Back to Journals » Journal of Inflammation Research » Volume 17
Identification of Neutrophil Extracellular Trap-Related Gene Expression Signatures in Ischemia Reperfusion Injury During Lung Transplantation: A Transcriptome Analysis and Clinical Validation
Authors Gao J , Zhang Z , Yu J, Zhang N, Fu Y , Jiang X, Xia Z, Zhang Q, Wen Z
Received 14 October 2023
Accepted for publication 1 February 2024
Published 12 February 2024 Volume 2024:17 Pages 981—1001
DOI https://doi.org/10.2147/JIR.S444774
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Adam D Bachstetter
Jiameng Gao,1,2,* Zhiyuan Zhang,1,2,* Jing Yu,1,2,* Nan Zhang,1,2 Yu Fu,1,2 Xuemei Jiang,1,2 Zheyu Xia,3 Qingqing Zhang,1,2 Zongmei Wen1,2
1Department of Anesthesiology, Shanghai Pulmonary Hospital, School of Medicine, Tongji University, Shanghai, People’s Republic of China; 2Shanghai Engineering Research Center of Lung Transplantation, Shanghai, People’s Republic of China; 3School of Medicine, Tongji University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zongmei Wen; Qingqing Zhang, Department of Anesthesiology, Shanghai Pulmonary Hospital, School of Medicine, Tongji University, 507 Zhengmin Road, Shanghai, 200433, People’s Republic of China, Tel +86 13761635280 ; +86 13764542055, Email [email protected]; [email protected]
Purpose: Ischemia reperfusion injury (IRI) unavoidably occurs during lung transplantation, further contributing to primary graft dysfunction (PGD). Neutrophils are the end effectors of IRI and activated neutrophils release neutrophil extracellular traps (NETs) to further amplify damage. Nevertheless, potential contributions of NETs in IRI remain incompletely understood. This study aimed to explore NET-related gene biomarkers in IRI during lung transplantation.
Methods: Differential expression analysis was applied to identify differentially expressed genes (DEGs) for IRI during lung transplantation based on matrix data (GSE145989, 127003) downloaded from GEO database. The CIBERSORT and weighted gene co-expression network analysis (WGCNA) algorithms were utilized to identify key modules associated with neutrophil infiltration. Moreover, the least absolute shrinkage and selection operator regression and random forest were applied to identify potential NET-associated hub genes. Subsequently, the screened hub genes underwent further validation of an external dataset (GSE18995) and nomogram model. Based on clinical peripheral blood samples, immunofluorescence staining and dsDNA quantification were used to assess NET formation, and ELISA was applied to validate the expression of hub genes.
Results: Thirty-eight genes resulted from the intersection between 586 DEGs and 75 brown module genes, primarily enriched in leukocyte migration and NETs formation. Subsequently, four candidate hub genes (FCAR, MMP9, PADI4, and S100A12) were screened out via machine learning algorithms. Validation using an external dataset and nomogram model achieved better predictive value. Substantial NETs formation was demonstrated in IRI, with more pronounced NETs observed in patients with PGD ≥ 2. PADI4, S100A12, and MMP9 were all confirmed to be up-regulated after reperfusion through ELISA, with higher levels of S100A12 in PGD ≥ 2 patients compared with non-PGD patients.
Conclusion: We identified three potential NET-related biomarkers for IRI that provide new insights into early detection and potential therapeutic targets of IRI and PGD after lung transplantation.
Keywords: ischemia reperfusion injury, neutrophil extracellular traps, lung transplantation, primary graft dysfunction, WGCNA, machine learning
Introduction
Lung transplantation has been established worldwide as the only effective remedy for end-stage lung diseases, including chronic obstructive pulmonary disease, interstitial lung disease, cystic fibrosis.1,2 Over the past few decades, the field of lung transplantation has witnessed remarkable advances, experiencing a substantial surge in lung transplantation procedures since the first successful operation in 1983.3 Annually, over 4000 lung transplantations are conducted worldwide.4 Despite, morbidity and mortality of lung transplant patients remain high compared with other solid organ transplants. IRI is an unavoidable occurrence following lung transplantation, which characterized by uncontrolled sterile inflammation, increased microvascular permeability, elevated pulmonary vascular resistance, pulmonary edema, impaired oxygenation, and pulmonary hypertension.5–7 And it is the main mechanism of primary graft dysfunction (PGD), one of the major clinical challenges for clinicians in the perioperative period of lung transplantation as well as a leading cause of early allograft failure.8,9 Since the exact underlying pathogenesis of IRI during lung transplantation remains unclear and the absence of effective therapies, further investigation is warranted to explore targeted treatments aimed at the pathogenetically altered expressed genes associated with IRI and PGD.
Neutrophils are the end effectors of innate immune response in IRI after lung transplant.10,11 Recipient derived neutrophils are recruited to lung allografts and subsequently infiltrate into alveolar space after reperfusion, triggering various damage associated effector events, including the expanded production of ROS, the release of azurophilic granules and metalloproteinase.12 Among the effector functions of neutrophils, the formation of neutrophil extracellular traps (NETs) generated by a regulated cell death program termed “NETosis” has been confirmed to be closely correlated to PGD following lung transplantation. NETs are composed of extracellular chromatin decorated with histones, antibacterial peptides, and serine proteases.13 And those components could contribute to tissue injury and vascular occlusion.14,15 NETs seemingly function as a biomarker of severity of inflammation. Perioperative elevation of NETs biomarkers has been demonstrated to be associated with PGD3 following lung transplantation, suggesting their potential utility in identifying recipients at risk of PGD3 and initiating preventive therapies.16 Multiple experimental models of lung transplantation have similarly demonstrated platelet-driven NETs formation in the pathogenesis of PGD.17 Consequently, the identification of NET-related gene biomarkers will enhance our mechanistic comprehension and offer potential targeted therapeutic strategies for PGD.
Previous transcriptome research of human lung allograft before and after transplantation was largely restricted as the deficiency of clinical data. Therefore, in our study, we used neutrophil infiltration levels to approximately evaluate the severity of IRI after lung transplant based on CIBERSORT algorithm. Subsequently, WGCNA and machine learning algorithms including least absolute shrinkage and selection operator (LASSO) regression and random forest (RF) screened out the most significant genes related to neutrophil infiltration and NETs formation, which may have a potential correlation with PGD. Finally, the expression of hub genes was validated in clinical samples. This is the first time that WGCNA and machine learning were used to explore NETs-related hub genes of IRI after lung transplantation, which could provide novel insight for the early diagnosis at the molecular level and treatment of PGD patients.
