Back to Journals » Journal of Inflammation Research » Volume 16
Identification of Shared Immune Cells and Immune-Related Co-Disease Genes in Chronic Heart Failure and Systemic Lupus Erythematosus Based on Transcriptome Sequencing
Authors Luo Z , Lu G , Yang Q , Ding J, Wang T, Hu P
Received 24 May 2023
Accepted for publication 22 June 2023
Published 30 June 2023 Volume 2023:16 Pages 2689—2705
DOI https://doi.org/10.2147/JIR.S418598
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Ziyue Luo,1,* Guifang Lu,2,* Qiang Yang,1 Juncan Ding,1 Tianyu Wang,1 Pengfei Hu3
1Second Clinical Medical College, Zhejiang Chinese Medical University, Hangzhou, Zhejiang Province, 310053, People’s Republic of China; 2Department of Rheumatology, The Second Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou, Zhejiang Province, 310005, People’s Republic of China; 3Department of Cardiology, The Second Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou, Zhejiang Province, 310005, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Pengfei Hu, Department of Cardiology, The Second Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou, Zhejiang Province, 310005, People’s Republic of China, Tel +86 15267037741, Email [email protected]
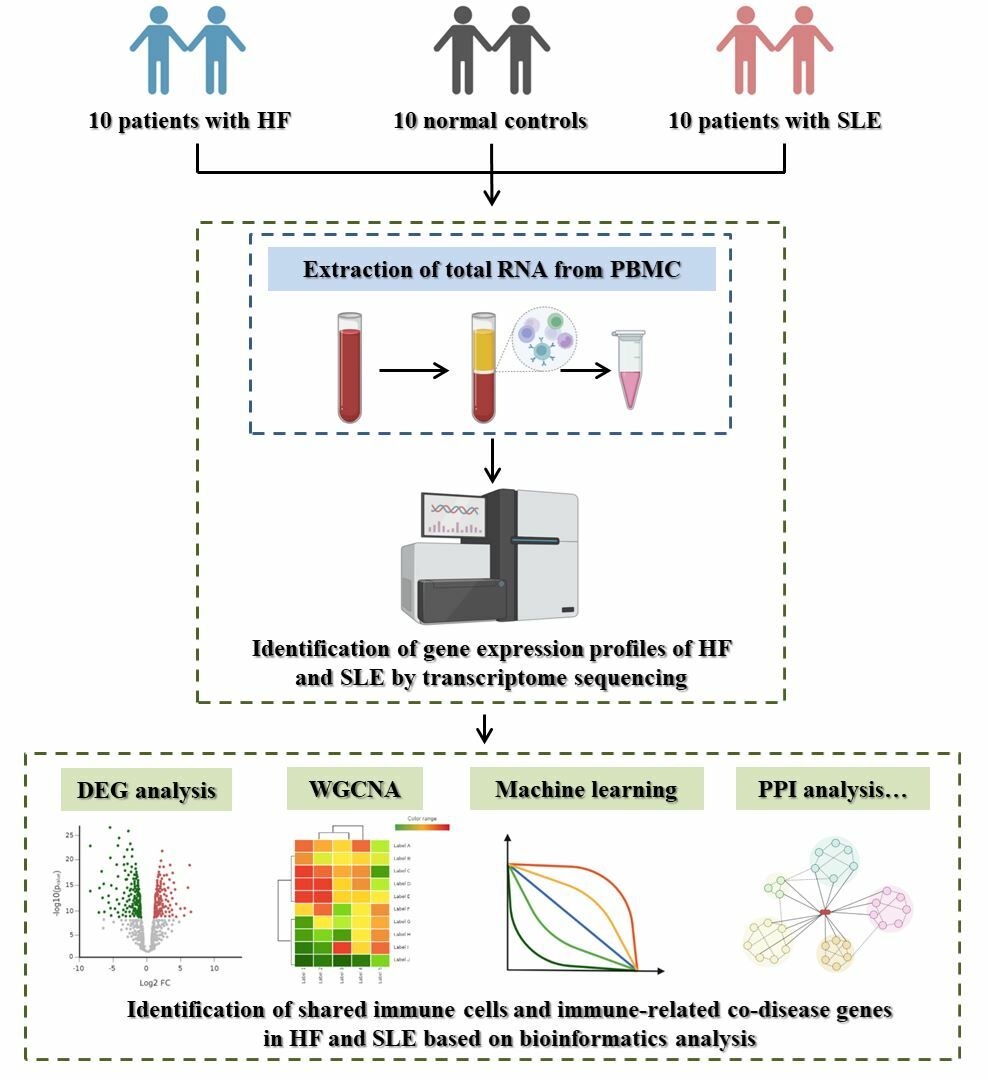
Purpose: The purpose was to identify shared immune cells and co-disease genes in chronic heart failure (HF) and systemic lupus erythematosus (SLE), as well as explore the potential mechanisms of action between HF and SLE.
Methods: A collection of peripheral blood mononuclear cells (PBMCs) from ten patients with HF and SLE and ten normal controls (NC) was used for transcriptome sequencing. Differentially expressed genes (DEGs) analysis, enrichment analysis, immune infiltration analysis, weighted gene co-expression network analysis (WGCNA), protein-protein interaction (PPI) analysis, and machine learning were applied for the screening of shared immune cells and co-disease genes in HF and SLE. Gene expression analysis and correlation analysis were used to explore the potential mechanisms of co-disease genes and immune cells in HF and SLE.
