, which is resistant to antibiotics such as meropenem, ceftazidime-avibactam and tetracycline. K9 carried two different plasmids that contained blaNDM− 5 and blaKPC− 2. Both plasmids were shown to be novel hybrid plasmids and IS 26 played an important role in generation of two plasmids. Gene blaKPC-2 was flanked by the NTEKPC-Ib-like genetic structure (IS 26-ΔTn 3-ISKpn8-blaKPC-2-ISKpn6-IS 26) and was located on a conjugative IncFII/R/N type hybrid plasmid.
Conclusion: The resistance gene blaNDM− 5 located on a region organized as IS 26-blaNDM– 5-ble-trpF-dsbD-ISCR1-sul1-aadA2-dfrA12-IntI1-IS 26 was carried by a phage-plasmid. We described a clinical CRKP co-producing KPC-2 and NDM-5 and emphasized an urgent need to control their further spread.
Keywords: carbapenem-resistant Klebsiella pneumoniae, KPC-2, NDM-5, beta-lactamases, hybrid plasmid, IS 26
Introduction
Carbapenem-resistant Klebsiella pneumoniae (CRKP) poses a serious threat to human health,1,2 which can cause serious infections in debilitated or immunocompromised patients, leading to prolonged hospital stay and increased mortality rates. According to the data from the China Antimicrobial Resistance Surveillance System (CARSS), the national average isolation rate of CRKP is increasing rapidly from 6.4% in 2014 to 10.9% in 2020. Notably, an extremely high incidence of CRKP was observed in Henan province in 2020 (30.2%), ranking first in China (http://www.carss.cn/Report/Details?aId=808). The main resistance mechanism of CRKP is the production of carbapenemases.3 K. pneumoniae carbapenemases (KPC) and New Delhi β-lactamases (NDMs) are the two common types of carbapenemases produced by CRKP, and KPC-2 is the most widely distributed KPC variant which is able to hydrolyze all β-lactam antibiotics but can be inhibited by avibactam, a new approved diazabicyclooctane non-β-lactam β-lactamase inhibitor for use in combination with ceftazidime.4 However, as metallo-β-lactamase belonged to class B1 β-lactamases, NDMs cannot be inhibited by avibactam. It is worth noting that the recent emergence of CRKP co-producing KPC-2 and NDMs in different regions of China will seriously threaten the application of regimen of ceftazidime/avibactam in clinical treatment.5–8 As the province with the highest isolation rate of CRKP in China, the prevalence and molecular characteristics of CRKP co-producing KPC and NDMs carbapenemases in Henan are still largely unknown.
Herein, we genetically characterized a KPC-2 and NDM-5 positive CRKP named K9 in Henan province. Unlike previously reported NDMs and KPC-2 co-producing CRKP strains that carried blaNDMs-harboring plasmids belonged to IncX3, N2, HI1B types and blaKPC-2-harboring plasmids mainly belonged to IncFII type,5–7 the blaNDM-5 and blaKPC-2 gene in K9 were carried by an untypeable phage-plasmid and an IncFII/R/N hybrid plasmid, respectively, indicating a different evaluation pathway in the generation of K9 co-producing two different types of carbapenemases.
Materials and Methods
Bacterial Isolates, Identification, and Antimicrobial Susceptibility Testing
A total of 27 non-repeating CRKP strains were recovered from clinical specimens in the Affiliated Cancer Hospital of Zhengzhou University from January 2019 to January 2021. All isolates were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Auto Bioengineering Co., Ltd. Zhengzhou, China) and the BD Phoenix™ M-50 instrument (BD Diagnostic Systems, Sparks, MD, USA). Minimal inhibitory concentrations (MICs) were initially determined by broth microdilution method using a BD Phoenix™ M-50 instrument, and MICs of imipenem, meropenem, ciprofloxacin, levofloxacin and tigecycline were validated using E-test strips (Auto Bioengineering Co., Ltd. Zhengzhou, China). The tigecycline breakpoint was determined using the FDA standard (susceptible, ≤2 mg/L; intermediate, 4 mg/L; resistant, ≥8 mg/L), the polymyxin breakpoint using the EU 2021 drug sensitivity test standard (https://www.eucast.org), and the remaining breakpoints were set according to the CLSI M100.9 Escherichia coli ATCC25922 was served as a quality control strain in the susceptibility testing assay.