Materials and Methods
Data Acquisition and Processing
The flowchart in Figure 1 depicts the methodology of this study. The GSE145989,18 GSE12700319 and GSE1899520 microarray data were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/).21 Table 1 presents detailed dataset information. The GSE145989 dataset comprises 67 reperfused lung allograft biopsy samples collected at 1 or 2 hours after reperfusion, extracted on the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array).18 Our study selectively included 51 reperfused lung allograft biopsy samples collected at 2 hours after reperfusion. All microarray data were normalized through R package “limma”.22 When multiple probes identified the same gene, the average value was calculated to determine its expression. The dataset GSE145989 and GSE127003 were then merged into a larger dataset via R for subsequent analysis. The R package “SVA”23 was applied to eliminate batch effects by Combat, which was employed to normalize the expression values from different batches or platforms. The GSE18995 dataset was used for validation of hub genes.
 |
Table 1 Basic Information of GEO Datasets Used in the Study |
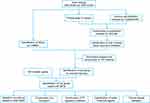 |
Figure 1 The workflow of the study. |
Identification of DEGs and Functional Enrichment Analyses
The normalized and batch-eliminated merged microarray expression dataset underwent differential expression analysis via the R package “limma” to identify differentially expressed genes (DEGs).22 False discovery rate-adjusted p values (FDR) < 0.05 and |log (fold-change)| values ≥0.5 were set as the threshold. The identified DEGs were subsequently subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses. The R package “org.Hs.eg.db.” was initially utilized to convert the hub gene symbols as “ENTREZID”. Subsequently, biological functions were assigned to these genes, including biological process (BP), cellular components (CC), molecular function (MF), and KEGG pathway enrichment, based on the R packages “clusterProfiler”,24 “org.Hs.eg.db”, “enrichplot” and “ggplot2”. A threshold of FDR <0.05 was utilized as the cutoff criterion.
Immune Cell Infiltration Analysis
The relative abundance of 22 immune cell subsets in normalized merged dataset were calculated based on the CIBERSORT gene expression deconvolution package.25 The Wilcoxon test was employed to assess the differences in 22 immune cells between ischemic and reperfused lung allograft biopsy samples. The comparison regarding the proportion of diverse types of immune cells between CIT and reperfusion groups was visualized via the boxplot. A heatmap depicting the correlation of 22 types of infiltrating immune cells was carried out using the R package “corrplot”.26
WGCNA
Since small variation of gene expression data often represents noise, we used Coefficient of Variation values >0.1 (CV genes) to select variant genes in 113 CIT lung samples and 97 reperfused lung samples. Then, the R package “WGCNA” was applied to construct the mRNA co-expression network based on the selected CV genes. Pearson’s correlation matrices were determined with the expression of individual transcripts converted to the similarity matrix. According to the principle of a scale-free network, soft thresholds (power = 5, R2 = 0.9) were selected to construct a scale-free co-expression network in turn, and the adjacency matrix was converted into a topological overlap matrix. Then, cluster analysis was performed to identify gene modules, with a minimum size of 30 for the genes dendrogram, and a threshold of 0.25 was set in order to merge identical modules. The module eigengenes were used for component analysis of each module. Pearson’s tests were subsequently used to compute the correlations between the infiltration levels of immune cells and the modules to identify significant modules, with a distinct module with p < 0.05 considered to be significantly correlated with immune cells infiltration. The module with the maximum correlation coefficients was identified as the hub module. Genes in the hub module were further selected with the threshold of membership values (MM) >0.8 and gene significance values (GS) >0.5.
Functional Enrichment Analyses and Construction of PPI Network of Hub Genes
DEGs and brown module genes were intersected to obtain differentially expressed brown module genes through an online Venn diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). Then these genes were used to perform GO and KEGG functional enrichment analyses. The STRING database (https://string-db.org/) was utilized to perform PPI network analysis and identify the interactions among proteins that were encoded by the hub genes with the threshold of interaction score >0.4.27 Then, the results downloaded from the STRING database were visualized through Cytoscape software (v3.9.1).
Machine Learning
Two machine learning algorithms, LASSO and RF, were employed to further filter candidate genes for biomarkers of IRI after lung transplantation based on differentially expressed brown module genes. LASSO, a regression method, serves for variable selection and regularization within statistical models, enhancing both predictive accuracy and comprehensibility.28 The binomial distribution variables are used in the LASSO classification, coupled with lambda. min value for the minimum criterion used to build the model. RF is a suitable approach that offers several advantages, including no restrictions on variable conditions and better accuracy, sensitivity, and specificity, which can be utilized for predicting continuous variables and generating forecasts without apparent variations.29 R packages “glmnet”30 and “randomForest”31 were employed for conducting LASSO regression and RF analysis. The intersection genes of LASSO, RF and NET-related genes from literature32 were then considered as hub genes for biomarkers of IRI after lung transplantation.
Differential Expression Analysis and ROC Curve Validation
R packages “limma” and “ggpubr” were used to analyze the differential expression levels of the final hub genes in CIT and reperfusion groups based on merged dataset and GSE18995. Meanwhile, receiver operating characteristic (ROC) curve analysis was performed for each hub gene using the R package “pROC”,33 and the area under the curve (AUC) with a 95% confidence interval (CI) was calculated. The significance of IRI was judged by the AUC value, with values close to 1 indicating higher accuracy of the model training.
Construction of Nomogram and Verification
Based on the four candidate genes, the R package “rms” was applied to construct the nomogram. “Points” indicates the score of candidate genes, and “Total Points” signifies the summation of all the scores of genes above. The ROC was subsequently established to assess the predictive value of the nomogram and candidate genes regarding IRI after lung transplantation. Calculation of AUC and 95% CI quantified their predictive capacity. AUC > 0.7 was considered as ideal predictive value. Then calibration curve was plotted to assess the predictive accuracy of the nomogram. Furthermore, the decision analysis (DCA) and clinical impact curve were plotted to evaluate the clinical value of the nomogram.
Construction of Potential TF-Target Gene Regulatory Networks and Small-Molecule Drug Prediction
DAVID database (https://david.ncifcrf.gov/) was employed to predict upstream transcription factors (TFs) based on DEGs. The results were then visualized using the Cytoscape software. The Broad Institutes Connectivity Map (CMap) database (https://clue.io/) was employed to identify potential small candidate molecules. We queried CMap with a signature containing the top 150 most upregulated genes in DEGs and obtained an inversely correlated connectivity score (CS) to IRI. Four small molecules selected according to CS could reverse biological states encoded in specific gene expression markers, which represents that these compounds might have a potential therapeutic effect in IRI after lung transplantation. Moreover, we obtained detailed information, including targeted genes, mechanism of action (MOA), and clinical phase, as well as 3D confirmation of the established small molecules extracted from PubChem (https://pubchem.ncbi.nml.nih.gov).
Clinical Samples Collection and Treatment
A total of 12 patients who underwent lung transplantation with PGD 2 grade or more, as well as 12 patients with no PGD, were selected and enrolled into this study from Shanghai Pulmonary Hospital between January 2022 and August 2022 according to 2016 consensus statement of ISHLT working group on PGD definition.34 Patients were stratified by the cut point of PaO2/FiO2 ratio of 300 at 48 hours after reperfusion and the presence of bilateral alveolar infiltrates on chest X ray. The use of postoperative extracorporeal life support (ECLS) should be explicitly reported and taken into account. Furthermore, utilizing atomized prostacyclin or other drugs that may improve oxygenation did not affect PGD classification.35 Human peripheral blood samples at cold ischemic time and 12 hours after reperfusion were obtained from these lung transplant patients, collected into citrated vacutainers and centrifuged at 2000 ×g for 20 min. And then the generated plasma was separated and preserved at −80°C for further experiments. The study was approved (No. K22-267Z) by the Research Ethics Commission of Shanghai Pulmonary Hospital (Shanghai, China), and written informed consent was obtained from the recipients involved in the study or their representatives.