Results: In this study, it was found that two immune cells, T cells CD4 naïve and Monocytes, displayed similar expression patterns in HF and SLE at the same time. By taking intersection of the above immune cell-associated genes with the DEGs common to both HF and SLE, four immune-associated co-disease genes, CCR7, RNASE2, RNASE3 and CXCL10, were finally identified. CCR7, as one of the four key genes, was significantly down-regulated in HF and SLE, while the rest three key genes were all significantly up-regulated in both diseases.
Conclusion: T cells CD4 naïve and Monocytes were first revealed as possible shared immune cells of HF and SLE, and CCR7, RNASE2, RNASE3 and CXCL10 were identified as possible key genes common to HF and SLE as well as potential biomarkers or therapeutic targets for HF and SLE.
Keywords: heart failure, systemic lupus erythematosus, immunology, transcriptome sequencing, machine learning
Graphical Abstract:
Introduction
Heart failure (HF) is a pathological condition of myocardial injury resulting from various etiologies, such as myocardial infarction, cardiomyopathy, haemodynamic overload, and inflammation, which ultimately results in ventricular pumping or filling hypoperfusion, generally representing the end-stage of cardiac disease.1 Chronic HF is a persistent state of HF and is characterized by symptoms of jugular venous filling, peripheral oedema, and dyspnoea.2 Despite improvements in the treatment of cardiovascular diseases, the prevalence and mortality of HF still remains high, causing it becoming one of the most serious public health problems.3
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease, characterized by the main symptoms of butterfly rash, arthralgia and photosensitivity.4 SLE can involve multiple systems and organs, including the cardiovascular, neurological, urological and haematological systems, and may progress to moderate to severe multi-organ damage, resulting in a poor prognosis.5 Despite, numerous studies having been conducted, the pathogenesis of SLE remain poorly understood, with genetic, endocrine, infectious, immune abnormalities and some environmental factors being implicated. As such, SLE have become a huge burden on public health.6,7
Recent studies have investigated the potential relationship between cardiovascular diseases, specifically HF and SLE.8 Evidence suggests that SLE patients may have a higher burden of cardiovascular risk factors, including an increased risk of HF, as compared with the general population.9 In addition, HF patients with concomitant SLE have a higher mortality rate compared to those without SLE.10 These findings suggest a possible link between HF and SLE, as supported by recent research.
Recent advancements in next-generation sequencing technologies and bioinformatics have allowed for the identification of potential key genes and targets for diseases. Transcriptome sequencing data is increasingly being utilized to investigate potential therapeutic targets and their potential mechanism of action in multiple diseases simultaneously.11–13 Although studies have explored the gene targets in HF and SLE individually, there remains a lack of research on the common targets and mechanisms of action of both diseases.14,15 Therefore, identifying effective therapeutic pathways and understanding the potential mechanisms of action for HF and SLE is crucial in combating the great threat they pose to human health. This study aims to identify shared immune cells and co-disease genes and to explore potential mechanisms of action for HF and SLE through transcriptome sequencing and bioinformatics analysis.
Methods
General Information on the Study Population
This study included patients who were treated at the Second Affiliated Hospital of Zhejiang Chinese Medical University from October 2022 to January 2023, based on the established inclusion criteria. A total of 10 patients with HF, 10 patients with SLE and 10 normal controls (NC) were included in this study.
Diagnostic Criteria
HF: Patients with chronic HF were diagnosed according to the 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines.16
SLE: Patients with SLE were diagnosed according to the 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus.17
Inclusion Criteria
Patients aged ≥ 18 years; with HF, SLE and other non-acute phase co-morbidities; they themselves or their family members were willing to participate in the study and agreed to sign an informed consent form.
Exclusion Criteria
Patients with comorbid psychiatric or oncological diseases or in the acute phase of various diseases; patients with critical or even life-threatening conditions such as cardiogenic shock, malignant arrhythmia, coagulation dysfunction, and serious infections; patients with other rheumatic diseases such as rheumatoid arthritis, dry syndrome, and gout; patients have participated in other clinical trials within the last 1 month; patients with impaired consciousness or lacking civil capacity.
Extraction of Total RNA from Peripheral Blood Mononuclear Cells (PBMCs)
Peripheral blood was collected from the peripheral veins of 30 cases. 10 mL of peripheral blood was collected in EDTA anticoagulated tubes and PBMCs were extracted within 2 hours of collection at room temperature. The anticoagulated whole blood diluted with an equal volume of PBS was slowly added to centrifuge tubes prepped with lymphocyte separation solution, 5 mL per tube. After centrifugation at 2500 rpm for 25 minutes at room temperature, a white film layer between the plasma layer and the lymphocyte isolate layer, known as the PBMC layer, was carefully aspirated with a pipette and placed in a clean centrifuge tube. Then, 1 × PBS was added to a full tube and the cells were centrifuged at 1500 rpm for 6 minutes and washed again. The supernatant was discarded, and the cells were resuspended with 5mL of PBS and washed twice, followed by addition of 1mL of Trizol. The samples were mixed and store at −80°C in the refrigerator for further use.
Construction of RNA Libraries
Total RNA was extracted using the Trizol method. cDNA was analysed for quantity and purity using Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit. Samples of high quality RNA with RIN number > 7.0 were used to construct sequencing libraries. The mRNA was purified using Dynabeads Oligo (dT). After purification, the mRNA was fragmented into short fragments using divalent cations at high temperature. The fragmented mRNA was used as a template to synthesise the first strand of cDNA in the M-MuLV reverse transcriptase system, followed by degradation of the RNA strand with RNaseH and synthesis of the second strand of cDNA in the DNA polymerase I system. The double-stranded cDNA was then end-repaired and screened for 370–420 bp. After library construction, qRT-PCR was performed to accurately quantify the effective concentration of the library and ensure the quality of the library.