Beta-Lactamase Phenotype and Carbapenemase Genes
Enzyme inhibitor enhancement tests were performed to confirm carbapenemase phenotype.10 This method was performed following the standard diffusion method.10 On Mueller–Hinton agar, one disc of meropenem alone and three discs of meropenem containing 400 µg of PBA, 292 µg of EDTA or both were used to differentiate class A and class B carbapenemase. The results were interpreted as previously described.10 Enzyme immunochromatographic assay (NG-Test® CARBA 5 kit, Fosun Long March Medical Science Co., Ltd., Shanghai, China) was used to detect NDM, KPC, VIM, IMP, or OXA-48 carbapenemases. NDM and KPC variants and other beta-lactamase were confirmed by PCR with primers in Table S1 and the related literature. PCR amplification products were sequenced on both strands (BGI, Shenzhen, China) and were compared using BLAST (https://blast.ncbi.nlm.nih.gov/Blast).
Plasmid Conjugation Experiments
Conjugation assay was conducted to evaluate the transferability of plasmids by using the method described previously.11 CRKP K9, coproducing KPC-2 and NDM-5, served as the donor strain and E. coli J53 was used as the recipient strain. The donor and recipient were mixed in a ratio of 10:1 and incubated statically in an LB broth at 35°C for 24 h. Transconjugants were selected by LB agar plates supplemented with 2 μg/mL meropenem, 100 μg/mL sodium azide and (or not) 76 μg/mL sulfamethoxazole. The presence of the blaKPC-2 or blaNDM-5 genes was identified by PCR amplification and the forms of plasmid DNA contained by the donor and recipient strains were analyzed by agarose gel electrophoresis (0.5% agarose in Tris-acetate EDTA buffer).
WGS and Gene Environment Analysis
The strain genome was sequenced using a PacBio RS II platform and Illumina HiSeq 4000 platform at the Beijing Genomics Institute (BGI, Shenzhen, China). Four SMRT cells (Zero-ModeWaveguide arrays of sequencing) were used by the PacBio platform to generate the long-subreads set. Sequencing data were assembled with FalconV0.3.0 and Proovread 2.12 software, quality control was performed with FastQC0.11.9 software, and the assembled data were submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/) to obtain sequence numbers and annotation information. MLST typing, drug resistance genes, and plasmid typing were obtained by searching against the database BacWGSTdb (http://bacdb.cn/BacWGSTdb/index.php). The MLST typing and serotype prediction were also confirmed by the Kleborate tool (https://github.com/katholt/Kleborate).12 Easyfig 2.2.3 software was used to align genomic and open reading frame sequences,13 and the BRIG v0.95 software was used to generate plasmid circle figures.14
Results and Discussion
Identification of a KPC-2 and NDM-5 Co-Producing Strain K9 Among 27 CRKP Isolates
Susceptibility testing showed that almost all strains were resistant to β-lactam antimicrobial agents including imipenem and aztreonam, but remained susceptible to polymyxin (Table 1). Furthermore, the enzyme inhibitor enhancement experiment indicated that all the strains produced class A carbapenemases (Table 1 and Figure S1). Notably, only one KPC and NDM co-producing K. pneumoniae isolate, namely K9, was selected according to the enzyme immunochromatographic assay results. This isolate was recovered from an abdominal pus sample of a 63-year-old male patient in the hematological malignancies of Affiliated Cancer Hospital of Zhengzhou University in 2021 (Table 1).