Neutrophils Isolation and Immunofluorescence Staining
Human peripheral neutrophils were isolated by discontinuous density gradient centrifugation with two solutions of differential density (Ficoll and Histopaque-1119, Sigma; St Louis, MO, USA), according to the manufacturer’s instructions. NET formation was examined by immunofluorescence staining. The Human peripheral neutrophils were first treated with SYTOX Green (Thermo Fisher Scientific, Waltham, MA, USA) for 15 min. Subsequently, the neutrophils were fixed in 4% paraformaldehyde and permeabilized with Triton X-100. Then, fixed neutrophils were stained with rabbit polyclonal anti-citrullinated histone H3 (CitH3) rabbit antibody (1:1000 dilution; Abcam, Cambridge, UK) overnight at 4°C. These slides were incubated with goat anti-rabbit IgG labeled-secondary antibodies (EarthOx, San Francisco, CA, USA) for 1 h and the DNA was stained with 4ʹ,6-diamidino-2-phenylindole (DAPI) at room temperature for 10 min. Last, the cells were washed twice with PBS and covered with an antifluorescence quencher. A fluorescence microscope was used to image NETs to assess their structure and location.
Quantification of Plasma dsDNA
We used the PicoGreen dsDNA Quantification Kit (Solarbio, Beijing, P.R. China) to quantify plasma NETs levels. All of the experimental manipulations were based on the instructions of manufacturers with minor adjustments. Each sample was analyzed twice.
Enzyme-Linked Immunosorbent Assay
Plasma levels of three proteins (S100A12, PADI4, and MMP9) were quantified using enzyme-linked immunosorbent assay (Cusabio Biotech Co., Wuhan, Hubei Province, P.R. China). ELISA kits for S100A12 (CSB-E13095h), PADI4(CSB-E16219h) and MMP9 (CSB-E08006h) were purchased from Cusabio, Wuhan, China (https://www.cusabio.com/). All of the experimental manipulations were based on the instructions of manufacturers with minor adjustments.
Statistical Analysis
All statistical analyses were conducted using R (version 4.2.1) or GraphPad Prism 9.5.1. Intergroup comparisons were performed by Student’s t-test for normal distributed variables and Mann–Whitney test for skewed distributed data. Correlations were assessed through Spearman correlation or Pearson correlation analysis. *p<0.05, **p<0.01, and ***p<0.001 were considered statistically significant.
Results
Data Preprocessing and DEG Screening
The dataset GSE145989 and GSE127003 downloaded from GEO were merged and batch-normalized for subsequent analysis, resulting in a larger merged dataset with 113 ischemic lung allograft biopsy samples and 97 reperfused lung allograft biopsy samples. A total of 586 DEGs were identified using the merged dataset based on the criteria of |logFC| > 0.5 and adjusted p< 0.05, with 457 genes up-regulated and 129 genes down-regulated presented in the volcano plots (Figure 2A). The top 30 genes from the upregulated and downregulated genes were selected to create a heat map, where red represents upregulated genes and blue represents downregulated genes (Figure 2B). GO and KEGG functional enrichment analysis was performed on the DEGs with top 10 enrichment terminologies presented in Figure 2C and D. The GO analysis revealed predominant enrichment of these genes in cytokine−mediated signaling pathway, leukocyte migration, receptor ligand activity, signaling receptor activator activity, chemokine activity, and chemokine receptor binding (Figure 2C). The KEGG pathway enrichment analysis identified the top four significantly enriched pathways as TNF signaling pathway, cytokine−cytokine receptor interaction, IL−17 signaling pathway, and NF−kappa B signaling pathway (Figure 2D). These results strongly indicated the involvement of immune response, especially in leukocyte migration and inflammatory response, in the development and occurrence of IRI during lung transplantation.
Estimation of the Immune-Infiltration Level in IRI After Lung Transplantation and Construction of Weighted Gene Co-Expression Network
CIBERSORT, a bioinformatics algorithm capable of estimating 22 immune cell types, was utilized to assess the composition of immune cells based on the gene expression matrix.
Using the CIBERSORT algorithm, the analysis evaluated the infiltration levels of 22 distinct types of immune cells in the ischemia and reperfused groups, presented in the boxplot (Figure 3A). The results of immune infiltration analysis indicated an increased abundance of immune cells in the allograft after reperfusion, including neutrophils, CD4+ T cells, NK cells, monocytes, dendritic cells, and mast cells. Especially, the abundance of neutrophil in the lung allograft exhibited a significant increase following reperfusion. The correlation of 22 types of immune cells revealed that neutrophils were positively associated with resting NK cells (r = 0.43) and Monocytes (r = 0.27), whereas negatively related to macrophages M1 (r = −0.48) and macrophages M2 (r = −0.58) (Figure 3B). The extent of immune cell infiltration in the allograft can be indicative of inflammatory damage to the lung allograft, especially the neutrophils serve as the end effector of IRI, and this was employed as a trait data for the subsequent construction of WGCNA.
To further precisely identify the central hub genes associated with IRI after lung transplantation, we constructed a gene co-expression network using the WGCNA algorithm. A total of 3779 CV genes were recognized for the construction of WGCNA (Supplementary Table S1). We choose immune cell infiltration extent in the CIBERSORT results as the trait data for WGCNA construction (Supplementary Table S2). The soft threshold was set to 5 to satisfy the scale-free topology of the network, resulting in an R² of approximately 0.9 and notably high average connectivity (Figure 3C). Using the average linkage hierarchical clustering method, we constructed a gene hierarchy clustering dendrogram, identifying 9 similar gene modules (Figure 3D). Among these 9 modules, the correlation (R2) of the brown module with neutrophils notably exceeded that of the other modules, reaching 0.73 (Figure 3E). Consequently, the brown module was identified as the hub module. The scatter plot (Figure 3F) showed a strong correlation between GS and MM in the brown module (Coy=0.91, p=3.2e-110). A total of 75 hub genes with module membership values >0.8 and gene significance values >0.5 were discerned from the brown module (Supplementary Table S3).
Functional Enrichment and PPI Network Analysis of the Hub Genes
For further exploration of IRI mechanism during lung transplantation, we intersected DEGs and brown module genes and obtained a total of 38 signature genes illustrated in Figure 4A (Supplementary Table S4). Subsequently, these signature genes were applied GO and KEGG pathway enrichment functional analysis (Figure 4B and C). GO analysis showed that these genes were mainly enriched in leukocyte migration, positive regulation of response to external stimulus, leukocyte chemotaxis, and cell chemotaxis, immune receptor activity, and cytokine receptor activity (Figure 4B). The KEGG pathway analysis indicated significant enrichment of the hub genes primarily in apoptosis, neutrophil extracellular trap formation, and cytokine receptor, and cytokine–cytokine receptor interaction pathways (Figure 4C). The protein–protein interaction (PPI) network was constructed to further assess the interactions among these genes using the STRING online tool (https://stringdb.org/) (Figure 4D). The Cytoscape software (v3.9.1) was applied to visualize the PPI network, comprising 28 nodes. All of these 38 hub genes were selected for subsequent analysis.