Sequencing Analysis
After library inspection, 2×150bp paired-end sequencing was performed on an Illumina Novaseq™ 6000. bcl file was converted to fastq file using bcl2fastq software (v 2.19) and default parameters. Quality control and filtering was performed using fastp software (v 0.20.1) and default parameters. The sequencing data were compared to the reference genome using hisat2 software (v 2.2.1) based on the GRCh38 genome file from the UCSC database. The sam files were converted to bam files using samtools software (v 1.13). The Rsubread package (v 2.12.3) of R software (v 4.2.3) was used to analyse the expression of the sequencing data based on the UCSC database Homo_sapiens.GRCh38.108.gtf annotation file, retaining the protein-coding gene expression files for subsequent analysis. The amount of sequencing data is detailed in Supplementary Table 1.
Differentially Expressed Genes (DEGs) Analysis
For expression files, genes with mean expression values > 0 were retained. DEG analysis was performed using the edgeR package (v 3.40.2), setting the threshold to |logFC|>1 and FDR<0.05, based on HF vs NC, SLE vs NC, respectively. Volcano map analysis and heatmap analyses were performed using the enhanced volcano package (v 1.16.0) and the pheatmap package (v 1.0.12), respectively. The Venn package (v 1.11) was used for Venn diagram analysis in order to obtain the intersected DEGs.
Enrichment Analysis
Enrichment analysis was conducted using several R packages, including org.Hs.eg.db (v 3.16.0), clusterProfiler (v 4.6.2), enrichplot (v 1.18.3) and ggplot2 (v 3.4.1), to perform Gene set enrichment analysis (GSEA) which based on Gene Ontology (GO)-biological process (BP) and Kyoto encyclopedia of genes and genomes (KEGG), setting a threshold of corrected P-value < 0.05, and the top 10 enriched pathways were mapped. GO and KEGG analyses were performed on common DEGs using the same packages and threshold, and the enrichment bubbles were plotted.
Immuno-Infiltration Analysis
The CIBERSORT package (v 1.0.4) was used to perform immuno-infiltration analysis, with a perm value of 1000. Samples with a P-value < 0.05 were selected for subsequent analysis based on the immuno-infiltration results. Histograms were plotted against the immuno-infiltrate results and immune cell correlations were analysed using the corrplot package (v 0.92). Differential expression between immune cell groups was analysed using violin plots generated by the vioplot package (v 0.4.0).
Weighted Gene Co-Expression Network Analysis (WGCNA)
WGCNA was performed separately for HF and SLE using common differential immune cells as the signature module. The top 15,000 genes with the largest variance values were selected for subsequent analysis using the WGCNA package (v 1.72-1). Samples were first clustered to remove outliers, and re-clustered and analysed in conjunction with clinical features. A soft threshold was selected using the pickSoftThreshold method based on the average connectivity R^2 greater than 0.85. Based on this soft threshold, set mergeCutHeight = 0.2 and minModuleSize = 30 to build the co-expression network, divide the modules, and merge similar modules. The correlation coefficient matrix between module feature vectors and clinical traits was calculated and a correlation heat map was drawn. Venn diagram analysis was performed using the venn package (v 1.11) to obtain interested genes.
Protein-Protein Interaction (PPI) Analysis
The STRING database was used for PPI analysis of intersecting genes. Key genes were identified from the PPI results using 5 algorithms (Degree, DMNC, EPC, MCC, MNC) in Cytoscape software (v 3.9.1) The results were visualized using the UpSet plots of the 5 algorithms using the UpSetR package (v 1.4.0).
Machine Learning
Machine learning analyses were respectively performed, including Lasso analysis using the glmnet package (v 4.1–7) for key genes; SVM analysis using the e1071 package (v 1.7–13) and caret package (v 6.0–94); and random forest analysis using the randomForest package (v 4.7–1.1). Setting ntree from 1 to 500, followed by setting ntree to the minimum error value for random forest analysis again. Key genes were obtained based on MeanDecreaseGini values > 1, and the final key genes were obtained by venn diagram analysis.
Receiver Operating Characteristic Curve (ROC) Analysis and Correlation Analysis
Expression box plotting was performed for the key genes and ROC analysis was performed using the pROC package (v 1.18). Spearman correlation analysis was performed to identify key genes specific to HF and SLE respectively, and correlation analysis and lollipop plots were conducted for key genes and immune cells using the ggpubr package.
Results
Basic Patient Data
The study population consisted of 10 patients with HF (age: 64.30±14.26), 10 patients with SLE (age: 62.50±11.65), and 10 NCs (age: 62.50±9.85). There was no significant difference in the basic information of the cases between groups (p > 0.05). General information of the enrolled cases was shown in Supplementary Table 2.
Screening for Shared DEGs in HF and SLE
DEG analysis identified 1276 genes in HF vs NC, including 491 up-regulated genes and 785 down-regulated genes. The above DEGs in HF were plotted for volcano map (Figure 1A) and heat map (Figure 1B). Similarly, a total of 541 genes were obtained in SLE vs NC, including 359 up-regulated genes and 182 down-regulated genes. The above DEGs in SLE were plotted for volcano map (Figure 1C) and heat map (Figure 1D). A total of 288 co-morbid genes were obtained by taking intersection of the DEGs of HF and SLE using Venn (Figure 1E).