 |
Table 1 Clinical Information of 27 CRKP Strains Along with Partial Susceptibility Profiles and Carbapenemase Phenotype and Genotype |
According to the WGS result, K9 was assigned to ST11 and KL47 based on the analysis of software Kleborate, and found to harbor one chromosome of 5,531,197 bp (CP074116) and three plasmids namely pKPN-hnqyy-ndm (CP074117), pKPN-hnqyy-kpc (CP074118), and pKPN-hnqyy-3 (CP074119) of 123,557 bp, 116,047 bp, and 10,060 bp, respectively. Associated with the multidrug-resistant (MDR) phenotype, a total of thirteen resistance determinants were identified in strain K9 (Table 2). These chromosomal and plasmid-carried determinants conferred resistance to β-lactams (blaSHV-182, blaNDM-5, blaCTX-M-65 and blaKPC-2), aminoglycosides (aadA2), quinolones (oqxAB), fosfomycin (fosA) and trimethoprim-sulfamethoxazole (qacE, sul1 and dfrA12). The blaKPC-2 and blaNDM-5 genes were found to be present on two distinct plasmids (Table 2). Virulome analysis showed that strain K9 carried a large number of virulence-associated factors, such as type 1 and type 3 fimbriae, capsule, iron uptake (Ent siderophore and yersiniabactin), type 6 secretion systems (T6SS-I, T6SS-II, T6SS-III) and lipopolysaccharide (LPS) biosynthetic locus (rfb) located on the chromosome. While, no typical pLVPK-like virulence plasmid carrying rmpA/A2 or iuc locus was detected in K9.15 Conjugative assay revealed that only the blaKPC-2 positive plasmid pKPN-hnqyy-kpc was successfully transferred to E. coli J53 at a frequency of 5.7×10−1 per donor cell (Figure S2).
 |
Table 2 Resistome of K. pneumoniae K9 |
Characterization of the Hybrid IncFII/R/N Type blaKPC-2-Bearing Plasmid
Plasmid pKPN-hnqyy-kpc has an average GC content of 52.94% and harbors 144 predicted open reading frames (ORFs). It is a multi-replicon plasmid possessing three plasmid replicons including IncFII, IncR, and IncN. A BLASTN search of this hybrid plasmid against the NCBI nucleotide database revealed that pKPN-hnqyy-kpc displayed 60% query coverage and 100% nucleotide identity to plasmid p69-2 (CP025458) from K. pneumoniae, 54% query coverage and 99.99% nucleotide identity to plasmid pJX2-2 (CP064248) from K. pneumoniae and 39% query coverage and 99.63% nucleotide identity to plasmid pL22-2 (CP031259) from K. pneumoniae (Figure 1). In plasmid pKPN-hnqyy-kpc, a total of 8 copies of IS26 were dispersed among the p69-2-like region, and the observation of an 8 bp target duplication repeats (TAGGGGAA) flanked p69-2-like region and pL22-2-like region indicated that this plasmid might be formed by the fusion of plasmids pL22-2 (IncN) and p69-2 (IncFII-IncR) mediated by IS26 elements. This observation provided more direct evidence for the important role of IS26 in the formation of hybrid blaKPC-bearing plasmids. It has been documented that the conjugative IncFII/R type plasmids were one of the key vectors mediating the transmission of blaKPC.15 However, IncFII/R/N type blaKPC-bearing plasmids were rarely reported to cointegrate with IncN type plasmid. Genetic comparative analysis between pKPN-hnqyy-kpc and a reported IncFII/R/N type blaKPC-bearing plasmid, namely pKPC-TVGHCRE225 (CP023725), showed only 62% query coverage and 99.75% nucleotide identity, and the blaCTX-M-65 gene and a gene cluster of type IV secretion system were absent in pKPC-TVGHCRE225, indicating the formation of the two plasmids might be underwent different evolutionary processes.