Identification of NET Formation-Related Biomarkers via Machine Learning
The LASSO regression and RF machine learning algorithms were applied to further streamline the important characteristic variables for the identification of hub genes. The LASSO regression algorithm identified 34 potential candidate biomarkers based on lambda. min (Figure 5A and B). The RF algorithm in combination with feature selection was used to determine the association between error rate, number of classification trees, and the top 25 genes by weight were selected (Figure 5C and D). Four genes (S100A12, DAPI4, FCAR, and MMP9) were ultimately obtained by intersecting the genes screened from the LASSO and RF algorithm, alongside 136 NETs related genes from literature, visualized represented in Venn diagram (Figure 5E). The four key genes (S100A12, DAPI4, FCAR, and MMP9) were identified as the final hub genes. Gene correlations, depicted in Figure 5F, indicated a positive correlation among the four genes, suggesting their substantial functional similarity.
Differential Expression Analysis and Validation of Hub Genes
The four final key genes were performed differential expression analysis based on the merged data (Figure 6A). Compared with CIT groups, the expression of the four final hub genes was upregulated in reperfusion groups (p < 0.001) (Figure 6A). Subsequently, further validation of the four final hub genes was conducted using the external dataset GSE18995. Similarly, all four hub genes exhibited higher expression levels in the reperfusion groups compared to the CIT groups (Figure 6B). The ROC curve analysis was utilized to evaluate the sensitivity and specificity of the four hub genes in external dataset for IRI assessment, revealing AUC values exceeding 0.70 for all four genes, indicating their significant representation for IRI (Figure 6C).
Construction and Assessment of a Nomogram for IRI
The R package “rms” was applied to establish a nomogram for IRI after lung transplantation based on the four candidate hub genes (Figure 7A). ROC curves were established to assess the predictive specificity and sensitivity of both the nomogram model (Figure 7B) and each individual gene (Figure 7C). The ROC curve analysis showed that the AUC value of the nomogram model was higher than any other candidate gene, signifying the accuracy of the nomogram model. Compared with single gene, this nomogram achieved a better predictive value. Additionally, the calibration curve confirmed the accuracy of the nomogram model by demonstrating minimal error between the actual and predicted risk (Figure 7D). DCA manifested that the “nomogram” curve exceeded the gray line, and the “PADI4, MMP9, S100A12, and FCAR” curve implied that patients could benefit from the nomogram model at a high-risk threshold of 0 to 0.9. The nomogram model provided a greater clinical benefit than the “PADI4, MMP9, S100A12, and FCAR” curve (Figure 7E). The clinical impact curve was plotted to evaluate the clinical value of the nomogram model (Figure 7F). These results implied that the four genes may play a key role in the process of IRI after lung transplantation.
Construction of Potential TF-Target Gene Regulatory Networks and Identification of Small Molecular Therapeutic Agents
The interaction network consisted of 82 genes and 2 TFs (Figure 8A). NFKAPPAB was identified to regulate 50 genes, including S100A12, and PADI4. AP1 could regulate 50 genes, including PADI4 and EREG. The CMap analysis was applied to search therapeutic compounds of which gene expression patterns were opposite to IRI. Figure 8B shows that 4 small molecules with the highest absolute CMap scores were eventually identified as therapeutic agents for IRI after lung transplantation. Their detailed information, including targeted genes, MOA, and clinical phase were shown in Table 2.
 |
Table 2 Detailed Information of the 4 Small Molecules with the Highest Absolute CMap Scores |
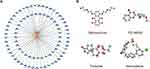 |
Figure 8 Construction of TF-Target Gene Regulatory Networks and Identification of small molecular therapeutic agents. (A) TF-target gene regulatory network of DEGs. (B) 3D conformers of the top 4 candidate therapeutic agents with the highest absolute CS scores. The DEGs and Broad Institutes Connectivity Map (cMAP) database (https://portals.broadinstitute.org/cmap) were used for the identification of small molecules. The threshold CS score chosen was set at < −95. Molecules displaying the most elevated absolute CS scores indicate gene expression pattern that stand in opposition to the specific expression patterns observed in IRI. This observation implies their promising therapeutic potential for mitigating IRI subsequent to lung transplantation. |
Baseline Characteristics of Lung Transplant Patients
The baseline characteristics of patients enrolled in this study are shown in Table 3. No differences were observed between groups concerning age, sex, body mass index (BMI), transplant diagnosis, pretransplant mean pulmonary artery pressures (mPAP), or transplant type. Patients with a PGD grade of 2 or more exhibited longer durations of mechanical ventilation compared to non-PGD patients.
 |
Table 3 Preoperative Characteristics of Lung Transplant Patients Stratified by Subsequent Development of Grade 2 or Above Primary Graft Dysfunction |
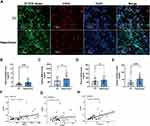
Plasma NET Formation Increased in Ischemia-Reperfusion During Lung Transplantation
Neutrophil immunofluorescence staining and plasma dsDNA quantification were employed to confirm the occurrence of NETs formation during ischemia-reperfusion in lung transplantation. The immunofluorescence image showed that NET component H3cit colocalized to extracellular chromatin fibers of neutrophils in reperfusion groups but hardly in CIT groups (Figure 9A). The levels of plasma dsDNA were significantly higher in reperfusion groups (Figure 9B). These results confirmed NETs formation in ischemia-reperfusion during lung transplantation. Furthermore, compared with non-PGD patients, both pre-reperfusion and 12 hours after reperfusion levels of plasma dsDNA in patients with PGD grade 2 or more were significantly higher (Table 4).
 |
Table 4 Mean MMP9, PADI4, dsDNA, and S100A12 Expression Levels in Sera of Lung Transplant Patients Stratified by Development of Grade 2 or Above Primary Graft Dysfunction |
Validation of the Hub Gene Expression in Clinical Samples via ELISA
To verify the changes in the expression levels of hub genes during the process of lung ischemia-reperfusion, we detected the expression of MMP9, PADI4, and S100A12 in plasma through ELISA. The results showed that MMP9, PADI4, and S100A12 expression was significantly upregulated after reperfusion in total of 24 patients underwent lung transplant (Figure 9C–E). Specifically, among 12 patients with PGD 2 grade or more, a significant increase of PADI4 and S100A12 expression was observed, whereas the MMP9 and S100A12 levels in plasma increased among 12 non-PGD patients after lung transplant. In addition, MMP9 and S100A12 had higher pre-reperfusion levels in patients with PGD 2 grade or more and only 12-hours S100A12 levels are significantly higher in patients with PGD 2 grade or above compared with non-PGD patients. Changes in dsDNA correlated with the changes in MMP9 and S100A12, yet no significant correlation was found between changes in dsDNA and PADI4 (Figure 9F–H).