Enrichment Analysis
GO-BP and KEGG-based GSEA were performed for all the genes in HF and SLE, respectively. Genes in HF were significantly enriched in BP such as defense response and innate immune response, as well as in Neutrophil extracellular trap formation, NOD-like receptor signaling pathway and other KEGG pathways (Figure 2A and B). Genes in SLE were significantly enriched in BP such as cellular response to biotic stimulus and humoral immune response, as well as in IL-17 signaling pathway, NOD-like receptor signaling pathway, Toll-like receptor signaling pathway and other KEGG pathways (Figure 2C and D). GO enrichment results showed significant enrichment of the common DEGs in response to virus, humoral immune response and other biological processes (BP); vesicle lumen, primary lysosome and other cellular components (CC); endopeptidase activity, sulfur compound binding and other molecular functions (MF) (Figure 2E–G). KEGG enrichment results showed that these common DEGs were significantly enriched in NOD-like receptor signaling pathway, coronavirus disease-COVID-19, IL-17 signaling pathway and other pathways (Figure 2H).
Identification of the Shared Immune Cells in HF and SLE
Immune infiltration analysis indicated significant differences in the expression of 3 immune cells, B cell naïve, T cell CD4 naïve, and Monocytes in HF vs NC. A high correlation was observed between T cell CD4 naïve and Monocytes (cor = −0.52) (Figure 3A–C). Similarly, in SLE vs NC, the expression of 4 immune cells, Plasma cells, T cell CD4 naïve, NK cells resting, and Monocytes, showed significant differences in expression in SLE vs NC, with a high correlation observed between T cell CD4 naïve and Monocytes (cor = −0.52) (Figure 3D–F).
Identification of the Shared Immune Cell-Related Genes in HF and SLE
The results of WGCNA showed that using the two common differential immune cells, T cells CD4 naïve and Monocytes, as characteristic modules in HF, three modules, lightyellow (92), darkred (63), turquoise (3381), were identified to be highly correlated with T cells CD4 naïve based on module correlations ≥ 0.6. We also obtained darkgrey (39), darkred (63), tan (153), which are three modules highly correlated with Monocytes based on module correlations ≥ 0.6 (Figure 4A–F). Similarly, in SLE, the yellow module (1123), was the only one module highly correlated with T cells CD4 naïve based on module correlations ≥ 0.6. Five modules, pink (456), magenta (389), yellow (1123), green (828), and turquoise (3204) modules, were highly correlated with Monocytes based on module correlations ≥ 0.6 (Figure 5A–F). In total, 4300 genes associated with T cells CD4 naïve and 5690 genes associated with Monocytes were obtained in both diseases after removing duplicates.
Identification of Co-Morbid Genes Associated with the Shared Immune Cells in HF and SLE and PPI Analysis
By taking intersections of the genes in the WGNCA key module (cor ≥ 0.6) with DEGs common to both diseases, a total of 207 intersected genes were obtained between T cells CD4 naïve and DEGs (Figure 6A). By applying PPI analysis for the analysis of the above genes using five algorithms, 22 T cells CD4 naïve-related co-morbid genes were obtained (Figure 6B and C). Similarly, 162 intersected genes were identified for Monocytes and DEGs (Figure 6D), PPI analysis was performed, and 26 Monocytes-associated co-morbid genes were finally obtained with the aid of the 5 algorithms (Figure 6E and F).
Machine Learning Analysis of the Key Genes Common to HF and SLE
From the 22 co-morbid genes associated with T cells CD4 naïve, five key genes were identified by lasso analysis (Figure 7A and B), four by SVM analysis (Figure 7C) and three by random forest analysis (Figure 7D and E). RNASE2 and CXCL10 were found to be the only two genes after intersection (Figure 7F). From the 26 co-morbid genes associated with Monocytes, five key genes were identified by lasso analysis (Figure 7G and H), four by SVM analysis (Figure 7I), and four by random forest analysis (Figure 7J and K). CCR7, RNASE2 and RNASE3 were finally obtained after taking intersection (Figure 7L).
Validation of the Differential Expression of the Key Genes
To validate the differential expression of key genes in HF vs SLE, corresponding expression box plots were plotted. CCR7 was shown to be significantly down-regulated in HF and SLE, while RNASE2, RNASE3 and CXCL10 were significantly up-regulated in HF and SLE (Figure 8A–H). The similar expression trend of these four key genes in HF vs SLE in part suggests that these key genes may be associated with similar underlying mechanisms of action in both diseases.
The Diagnostic Value of the Key Genes
The ROC analysis showed that in HF, the AUC of CCR7, RNASE2, RNASE3, and CXCL10 was 1.00, 0.96, 0.94, and 0.99, respectively (Figure 9A–D). In the SLE, the value of their AUCs was 0.93, 0.94, 1.00, and 0.99, respectively (Figure 9E–H). The above results confirm the good diagnostic value of these key genes in both HF and SLE.