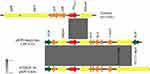
Tn3-based transposon-Tn4401 and non-Tn4401 elements (NTEKPC) were regarded as two most common blaKPC-containing mobile elements. Unlike Tn4401 structure that was mainly detected in KPC-producing strains from France, Greece and America, NTEKPC elements were relatively prevalent among clinical strains in China, Singapore, and Brazil.16 NTEKPCs were divided into three groups (NTEKPC-I, NTEKPC-II, and NTEKPC-III) based on the absence or presence of blaTEM, and NTEKPC-I and NTEKPC-II might have been evolved from Tn4401 by genetic recombination.15 In plasmid pKPN-hnqyy-kpc, a 10,215-bp non-Tn4401 element highly homologous to the NTEKPC-Ib in plasmid pKPC-LKEc (KC788405),17 with the structure of IS26-ΔTn3-ISKpn8-blaKPC-2-ISKpn6-IS26 (Figure 2) was located downstream of gene encoding DEAD/DEAH box helicase. Compared with NTEKPC-Ib in pKPC-LKEc, the tnpA of Tn1721 was truncated by IS26 in pKPN-hnqyy-kpc. The observations that IS26 was involved in the formation of recombinant plasmid and insertion in NTEKPC-Ib element indicating IS26 promoted the evolution and variation of plasmid and resistance transmission unit, which needs to pay more attention.18
Characterization of a Novel Hybrid blaNDM-5-Carrying Plasmid
Plasmid pKPN-hnqyy-ndm was 123,557 bp in length, which belonged to an untypable group by BIGSdb (GC content of 52.83%) (http://bigsdb.readthedocs.io) and harbored 126 predicted open reading frames. Unlike well-reported IncX3 and IncFII type blaNDM-carrying plasmids, this untypable plasmid only shared 34% query coverage and 99.49% nucleotide identity with IncFII plasmid pYJ6-NDM5 (AP023236) from an E. coli strain (Figure 3). In addition, plasmid pKPN-hnqyy-ndm contained an 80 kb fragment originated from typical phage-like plasmids, which shared 61% query coverage and 98.97% nucleotide identity with Klebsiella phage 020009 (CP038007) and 64% query coverage and 99.26% nucleotide identity with plasmid pJX2-3 (CP064249) from a carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) strain. Low homology to reported blaNDM-positive plasmids and characterization of phage-derived fragment suggested that pKPN-hnqyy-ndm was a novel cointegrated plasmid.
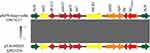
Similar to the hybrid plasmid pKPN-hnqyy-kpc, pKPN-hnqyy-ndm could be divided into two genetically and physically distinct modules: a ~43-kb composite transposon carrying resistance determinants (blaNDM-5, aadA2, dfrA12, sul, and qacE) and a ~80-kb skeleton region essentially homologous to phage-like plasmids. The ~43 kb pYJ6-NDM5-like plasmid-derived module which was flanked by two IS26 elements in opposite orientation contained genes responsible for plasmid maintenance and stability including parM (plasmid partition), sok-hok (postsegregation killing), and psiAB (plasmid SOS inhibition protein), a tra region and a multidrug resistance region organized as IS26-blaNDM–5-ble-trpF-dsbD-ISCR1-sul1-aadA2-dfrA12-IntI1-IS26 (Figures 3 and 4). The ~80-kb phage-derived module contained genes encoding phage associated proteins including phage tail assembly protein JIKLM, head-tail adaptor proteins, phage major capsid protein, phage portal protein, and phage DNA packaging protein A.