Discussion
In the current study, we identified four novel NET-related biomarkers during lung ischemia-reperfusion through transcriptome analysis, and further validated their expression in clinical samples. NETs, considered to be web-like structures composed of DNA and granule proteins released after cell death, were found induced in mouse models of pulmonary IRI and human lung allograft after transplantation, with the pathogenicity in PGD.16,17 NETs released from activated neutrophils contribute to inflammatory process and already formed NETs further recruit neutrophils, creating a feedforward cycle.36 Elevated perioperative NETs biomarkers, including citrullinated H3R8 nucleosome histones and circulating H3.1 nucleosomes, indirectly measuring NET formation, have been demonstrated to be related to PGD grade 3.16,37 Considering the particular sensitivity to DNAse-1 mediated digestion,38 researchers attempted to attain the breakdown of NETs by DNAse-1 to restore lung function. Despite the rapidly degraded NETs in allograft by DNAse-1 treatment, the consequential release of NET fragments contributes to consistent interactions between infiltrating CD4+ T cells and donor‐derived antigen presenting cells as well as the activation of toll‐like receptor (TLR)-myeloid differentiation factor 88 (MyD88) signaling pathways, which subsequently promotes human alveolar macrophage inflammatory cytokine production.39 A recent study showed the use of a cytokine adsorber during lung transplantation reduced NETs and the incidence of PGD at 1-month post-transplant, indicating the availability of NETs reduction in alleviating PGD.37 Nevertheless, gene-targeted blocking of NET formation presents another highly valuable approach in mitigating lung IRI.
Further analysis via LASSO and RF identified 4 key genes significantly associated with NET formation: FCAR, MMP9, PADI4, and S100A12. All of these key genes upregulated in the lung allograft 2h after reperfusion, indicating the initiation of the programmed NETosis launched in the early phase after reperfusion. Elevated plasma dsDNA levels in lung transplant patients indicated abundant NET formation, with severe NETs formation observed in patients with PGD 2 grade or above. Interestingly, higher pre-reperfusion dsDNA levels were observed in patients with a PGD grade of 2 or higher, indicating a potentially elevated pre-transplant NETosis degree and inflammatory status correlating with PGD incidence.
In addition, we applied CMap database to identify distinct small molecules effective in IRI. Mitoxantrone, approved for treating aggressive relapsing and progressive multiple sclerosis, is a topoisomerase inhibitor exhibiting anti-inflammatory and antioxidant properties.40 PD-98059, an inhibitor of the MAPK/ERK signaling pathway, plays a pivotal role in cell survival and inflammation regulation.41 PD-98059 confers a protection against ERK-mediated apoptosis.42 Nevertheless, its involvement in NETosis remains ambiguous. Triptolide, derived from Thunder God Vine, is recognized for its anti-inflammatory, immunosuppressive, and antioxidative properties.43 Vemurafenib is a B-RAF inhibitor primarily used for treating malignant melanoma.44 Although its primary application is in cancer therapy, studies indicate the involvement of the B-RAF pathway in inflammation and oxidative stress.45 IRI in lung transplantation represents a multifaceted and intricate pathology, characterized by the intricate interplay of oxidative stress, metabolic disruptions, immune activation, and cellular demise.46 Hence, our discoveries emphasize the potential benefits of targeting specific pathways or receptors to regulate immune cell function and mitigate inflammation in various aspects of lung transplantation.
FCAR, encoding FcαRI (CD89), the human Fc receptor for IgA, is highly expressed on neutrophils.47 Previous studies demonstrated that FCAR shapes neutrophil response and innate immunity, inducing rapid NET formation and NETosis by IgA-opsonized particles in the presence of bacteria.48 Additionally, a study demonstrated the upregulation of FcαRI surface expression in neutrophils following exposure to inflammatory mediators like GM-CSF, the TLR4 agonist LPS, and the cytokines G-CSF and TNF-α,49 which is consistent with the upregulation of FCAR after reperfusion during lung transplantation in our current study. However, Wehrli et al previously reported that FcαRI induces distinct forms of neutrophil death, depending on the inflammatory microenvironment. FcαRI-mediated neutrophil death is apoptotic in the absence of bacteria, but under GM-CSF or LPS stimulation it still includes the partially nonapoptotic death, involved caspases, ROS, PI3K, and MAPKs.49 The observed upregulation of FCAR in our study suggests a potential association with neutrophil activity, including NETs formation. Further research is necessary to elucidate the role of FCAR in neutrophil response and NETs formation during the sterile inflammation of IRI after lung transplantation.
S100 calcium-binding protein A12 (S100A12), a member of the S100 family of EF-hand calcium-binding proteins, is predominantly expressed and secreted by activated neutrophils, accounting for approximately 5% of the total amount of cytosolic proteins in neutrophil granulocytes.50 Extracellular S100A12 shows cytokine-like characteristics and serum S100A12 serves as a marker of various inflammatory diseases.51–53 Various clinical evidence demonstrated that S100A12 can be markedly upregulated in diverse ischemic inflammatory conditions, suggesting its potential as a sensitive and specific diagnostic and prognostic marker of ischemic diseases.54,55 Hongxing Lei recently found that activation of S100A12 was observed in COVID-19 patients with its activation levels correlated with COVID-19 severity based on transcriptome datasets derived from the blood of COVID-19 patients.56 PGD shares similar pathological features as COVID-19, characterized by uncontrolled inflammatory response leading to diffuse alveolar injury and progressive hypoxemia. Consistent with COVID-19 results, our transcriptome results indicated a significant upregulation of S100A12 expression following lung ischemia-reperfusion, as well as the elevated S100A12 plasma levels associated with PGD progression. Mechanistically, S100A12 interacts with the multiligand receptor for advanced glycation end products (RAGE) and toll-like receptor 4 (TLR4), pivotal in activating inflammatory cells like macrophages and lymphocytes, thereby promoting inflammation.57,58 Animal models showed that S100A12 promotes inflammation and cell apoptosis in sepsis-induced ARDS via activation of NLRP3 inflammasome signaling.59 Zhang et al demonstrated that S100A12 promotes inflammation and apoptosis in ischemia/reperfusion injury via ERK signaling through in vitro experiments.60 These findings suggest a potential role for S100A12 in lung ischemia-reperfusion, and plasma levels might serve as biomarkers of PGD progression, requiring further mechanistic investigation.
Proteins of the matrix metalloproteinase (MMP) family can degrade almost all extracellular matrix (ECM) components derived from inactive precursor zymogens.61 MMP9, a representative member of MMPs, degrades the ECM components of the basement membrane to allow neutrophil migration in the early phase of IRI. Yano et al observed elevated MMP9 activity and gene expression in IRI using rat lung transplantation models consistent with our findings in human lung transplantation transcriptome analysis, as well as a decreased level of tissue inhibitors of metalloproteinase (TIMP)-1, which is ubiquitous natural inhibitor and form complexes with MMP-9.62 The MMP9 mRNA to TIMP-1 mRNA ratio peaked in the early reperfusion phase, notably higher than in other phases, suggesting its potential as an early biomarker for IRI.63 In addition, increased activity of Caspase-1 and IL-1β were observed after IRI, and the lung pyroptosis and injury alleviated when treatment the mice with MMP9 selective inhibitor SB-3CT, suggesting MMP9 involvement in IRI after lung transplant, potentially associated with promoting lung pyroptosis.64 The transcriptome analysis in this study showed elevated MMP9 mRNA levels after reperfusion, and we observed a significant difference in MMP9 plasma levels between pre- and post-transplant of total 24 lung transplant patients. Nonetheless, no significant peri-reperfusion differences were observed in patients with PGD 2 grade or above, possibly due to the limited sample volume. The correlation analysis showed the changes in plasma MMP9 levels were correlated with NET formation, but the specific mechanism in PGD requires further research.