Correlation Analysis
The correlation analysis revealed significant associations between the four key genes and immune cells in HF, CCR7 was significantly correlated with T cells CD4 naïve and B cells naïve (Figure 10A); RNASE2 was significantly correlated with T cells CD4 naïve and B cells naïve (Figure 10B); RNASE3 was correlated with T cells CD4 naïve significantly (Figure 10C); CXCL10 was correlated with T cells CD4 naïve, Monocytes, B cells naïve, NK cells activated, T cells CD4 memory activated significantly (Figure 10D). In SLE, CCR7 was correlated with T cells CD4 naïve and Monocytes significantly (Figure 10E); RNASE2 was correlated with T cells CD4 naïve and T cells CD4 memory resting significantly (Figure 10F); RNASE3 was correlated with T cells CD4 naïve, Monocytes, and T cells CD4 memory resting significantly (Figure 10G); CXCL10 was correlated with Monocytes, T cells CD4 naïve, Mast cells activated, B cells naïve significantly (Figure 10H). The above findings were also consistent with the immuno-infiltration analysis and WGCNA results of this study. In addition, correlation analysis was performed between key genes. In HF, RNASE2 had a high correlation with RNASE3 (cor = 0.8) and CCR7 had a high correlation with CXCL10 (cor = −0.76) (Figure 10I). In SLE, RNASE2 had a high correlation with RNASE3 (cor = 0.79), CXCL10 had a high correlation with RNASE3 (cor = 0.67); CCR7 had an equally significant correlation with CXCL10 (cor = −0.63), and a high correlation with RNASE3 (cor = −0.62) (Figure 10J). A line graph of the 4 key genes and immune cells was shown in Supplementary Figures 1 and 2.
Discussion
HF is a growing global epidemic affecting at least 260,000 individuals worldwide and has now become increasingly common.18 Despite advances in treatment strategies for chronic HF, the survival rate of patients with HF remains very low.19 SLE is a global health problem with prevalence, morbidity and mortality increasing in Asia according to recent epidemiological reports.20 There is no denying that HF and SLE pose significant threats to human health. Previous studies have indicated that acute cardiac manifestations of SLE can result in cardiac dysfunction and increase the risk of HF in SLE.21 Moreover, numerous studies have confirmed the essential involvement of the immune response in the pathogenesis of HF or SLE.22,23 In this study, transcriptome sequencing was used to identify the shared immune cells and co-morbid genes between HF and SLE, aiming to elucidate the potential mechanisms of action underlying these conditions.
This study identified 288 common DEGs in HF and SLE through DEG analysis. Enrichment analysis indicated that these DEGs were significantly associated with inflammatory signalling pathways, such as NOD-like receptor signaling pathway, and IL-17 signaling pathway. NOD-like Receptors (NLRs) are known to regulate inflammasome formation, which in turn play a role in inflammatory responses.24 NOD-like Receptor Pyrin Domain-containing Protein 3 (NLRP3) has been extensively studied and found to regulate the synthesis and secretion of inflammatory factors, making it crucial in the development of intrinsic immunity and inflammatory diseases.25 On the other hand, studies related to SLE suggest that NLRP3 inflammatory vesicles are important in the pathogenesis of cardiovascular diseases, including HF, and inhibition of NLRP3 inflammatory vesicle-related signalling pathways is predicted to provide new interventional mediators for treating HF.26,27 In contrast, in studies related to SLE, NLRP3 may be closely associated with the progression of Lupus Nephritis (LN), and inhibition of the activated NLRP3 inflammatory vesicle pathway may be a novel therapeutic approach for SLE and LN.28,29 IL-17, produced mainly by activated T cells, is an inflammatory cytokine involved in the inflammatory processes.30 Numerous studies have confirmed the association of IL-17 with the pathogenesis of inflammatory diseases such as HF and autoimmune diseases such as SLE.31,32 Based on the results of the present study, immune dysfunction was revealed as a common pathophysiological feature of HF and SLE, with the NOD-like receptor and IL-17 as potential mechanisms of action in HF and SLE.
An immune infiltration analysis was subsequently performed to investigate the involvement of immune cells in HF and SLE. The results showed that both diseases exhibited aberrant expression of T cell CD4 naïve and Monocytes, which displayed a high correlation. T cell CD4 naïve can differentiate into different effector cell subpopulations, such as Th17 and Treg in response to different cytokines and environments, thus regulating the Th17/Treg ratio.33 Furthermore, the expression of T cell CD4 naïve is significantly reduced in both HF and SLE,34,35 and that Th17/Treg balance is impaired in rats with congestive HF and can affect the progression of HF in rats by regulating LOX expression.36–38 Monocytes, on the other hand, are a subpopulation of leukocytes that play a key role in maintaining balance, pathogen recognition and clearance, and inflammation.39 The subpopulations of Monocytes are key to the inflammatory cascade response in HF, and targeting inflammation-associated immune cells such as Monocytes would hold promise for the treatment of HF.40,41 In addition, Monocytes are also involved in the progression of inflammation in SLE, and have been suggested as a potential biomarker for cardiovascular risk in SLE.42,43 Overall, it is undeniable that both T cell CD4 naïve and Monocytes are possible shared immune cells in HF and SLE, and may even be involved in similar mechanisms of action.