Phages have been reported to be involved in the evolution of plasmids and mediate their spread.19 Phage-plasmids (PPs), which are known to be both plasmids and phages, have been reported to promote the spread of antibiotic resistance genes (ARGs) by infection and lysogenic conversion.19,20 A bioinformatics analysis showed that PPs are mainly prevalent in Escherichia and Klebsiella species, and β-lactamase encoding genes such as blaCTX-M-55, blaOXA-48, blaTEM, and blaNDM carried by PPs were only detected in E. coli strains. These PPs are mainly belonging to Phage P1-like and Phage P7-like types.20 Notably, the phage-plasmid pKPN-hnqyy-ndm in our study with a novel phage backbone showed very low homology with those reported PPs. To the best of our knowledge, this is the first report on clinical K. pneumoniae isolate carrying blaNDM-5-positive phage-plasmid. While, whether plasmid pKPN-hnqyy-ndm has the ability to lytic capacities or transfer blaNDM requires further investigation.
CRKP isolates co-producing KPC and NDM have been reported in different countries such as India, Pakistan, and China.5–8,21,22 In these isolates, the blaKPC-2 gene was mainly carried by IncFII, IncFII/IncR and IncFIA/IncFII type plasmids, with sizes ranging from 97-kb to 175-kb. While, the blaNDM gene was mainly detected in IncX3, IncN, and IncHI1B type plasmids. It is worth noting that two recent reports revealed either blaNDM or blaKPC gene was identified in cointegrated plasmids in KPC and NDM co-producing CRKP.5,22 Moreover, our study found that both blaNDM and blaKPC carrying plasmids were hybrid plasmids, indicating further evolution of these prevalent plasmids with single replicon. IS26 which has been reported to contribute to the generation of hybrid plasmid co-harboring blaKPC-2 and virulence factors might play an important role in generation of blaNDM, blaKPC-bearing MDR cointegrate plasmids in K9. Based on the unique structure of the two carbapenemase-encoding hybrid plasmids, we speculated the evolutionary paths of NDM and KPC plasmids in K9 were differed significantly from those previously reported.
Conclusion
Here, we describe the genetic characteristics of CRKP strain K9 co-harbouring blaNDM-5 and blaKPC-2 with capsular serotype KL47 belonging to ST11. The strain carried multiple resistance and virulence genes. The blaKPC-2 gene located on a hybrid plasmid and was present in a NTEKPC-Ib-like structure. The blaNDM-5 gene was located on the non-conjugative plasmid-phage complexes and was present in a tandem resistance gene cassette with the IS26 transposable element at both ends. Notably, IS26 played an important role in generation of blaKPC-2-harboring plasmid and blaNDM-5-harboring plasmid. And we provided a direct evidence in the formation of the blaKPC-2-harboring IncFII/R/N plasmids. The KPC-NDM co-producing CRKP as an emerging and growing threat in the hospital and the community which should be further monitored.
Data Sharing Statement
The datasets presented in this study can be found in online repositories. The complete chromosome genome sequence of CRKP strains K9 was deposited in GenBank with accession numbers CP074116. The complete sequences of blaNDM-5-harboring plasmid and blaKPC-2-harboring plasmid were submitted to GenBank under accession numbers CP074117 and CP074118, respectively.