Peptidyl arginine deiminase 4 (PAD4), encoded by PADI4 and predominantly expressed in granulocytes, is a crucial post-translational modifying enzyme that converts arginine residues into citrulline residues in the presence of calcium ions. It has been demonstrated that PAD4-mediated histone citrullination via converting arginine to citrulline is the prerequisite and trigger for NETs formation. This process reduces charge-based interactions with DNA, promoting chromatin decondensation.65 The observed positive correlation of plasma PAD4 levels and plasma dsDNA levels in our study future indicates the potential relevance between PAD4 and NETs. NET formation involves two mechanisms: neutrophil elastase and other proteases in granules possibly cleaving histones, and PAD4 converting arginine to citrulline, leading to histone release from DNA, essential for NET release.66 Studies have revealed that efficient DNA decondensation, nuclear lamin meshwork, and nuclear envelope rupture and extracellular DNA release in NETosis require PAD4 enzymatic activity and nuclear localization signal.67 Moreover, animal studies showed that PADI4 deficiency could ameliorate NET formation and inflammatory response induced by NETs.68,69 Our transcriptome analysis revealed a significant increase in PADI4 expression, particularly in patients with PGD 2 grade or more, and the upregulation of PADI4 may contribute to the elevated NET formation. Nevertheless, our results indicate no significant alteration in PADI4 expression levels after reperfusion in non-PGD patients. Correlation analysis showed a deficient correlation between PADI4 and dsDNA levels, possibly influenced by the baseline status in lung transplant patients. Further multi-central and high-volume clinical research could resolve this issue. In addition, higher post-reperfusion PADI4 expression in plasma of patients with PGD grade 2 or more compared with non-PGD patients indicates that NET formation is a vital segment of the pathogenesis of IRI in PGD. Therefore, prevention of NET formation by inhibition of PADI4 to block histone release maybe an attractive strategy to prevent NET‐induced tissue damage in lung transplantation.
However, our study still has a few limitations. First, the limited sample size in our validation study might restrict the generalizability of the results. Therefore, larger-sample studies and functional studies of key genes in lung IRI are warranted. Besides, the final hub genes just were validated at the protein level and a PCR validated at the gene level using allograft tissue is required. Moreover, in addition to clinical samples, animal models of IRI and lung transplantation are necessary for the further validation of final hub genes. Future research requires further exploration of the mechanisms underlying these hub genes based on larger size clinical samples and animal models. Lastly, the NETs related genes were based on current literature, requiring continuous exploration and updates. The limitations of our research also require attention.
Our research provides evidence for peripheral blood biomarkers to predict inflammatory injury after lung transplantation. More importantly, peripheral blood samples are easily obtainable, repeatable, and minimally invasive for lung transplant patients. The results of the current study may catalyze further exploration of immune-associated mechanisms in lung transplantation in the future. With the utilization of bulk and single-cell RNA sequencing in broadening research as the decreasing cost, more specific factors which remarkably affect transplant outcomes will be allowed to identify.
Conclusion
In conclusion, we screened out four final hub genes as NETs-related biomarkers of IRI during lung transplantation based on WGCNA and machine learning algorithms. S100A12, PADI4, and MMP9 were validated to be highly expressed after reperfusion via clinical samples, and have been further confirmed to be related with the development of PGD. Our study provides novel insight into IRI during lung transplantation at the immune and molecular levels; nevertheless, the potential NETs related biomarkers identified in our study require more laboratory data for further validation.
Abbreviations
IRI, ischemia reperfusion injury; PGD, primary graft dysfunction; NETs, neutrophil extracellular traps; DEGs, differentially expressed genes; WGCNA, weighted gene co-expression network analysis; LASSO, least absolute shrinkage and selection operator; RF, random forest; FDR, false discovery rate; GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; BP, biological process; CC, cellular components; MF, molecular function; MM, membership values; GS, gene significance values; ROC, receiver operating characteristic; AUC, area under the curve; CI, confidence interval; DCA, decision analysis; TFs, transcription factors; CMap, connectivity map; CS, connectivity score; MOA, mechanism of action; ECLS, extracorporeal life support; CitH3, citrullinated histone H3; PPI, protein–protein interaction; BMI, body mass index; mPAP, mean pulmonary artery pressures; LAM, lymphangioleiomyomatosis; COPD, chronic obstructive pulmonary dysfunction; ILD, interstitial lung disease; IPF, idiopathic pulmonary fibrosis; MV, mechanical ventilation; CIT, cold ischemia time; SD, standard deviations; IQR, interquartile ranges; TLR, toll‐like receptor; MyD88, myeloid differentiation factor 88; S100A12, S100 calcium-binding protein A12; RAGE, receptor for advanced glycation end products; TLR4, toll-like receptor 4; MMP, matrix metalloproteinase; ECM, extracellular matrix; PAD4, peptidyl arginine deiminase 4.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author and GEO database (https://www.ncbi.nlm.nih.gov/geo).
Ethics Approval and Consent to Participate
The study was approved by the Research Ethics Commission of Shanghai Pulmonary Hospital (No. K22-267Z). The use of recipient peripheral blood conformed to the guidelines of the Declaration of Helsinki. Written informed consent was obtained from the enrolled recipients or their representatives in case their physical state did not permit before surgery, such as coma, extracorporeal circulation, or mechanical ventilation. The donated organs in this study were obtained from listed donors of brain death (DBD) and/or cardiac death (DCD) through the professional Organ Procurement Organization (OPO). All of the organs were donated voluntarily with written informed consent, and this was conducted in accordance with the Declaration of Istanbul.