To further identify key genes associated with the shared immune cells in HF and SLE, WGCNA, PPI, and machine learning were collectively employed. CCR7, RNASE2, RNASE3, and CXCL10 were finally screened out as co-morbid genes closely related to shared immune cells. These genes have been shown to be involved in the pathological processes that exacerbate the inflammatory state of diseases such as autoimmune diseases, chronic inflammation and cancer.44 Specifically, CCL21 has been found to directly influence the prognosis of HF after myocardial infarction by interfering with its expression, thus highlighting its potential as a prognostic biomarker for HF.45 Furthermore, CCR7 expression on Monocytes has been reported to represent the disease activity score of rheumatoid arthritis (RA),46 and in SLE, CCR7 has also been shown to be regulated by lncRNA-AC007278.2, which affects the auto immune activity.47 RNASE2 and RNASE3 encode proteins that belong to the same family of ribonucleases and can be involved in the innate immune response of the body.48 RNASE2 has been identified as a potential key gene in ischaemic cardiomyopathy, which can progress to chronic HF, and may have relevance to HF.49 Overexpression of RNASE2 in SLE patients have also been found to influence the processes of SLE by mediating the secretion of cytokines and chemokines from Monocytes.50 In contrast, although less studied in HF and SLE, RNASE3 has been shown to be differentially expressed in a variety of inflammatory diseases such as asthma and allergic contact dermatitis.51,52 CXCL10 encodes a pro-inflammatory cytokine that plays a key regulatory role in immune responses by regulating T cell migration.53 Elevated circulating levels of CXCL10 have been associated with the progression of HF, mainly expressed on CD4+ Th1 cells,54 and its presence may predominate in patients with advanced HF.55 CXCL10 is also important in the mechanism of action of SLE and its associated complications.56 Serum and urine CXCL10 levels may be useful in monitoring disease activity in SLE and LN.57 In conclusion, the identification of CCR7, RNASE2, RNASE3, and CXCL10 as shared genes between HF and SLE highlights their involvement in the similar immune mechanisms of action underlying the two diseases. These findings provide novel ideas for the development of targeted therapies to improve disease outcomes of HF and SLE, reducing the incidence of relevant complications, and improving disease prognosis.
Despite the findings presented in the current study, several limitations should be acknowledged. Firstly, transcriptome sequencing in this study was only performed on 30 included samples, thus the results needs to be validated in a larger cohort. Secondly, this study only performed bioinformatics analysis on the transcriptome data, with no in-depth investigation into the mechanism of action of HF and SLE. Further studies are needed to provide a more comprehensive understanding of the shared immune mechanisms between these two diseases.
Conclusion
The present study firstly revealed T cells CD4 naïve and Monocytes as potential shared immune cells in HF and SLE, and identified candidate genes such as CCR7, RNASE2, RNASE3 and CXCL10 as possible key genes common to HF and SLE that may serve as biomarkers or potential therapeutic targets for both diseases.
Ethics Approval and Informed Consent
This study was approved by the Institutional Ethics Committee of The Second Affiliated Hospital, Zhejiang Chinese Medical University (Ethical Review 2022 Research No. 097-01). All enrolled patients received informed consent before this study. This study strictly adhered to the Declaration of Helsinki.
Funding
This research was supported by National Chinese Medicine Clinical Research Base Support Discipline (Immune-related Cardiovascular Diseases, 2020-JDXK-ZC04) and Zhejiang University of Chinese Medicine Young and Middle-aged Research Innovation Fund Project (KC201938).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Baman JR, Ahmad FS. Heart failure. JAMA. 2020;324(10):1015. doi:10.1001/jama.2020.13310
2. Mascolo A, Di Mauro G, Cappetta D, et al. Current and future therapeutic perspective in chronic heart failure. Pharmacol Res. 2022;175:106035. doi:10.1016/j.phrs.2021.106035
3. Roger VL. Epidemiology of heart failure: a contemporary perspective. Circ Res. 2021;128(10):1421–1434. doi:10.1161/CIRCRESAHA.121.318172
4. Pan L, Lu MP, Wang JH, et al. Immunological pathogenesis and treatment of systemic lupus erythematosus. World J Pediatr. 2020;16(1):19–30. doi:10.1007/s12519-019-00229-3
5. Barber MRW, Drenkard C, Falasinnu T, et al. Global epidemiology of systemic lupus erythematosus. Nat Rev Rheumatol. 2021;17(9):515–532. doi:10.1038/s41584-021-00668-1
6. Owen KA, Grammer AC, Lipsky PE. Deconvoluting the heterogeneity of SLE: the contribution of ancestry. J Allergy Clin Immunol. 2022;149(1):12–23. doi:10.1016/j.jaci.2021.11.005
7. Tenbrock K, Rauen T. T cell dysregulation in SLE. Clin Immunol. 2022;239:109031. doi:10.1016/j.clim.2022.109031