Ethics Statement
The study protocol was approved by the Ethics Committee of Henan Cancer Hospital (NO:2022-KY-0036). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Funding
This study was supported by the Joint Construction Project of the Henan Provincial Health Commission (Grant No. LHGJ20210218, LHGJ20220209, SBGJ202102070).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Shrivastava SR, Shrivastava PS, Ramasam JJ. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J Med Soc. 2018;32(1):76. doi:10.4103/jms.jms_25_17
2. Yang Y, Yang Y, Chen G, et al. Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg Microbes Infect. 2021;10(1):1–42. doi:10.1080/22221751.2020.1868951
3. Leon-Sampedro R, DelaFuente J, Diaz-Agero C, et al. Pervasive transmission of a carbapenem resistance plasmid in the gut microbiota of hospitalized patients. Nat Microbiol. 2021;6(5):606–616. doi:10.1038/s41564-021-00879-y
4. Bush K, Bradford PA. Epidemiology of beta-lactamase-producing pathogens. Clin Microbiol Rev. 2020;33(2). doi:10.1128/CMR.00047-19
5. Gao H, Liu Y, Wang R, Wang Q, Jin L, Wang H. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine. 2020;51:102599. doi:10.1016/j.ebiom.2019.102599
6. Hao J, Zhang B, Deng J, Wei Y, Xiao X, Liu J. Emergence of a hypervirulent tigecycline-resistant Klebsiella pneumoniae Strain Co-producing bla NDM-1 and bla KPC-2 With an Uncommon Sequence Type ST464 in Southwestern China. Front Microbiol. 2022;13:868705. doi:10.3389/fmicb.2022.868705
7. Zou H, Jia X, Liu H, Li S, Wu X, Huang S. Emergence of NDM-5-producing Escherichia coli in a Teaching Hospital in Chongqing, China: incF-Type plasmids may contribute to the prevalence of bla NDM- 5. Front Microbiol. 2020;11:334. doi:10.3389/fmicb.2020.00334
8. Zheng B, Xu H, Yu X, et al. Identification and genomic characterization of a KPC-2-, NDM-1- and NDM-5-producing Klebsiella michiganensis isolate. J Antimicrob Chemother. 2018;73(2):536–538. doi:10.1093/jac/dkx415
9. Clinical and Laboratory Standards Institute. CLSI Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. CLSI Document M100–Ed31. Wayne, PA: Clinical and Laboratory Standards Institute; 2021.
10. Liao Q, Yuan Y, Li Q, et al. Comparing three different phenotypic methods for accurate detection of carbapenemase-producing Enterobacterales. J Infect Chemother. 2021;27(6):794–799. doi:10.1016/j.jiac.2021.01.003
11. Dong H, Li Y, Cheng J, et al. Genomic epidemiology insights on NDM-producing pathogens revealed the pivotal role of plasmids on blaNDM transmission. Microbiol Spectr. 2022;10(2):e0215621. doi:10.1128/spectrum.02156-21
12. Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12(1):4188. doi:10.1038/s41467-021-24448-3
13. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. doi:10.1093/bioinformatics/btr039
14. Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12(1):1–10. doi:10.1186/1471-2164-12-402
15. Yang X, Dong N, Chan EW, Zhang R, Chen S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 2021;29(1):65–83. doi:10.1016/j.tim.2020.04.012
16. Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686–696. doi:10.1016/j.tim.2014.09.003
17. Huang Q-S, Liao W, Xiong Z, et al. Prevalence of the NTEKPC-I on IncF plasmids among Hypervirulent Klebsiella pneumoniae isolates in Jiangxi Province, South China. Front Microbiol. 2021;12:1338.
18. He S, Hickman AB, Varani AM, et al. Insertion Sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio. 2015;6(3):e00762. doi:10.1128/mBio.00762-15
19. Pfeifer E, Bonnin RA, Rocha EPC, Goldman GH. Phage-plasmids spread antibiotic resistance genes through infection and lysogenic conversion. mBio. 2022;13(5):e0185122. doi:10.1128/mbio.01851-22
20. Wang M, Jiang L, Wei J, et al. Similarities of P1-like phage plasmids and their role in the dissemination of bla(CTX-M-55). Microbiol Spectr. 2022;10(5):e0141022. doi:10.1128/spectrum.01410-22
21. Wei DD, Wan LG, Liu Y. Draft genome sequence of an NDM-1- and KPC-2-coproducing hypervirulent carbapenem-resistant Klebsiella pneumoniae strain isolated from burn wound infections. Genome Announc. 2018;6(13). doi:10.1128/genomeA.00192-18
22. Hu R, Li Q, Zhang F, Ding M, Liu J, Zhou Y. Characterisation of blaNDM-5 and blaKPC-2 co-occurrence in K64-ST11 carbapenem-resistant Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;27:63–66. doi:10.1016/j.jgar.2021.08.009
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.