Acknowledgments
The authors gratefully acknowledge the data provided by patients and researchers participating in GEO. Jiameng Gao, Zhiyuan Zhang, and Jing Yu are co-first authors for this study. Qingqing Zhang and Zongmei Wen are co-correspondence authors for this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China [grant numbers: 82170107], Shanghai Municipal Health Commission (202140406), Excellent Subject Leader Program of Shanghai Municipal Health Commission (2022XD007), and Foundation of the Science and Technology Commission of Shanghai Municipality (19ZR1443000, 23ZR1453300).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Schaffer JM, Singh SK, Reitz BA, Zamanian RT, Mallidi HR. Single- vs double-lung transplantation in patients with chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis since the implementation of lung allocation based on medical need. JAMA. 2015;313(9):936–948. doi:10.1001/jama.2015.1175
2. Lahzami S, Bridevaux PO, Soccal PM, et al. Survival impact of lung transplantation for COPD. Eur Respir J. 2010;36(1):74–80. doi:10.1183/09031936.00087809
3. Toronto Lung Transplant Group*. Unilateral lung transplantation for pulmonary fibrosis. New Engl J Med. 1986;314(18):1140–1145. doi:10.1056/nejm198605013141802
4. Chambers DC, Cherikh WS, Harhay MO, et al. The International Thoracic Organ Transplant Registry of the International Society for heart and lung transplantation: thirty-sixth adult lung and heart-lung transplantation report-2019; focus theme: donor and recipient size match. J Heart Lung Transplant. 2019;38(10):1042–1055. doi:10.1016/j.healun.2019.08.001
5. Shepherd HM, Gauthier JM, Li W, Krupnick AS, Gelman AE, Kreisel D. Innate immunity in lung transplantation. J Heart Lung Transplant. 2021;40(7):562–568. doi:10.1016/j.healun.2021.03.017
6. Capuzzimati M, Hough O, Liu M. Cell death and ischemia-reperfusion injury in lung transplantation. J Heart Lung Transplant. 2022;41(8):1003–1013. doi:10.1016/j.healun.2022.05.013
7. Chen-Yoshikawa TF. Ischemia-reperfusion injury in lung transplantation. Cells. 2021;10(6). doi:10.3390/cells10061333
8. Grimm JC, Valero V, Kilic A, et al. Association between prolonged graft ischemia and primary graft failure or survival following lung transplantation. JAMA Surgery. 2015;150(6):547–553. doi:10.1001/jamasurg.2015.12
9. Kreisel D, Krupnick AS, Puri V, et al. Short- and long-term outcomes of 1000 adult lung transplant recipients at a single center. J Thoracic Cardiovasc Surg. 2011;141(1):215–222. doi:10.1016/j.jtcvs.2010.09.009
10. Li W, Terada Y, Tyurina YY, et al. Necroptosis triggers spatially restricted neutrophil-mediated vascular damage during lung ischemia reperfusion injury. Proc Natl Acad Sci USA. 2022;119(10):e2111537119. doi:10.1073/pnas.2111537119
11. Meyer KC, Nunley DR, Dauber JH, et al. Neutrophils, Unopposed Neutrophil Elastase, and Alpha 1-Antiprotease defenses following human lung transplantation. Am J Respir Crit Care Med. 2001;164(1):97–102. doi:10.1164/ajrccm.164.1.2006096
12. Gelman AE, Fisher AJ, Huang HJ, et al. Report of the ISHLT Working Group on Primary Lung Graft Dysfunction Part III: mechanisms: a 2016 Consensus Group Statement of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2017;36(10):1114–1120. doi:10.1016/j.healun.2017.07.014
13. Jorch SK, Kubes P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nature Med. 2017;23(3):279–287. doi:10.1038/nm.4294
14. Campos J, Ponomaryov T, De Prendergast A, et al. Neutrophil extracellular traps and inflammasomes cooperatively promote venous thrombosis in mice. Blood Adv. 2021;5(9):2319–2324. doi:10.1182/bloodadvances.2020003377
15. Sharma S, Hofbauer TM, Ondracek AS, et al. Neutrophil extracellular traps promote fibrous vascular occlusions in chronic thrombosis. Blood. 2021;137(8):1104–1116. doi:10.1182/blood.2020005861
16. Bonneau S, Landry C, Bégin S, et al. Correlation between Neutrophil Extracellular Traps (NETs) expression and primary graft dysfunction following human lung transplantation. Cells. 2022;11(21). doi:10.3390/cells11213420
17. Sayah DM, Mallavia B, Liu F, et al. Neutrophil extracellular traps are pathogenic in primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med. 2015;191(4):455–463. doi:10.1164/rccm.201406-1086OC
18. Baciu C, Shin J, Hsin M, Cypel M, Keshavjee S, Liu M. Altered purine metabolism at reperfusion affects clinical outcome in lung transplantation. Thorax. 2023;78(3):249–257. doi:10.1136/thoraxjnl-2021-217498
19. Wong A, Zamel R, Yeung J, et al. Potential therapeutic targets for lung repair during human ex vivo lung perfusion. Eur Respir J. 2020;55(4). doi:10.1183/13993003.02222-2019
20. Kang CH, Anraku M, Cypel M, et al. Transcriptional signatures in donor lungs from donation after cardiac death vs after brain death: a functional pathway analysis. J Heart Lung Transplant. 2011;30(3):289–298. doi:10.1016/j.healun.2010.09.004
21. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Databaseissue):D991–D995. doi:10.1093/nar/gks1193
22. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
23. Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi:10.1093/bioinformatics/bts034
24. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
25. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nature Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
26. Hu K. Become competent within one day in generating boxplots and violin plots for a novice without prior R experience. Meth Protocols. 2020;3(4). doi:10.3390/mps3040064
27. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi:10.1093/nar/gky1131
28. Yang C, Delcher C, Shenkman E, Ranka S. Machine learning approaches for predicting high cost high need patient expenditures in health care. Biomed Eng Online. 2018;17(Suppl 1):131. doi:10.1186/s12938-018-0568-3
29. Ellis K, Kerr J, Godbole S, Lanckriet G, Wing D, Marshall S. A random forest classifier for the prediction of energy expenditure and type of physical activity from wrist and Hip accelerometers. Physiol Meas. 2014;35(11):2191–2203. doi:10.1088/0967-3334/35/11/2191
30. Blanco JL, Porto-Pazos AB, Pazos A, Fernandez-Lozano C. Prediction of high anti-angiogenic activity peptides in silico using a generalized linear model and feature selection. Sci Rep. 2018;8(1):15688. doi:10.1038/s41598-018-33911-z
31. Alderden J, Pepper GA, Wilson A, et al. Predicting pressure injury in critical care patients: a machine-learning model. Am J Crit Care. 2018;27(6):461–468. doi:10.4037/ajcc2018525
32. Wu J, Zhang F, Zheng X, et al. Identification of renal ischemia reperfusion injury subtypes and predictive strategies for delayed graft function and graft survival based on neutrophil extracellular trap-related genes. Front Immunol. 2022;13(1047367). doi:10.3389/fimmu.2022.1047367
33. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12(77). doi:10.1186/1471-2105-12-77
34. Snell GI, Yusen RD, Weill D, et al. Report of the ISHLT Working Group on Primary Lung Graft Dysfunction, part I: definition and grading-A 2016 Consensus Group statement of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2017;36(10):1097–1103. doi:10.1016/j.healun.2017.07.021
35. Van Raemdonck D, Hartwig MG, Hertz MI, et al. Report of the ISHLT Working Group on primary lung graft dysfunction Part IV: prevention and treatment: a 2016 Consensus Group statement of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2017;36(10):1121–1136. doi:10.1016/j.healun.2017.07.013
36. Merza M, Hartman H, Rahman M, et al. Neutrophil extracellular traps induce trypsin activation, inflammation, and tissue damage in mice with severe acute pancreatitis. Gastroenterology. 2015;149(7):1920–1931.e1928. doi:10.1053/j.gastro.2015.08.026
37. Lindstedt S, Niroomand A, Mittendorfer M, et al. Nothing but NETs: cytokine adsorption correlates with lower circulating nucleosomes and is associated with decreased primary graft dysfunction. J Heart Lung Transplant. 2023. doi:10.1016/j.healun.2023.06.011
38. Hakkim A, Fürnrohr BG, Amann K, et al. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc Natl Acad Sci USA. 2010;107(21):9813–9818. doi:10.1073/pnas.0909927107
39. Scozzi D, Wang X, Liao F, et al. Neutrophil extracellular trap fragments stimulate innate immune responses that prevent lung transplant tolerance. Am J Transplant. 2019;19(4):1011–1023. doi:10.1111/ajt.15163
40. Parker C, Waters R, Leighton C, et al. Effect of mitoxantrone on outcome of children with first relapse of acute lymphoblastic leukaemia (ALL R3): an open-label randomised trial. Lancet. 2010;376(9757):2009–2017. doi:10.1016/S0140-6736(10)62002-8
41. Pathania AS, Joshi A, Kumar S, et al. Reversal of boswellic acid analog BA145 induced caspase dependent apoptosis by PI3K inhibitor LY294002 and MEK inhibitor PD98059. Apoptosis. 2013;18(12):1561–1573. doi:10.1007/s10495-013-0889-4
42. Dhanalakshmi S, Agarwal C, Singh RP, Agarwal R. Silibinin up-regulates DNA-protein kinase-dependent p53 activation to enhance UVB-induced apoptosis in mouse epithelial JB6 cells. J Biol Chem. 2005;280(21):20375–20383.