8. Mak A, Chan JKY. Endothelial function and endothelial progenitor cells in systemic lupus erythematosus. Nat Rev Rheumatol. 2022;18(5):286–300. doi:10.1038/s41584-022-00770-y
9. Oliveira CB, Kaplan MJ. Cardiovascular disease risk and pathogenesis in systemic lupus erythematosus. Semin Immunopathol. 2022;44(3):309–324. doi:10.1007/s00281-022-00922-y
10. Yafasova A, Fosbøl EL, Schou M, et al. Long-term cardiovascular outcomes in systemic lupus erythematosus. J Am Coll Cardiol. 2021;77(14):1717–1727. doi:10.1016/j.jacc.2021.02.029
11. Yao M, Zhang C, Gao C, et al. Exploration of the shared gene signatures and molecular mechanisms between systemic lupus erythematosus and pulmonary arterial hypertension: evidence from transcriptome data. Front Immunol. 2021;12:658341. doi:10.3389/fimmu.2021.658341
12. Zhang T, Feng H, Zou X, et al. Integrated bioinformatics to identify potential key biomarkers for COVID-19-related chronic urticaria. Front Immunol. 2022;13:1054445. doi:10.3389/fimmu.2022.1054445
13. Li T, Qu J, Xu C, et al. Exploring the common gene signatures and pathogeneses of obesity with Alzheimer’s disease via transcriptome data. Front Endocrinol. 2022;13:1072955. doi:10.3389/fendo.2022.1072955
14. Castiglione V, Aimo A, Vergaro G, et al. Biomarkers for the diagnosis and management of heart failure. Heart Fail Rev. 2022;27(2):625–643. doi:10.1007/s10741-021-10105-w
15. Zhao X, Zhang L, Wang J, et al. Identification of key biomarkers and immune infiltration in systemic lupus erythematosus by integrated bioinformatics analysis. J Transl Med. 2021;19(1):35. doi:10.1186/s12967-020-02698-x
16. Heidenreich PA, Bozkurt B, Aguilar D, et al. 2022 AHA/ACC/HFSA guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association Joint Committee on clinical practice guidelines. Circulation. 2022;145(18):e895–e1032. doi:10.1161/CIR.0000000000001063
17. Aringer M, Costenbader K, Daikh D, et al. 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Ann Rheum Dis. 2019;78(9):1151–1159. doi:10.1136/annrheumdis-2018-214819
18. Sharma M, Bhatt LK. Emerging therapeutic targets for heart failure. Curr Cardiol Rep. 2022;24(11):1737–1754. doi:10.1007/s11886-022-01789-z
19. Crea F. Acute and chronic heart failure: exciting therapeutic perspectives. Eur Heart J. 2023;44(1):1–4. doi:10.1093/eurheartj/ehac767
20. Barber MRW, Falasinnu T, Ramsey-Goldman R, et al. The global epidemiology of SLE: narrowing the knowledge gaps. Rheumatology. 2023;62(Supplement_1):i4–i9. doi:10.1093/rheumatology/keac610
21. Chang JC, Xiao R, Knight AM, et al. A population-based study of risk factors for heart failure in pediatric and adult-onset systemic lupus erythematosus. Semin Arthritis Rheum. 2020;50(4):527–533. doi:10.1016/j.semarthrit.2020.03.019
22. Lu Y, Xia N, Cheng X. Regulatory T cells in chronic heart failure. Front Immunol. 2021;12:732794. doi:10.3389/fimmu.2021.732794
23. Li A, Guo F, Pan Q, et al. Mesenchymal stem cell therapy: hope for patients with systemic lupus erythematosus. Front Immunol. 2021;12:728190. doi:10.3389/fimmu.2021.728190
24. Platnich JM, Muruve DA. NOD-like receptors and inflammasomes: a review of their canonical and non-canonical signaling pathways. Arch Biochem Biophys. 2019;670:4–14. doi:10.1016/j.abb.2019.02.008
25. Li Z, Guo J, Bi L. Role of the NLRP3 inflammasome in autoimmune diseases. Biomed Pharmacother. 2020;130:110542. doi:10.1016/j.biopha.2020.110542
26. Pellegrini C, Martelli A, Antonioli L, et al. NLRP3 inflammasome in cardiovascular diseases: pathophysiological and pharmacological implications. Med Res Rev. 2021;41(4):1890–1926. doi:10.1002/med.21781
27. Wang Y, Li Y, Zhang W, et al. NLRP3 inflammasome: a novel insight into heart failure. J Cardiovasc Transl Res. 2023;16(1):166–176. doi:10.1007/s12265-022-10286-1
28. Oliveira CB, Lima CAD, Vajgel G, et al. The role of NLRP3 inflammasome in lupus nephritis. Int J Mol Sci. 2021;22(22):12476. doi:10.3390/ijms222212476
29. Guo C, Fu R, Zhou M, et al. Pathogenesis of lupus nephritis: RIP3 dependent necroptosis and NLRP3 inflammasome activation. J Autoimmun. 2019;103:102286. doi:10.1016/j.jaut.2019.05.014
30. Majumder S, McGeachy MJ. IL-17 in the pathogenesis of disease: good intentions gone awry. Annu Rev Immunol. 2021;39:537–556. doi:10.1146/annurev-immunol-101819-092536
31. Xue GL, Li DS, Wang ZY, et al. Interleukin-17 upregulation participates in the pathogenesis of heart failure in mice via NF-κB-dependent suppression of SERCA2a and Cav1.2 expression. Acta Pharmacol Sin. 2021;42(11):1780–1789. doi:10.1038/s41401-020-00580-6
32. Yang Y, Yan C, Yu L, et al. The star target in SLE: IL-17. Inflamm Res. 2023;72(2):313–328. doi:10.1007/s00011-022-01674-z
33. Long X, Luo C, Zhu Z. Role of CNSs conserved distal cis-regulatory elements in CD4 + T cell development and differentiation. Front Immunol. 2022;13:919550. doi:10.3389/fimmu.2022.919550