43. Patil S, Lis LG, Schumacher RJ, et al. Phosphonooxymethyl prodrug of triptolide: synthesis, physicochemical characterization, and efficacy in human colon adenocarcinoma and ovarian cancer xenografts. J Med Chem. 2015;58(23):9334–9344. doi:10.1021/acs.jmedchem.5b01329
44. Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467(7315):596–599. doi:10.1038/nature09454
45. Eder SK, Schwentner R, Ben Soussia P, et al. Vemurafenib acts as a molecular on-off switch governing systemic inflammation in Langerhans cell histiocytosis. Blood Adv. 2022;6(3):970–975. doi:10.1182/bloodadvances.2021005442
46. den Hengst WA, Gielis JF, Lin JY, Van Schil PE, De Windt LJ, Moens AL. Lung ischemia-reperfusion injury: a molecular and clinical view on a complex pathophysiological process. Am j Physiol Heart Circulatory Physiol. 2010;299(5):H1283–H1299. doi:10.1152/ajpheart.00251.2010
47. Herr AB, Ballister ER, Bjorkman PJ. Insights into IgA-mediated immune responses from the crystal structures of human FcalphaRI and its complex with IgA1-Fc. Nature. 2003;423(6940):614–620. doi:10.1038/nature01685
48. Aleyd E, van Hout MW, Ganzevles SH, et al. IgA enhances NETosis and release of neutrophil extracellular traps by polymorphonuclear cells via Fcα receptor I. J Immunol. 2014;192(5):2374–2383. doi:10.4049/jimmunol.1300261
49. Wehrli M, Cortinas-Elizondo F, Hlushchuk R, et al. Human IgA Fc receptor FcαRI (CD89) triggers different forms of neutrophil death depending on the inflammatory microenvironment. J Immunol. 2014;193(11):5649–5659. doi:10.4049/jimmunol.1400028
50. Vogl T, Pröpper C, Hartmann M, et al. S100A12 is expressed exclusively by granulocytes and acts independently from MRP8 and MRP14. J Biol Chem. 1999;274(36):25291–25296. doi:10.1074/jbc.274.36.25291
51. Pietzsch J, Hoppmann S. Human S100A12: a novel key player in inflammation? Amino Acids. 2009;36(3):381–389. doi:10.1007/s00726-008-0097-7
52. Jiang X, Huang CM, Feng CM, Xu Z, Fu L, Wang XM. Associations of serum S100A12 with severity and prognosis in patients with community-acquired pneumonia: a prospective cohort study. Front Immunol. 2021;12(714026). doi:10.3389/fimmu.2021.714026
53. Li Y, He Y, Chen S, et al. S100A12 as biomarker of disease severity and prognosis in patients with idiopathic pulmonary fibrosis. Front Immunol. 2022;13(810338). doi:10.3389/fimmu.2022.810338
54. Amini H, Knepp B, Rodriguez F, et al. Early peripheral blood gene expression associated with good and poor 90-day ischemic stroke outcomes. J Neuroinflammation. 2023;20(1):13. doi:10.1186/s12974-022-02680-y
55. Wakisaka Y, Ago T, Kamouchi M, et al. Plasma S100A12 is associated with functional outcome after ischemic stroke: research for biomarkers in ischemic stroke. J Neurol Sci. 2014;340(1–2):75–79. doi:10.1016/j.jns.2014.02.031
56. Lei H. A two-gene marker for the two-tiered innate immune response in COVID-19 patients. PLoS One. 2023;18(1):e0280392. doi:10.1371/journal.pone.0280392
57. Foell D, Wittkowski H, Kessel C, et al. Proinflammatory S100A12 can activate human monocytes via Toll-like receptor 4. Am J Respir Crit Care Med. 2013;187(12):1324–1334. doi:10.1164/rccm.201209-1602OC
58. Ingels C, Derese I, Wouters PJ, Van den Berghe G, Vanhorebeek I. Soluble RAGE and the RAGE ligands HMGB1 and S100A12 in critical illness: impact of glycemic control with insulin and relation with clinical outcome. Shock. 2015;43(2):109–116. doi:10.1097/shk.0000000000000278
59. Zhang Z, Han N, Shen Y. S100A12 promotes inflammation and cell apoptosis in sepsis-induced ARDS via activation of NLRP3 inflammasome signaling. Mol Immunol. 2020;122:38–48. doi:10.1016/j.molimm.2020.03.022
60. Zhang X, Shen R, Shu Z, Zhang Q, Chen Z. S100A12 promotes inflammation and apoptosis in ischemia/reperfusion injury via ERK signaling in vitro study using PC12 cells. Pathol Int. 2020;70(7):403–412. doi:10.1111/pin.12924
61. Hübner RH, Meffert S, Mundt U, et al. Matrix metalloproteinase-9 in bronchiolitis obliterans syndrome after lung transplantation. Eur Respir J. 2005;25(3):494–501. doi:10.1183/09031936.05.00091804
62. Gomez DE, Alonso DF, Yoshiji H, Thorgeirsson UP. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur J Cell Biol. 1997;74(2):111–122.
63. Yano M, Omoto Y, Yamakawa Y, et al. Increased matrix metalloproteinase 9 activity and mRNA expression in lung ischemia-reperfusion injury. J Heart Lung Transplant. 2001;20(6):679–686. doi:10.1016/s1053-2498(01)00250-9
64. Zhou P, Song NC, Zheng ZK, Li YQ, Li JS. MMP2 and MMP9 contribute to lung ischemia-reperfusion injury via promoting pyroptosis in mice. BMC Pulm Med. 2022;22(1):230. doi:10.1186/s12890-022-02018-7
65. Wang Y, Li M, Stadler S, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009;184(2):205–213. doi:10.1083/jcb.200806072
66. Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. 2010;191(3):677–691. doi:10.1083/jcb.201006052
67. Thiam HR, Wong SL, Qiu R, et al. NETosis proceeds by cytoskeleton and endomembrane disassembly and PAD4-mediated chromatin decondensation and nuclear envelope rupture. Proc Natl Acad Sci USA. 2020;117(13):7326–7337. doi:10.1073/pnas.1909546117
68. Suzuki M, Ikari J, Anazawa R, et al. PAD4 deficiency improves bleomycin-induced neutrophil extracellular traps and fibrosis in mouse lung. Am J Respir Cell Mol Biol. 2020;63(6):806–818. doi:10.1165/rcmb.2019-0433OC
69. Biron BM, Chung CS, Chen Y, et al. PAD4 deficiency leads to decreased organ dysfunction and improved survival in a dual insult model of hemorrhagic shock and sepsis. J Immunol. 2018;200(5):1817–1828. doi:10.4049/jimmunol.1700639
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.