34. Xydonas S, Parissis J, Lioni L, et al. Immunosenescence in patients with chronic systolic heart failure. J Cardiovasc Med. 2016;17(8):624–630. doi:10.2459/JCM.0000000000000372
35. Yuan S, Zeng Y, Li J, et al. Phenotypical changes and clinical significance of CD4 + /CD8 + T cells in SLE. Lupus Sci Med. 2022;9(1):e000660. doi:10.1136/lupus-2022-000660
36. Chen Z, Yan W, Mao Y, et al. Effect of aerobic exercise on treg and Th17 of rats with ischemic cardiomyopathy. J Cardiovasc Transl Res. 2018;11(3):230–235. doi:10.1007/s12265-018-9794-0
37. Lu M, Qin X, Yao J, et al. Th17/Treg imbalance modulates rat myocardial fibrosis and heart failure by regulating LOX expression. Acta Physiol. 2020;230(3):e13537. doi:10.1111/apha.13537
38. Shan J, Jin H, Xu Y. T cell metabolism: a new perspective on Th17/Treg cell imbalance in systemic lupus erythematosus. Front Immunol. 2020;11:1027. doi:10.3389/fimmu.2020.01027
39. Narasimhan PB, Marcovecchio P, Hamers AAJ, et al. Nonclassical monocytes in health and disease. Annu Rev Immunol. 2019;37:439–456. doi:10.1146/annurev-immunol-042617-053119
40. Dounousi E, Duni A, Naka KK, et al. The innate immune system and cardiovascular disease in ESKD: monocytes and natural killer cells. Curr Vasc Pharmacol. 2021;19(1):63–76. doi:10.2174/18756212MTA3yNzEe1
41. Rhee AJ, Lavine KJ. New approaches to target inflammation in heart failure: harnessing insights from studies of immune cell diversity. Annu Rev Physiol. 2020;82:1–20. doi:10.1146/annurev-physiol-021119-034412
42. Liu Y, Luo S, Zhan Y, et al. Increased expression of PPAR-γ modulates monocytes into a M2-like phenotype in SLE patients: an implicative protective mechanism and potential therapeutic strategy of systemic lupus erythematosus. Front Immunol. 2020;11:579372. doi:10.3389/fimmu.2020.579372
43. López P, Rodríguez-Carrio J, Martínez-Zapico A, et al. Low-density granulocytes and monocytes as biomarkers of cardiovascular risk in systemic lupus erythematosus. Rheumatology. 2020;59(7):1795. doi:10.1093/rheumatology/keaa225
44. Brandum EP, Jørgensen AS, Rosenkilde MM, et al. Dendritic cells and CCR7 expression: an important factor for autoimmune diseases, chronic inflammation, and cancer. Int J Mol Sci. 2021;22(15):8340. doi:10.3390/ijms22158340
45. Yndestad A, Finsen AV, Ueland T, et al. The homeostatic chemokine CCL21 predicts mortality and may play a pathogenic role in heart failure. PLoS One. 2012;7(3):e33038. doi:10.1371/journal.pone.0033038
46. Van Raemdonck K, Umar S, Shahrara S. The pathogenic importance of CCL21 and CCR7 in rheumatoid arthritis. Cytokine Growth Factor Rev. 2020;55:86–93. doi:10.1016/j.cytogfr.2020.05.007
47. You Y, Zhao X, Wu Y, et al. Integrated transcriptome profiling revealed that elevated long non-coding RNA-AC007278.2 expression repressed CCR7 transcription in systemic lupus erythematosus. Front Immunol. 2021;12:615859. doi:10.3389/fimmu.2021.615859
48. Wu L, Xu Y, Zhao H, et al. RNase T2 in inflammation and cancer: immunological and biological views. Front Immunol. 2020;11:1554. doi:10.3389/fimmu.2020.01554
49. Zheng PF, Liu F, Zheng ZF, et al. Identification MNS1, FRZB, OGN, LUM, SERP1NA3 and FCN3 as the potential immune-related key genes involved in ischaemic cardiomyopathy by random forest and nomogram. Aging. 2023;15(5):1475–1495. doi:10.18632/aging.204547
50. Zhu Y, Tang X, Xu Y, et al. RNASE2 mediates age-associated B cell expansion through monocyte derived IL-10 in patients with systemic lupus erythematosus. Front Immunol. 2022;13:752189. doi:10.3389/fimmu.2022.752189
51. Vernet R, Matran R, Zerimech F, et al. Identification of novel genes influencing eosinophil-specific protein levels in asthma families. J Allergy Clin Immunol. 2022;150(5):1168–1177. doi:10.1016/j.jaci.2022.05.017
52. Kim J, Kim S, Lee YI, et al. Eosinophil cationic protein is a potential surrogate marker of allergic contact dermatitis: a single-center, retrospective study of 216 patients. J Am Acad Dermatol. 2020;83(6):1819–1821. doi:10.1016/j.jaad.2020.05.070
53. Karin N, Razon H. Chemokines beyond chemo-attraction: CXCL10 and its significant role in cancer and autoimmunity. Cytokine. 2018;109:24–28.
54. Ngwenyama N, Salvador AM, Velázquez F, et al. CXCR3 regulates CD4+ T cell cardiotropism in pressure overload-induced cardiac dysfunction. JCI Insight. 2019;4(7). doi:10.1172/jci.insight.125527
55. Altara R, Manca M, Hessel MH, et al. CXCL10 is a circulating inflammatory marker in patients with advanced heart failure: a pilot study. J Cardiovasc Transl Res. 2016;9(4):302–314. doi:10.1007/s12265-016-9703-3
56. Ghafouri-Fard S, Shahir M, Taheri M, et al. A review on the role of chemokines in the pathogenesis of systemic lupus erythematosus. Cytokine. 2021;146:155640. doi:10.1016/j.cyto.2021.155640
57. Puapatanakul P, Chansritrakul S, Susantitaphong P, et al. Interferon-inducible protein 10 and disease activity in systemic lupus erythematosus and lupus nephritis: a systematic review and meta-analysis. Int J Mol Sci. 2019;20(19):4954. doi:10.3390/ijms20194954
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.










