Back to Journals » Journal of Inflammation Research » Volume 14
Identification of Key Candidate Genes Related to Inflammatory Osteolysis Associated with Vitamin E-Blended UHMWPE Debris of Orthopedic Implants by Integrated Bioinformatics Analysis and Experimental Confirmation
Authors Liu F , Dong J, Zhou D, Zhang Q
Received 25 May 2021
Accepted for publication 12 July 2021
Published 26 July 2021 Volume 2021:14 Pages 3537—3554
DOI https://doi.org/10.2147/JIR.S320839
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Video abstract of "DEGs in macrophages exposed to UHMWPE or VE-UHMWPE particles" [ID 320839].
Views: 88
Fanxiao Liu, Jun Dong, Dongsheng Zhou, Qingyu Zhang
Department of Orthopaedics, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong, People’s Republic of China
Correspondence: Qingyu Zhang
Department of Orthopaedics, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong, People’s Republic of China
Tel/Fax +86-0531-68773201
Email [email protected]
Purpose: This study aims to identify differentially expressed genes (DEGs) in macrophages exposed to ultra-high-molecular-weight polyethylene (UHMWPE) or vitamin E-blended UHMWPE (VE-UHMWPE) particles, thereby providing potential targets for the treatment of inflammatory osteolysis.
Methods: The GSE104589 dataset of genome expression in macrophages exposed to UHMWPE and VE-UHMWPE was downloaded from the Gene Expression Omnibus database to identify DEGs. Functional enrichment analysis was performed using DAVID, and the corresponding protein–protein interaction (PPI) network was constructed from the STRING database. Important modules were selected using the molecular complex detection algorithm, and hub genes were identified in cytoHubba. MicroRNAs targeting these DEGs were obtained from the TarBase, miRTarBase, and miRecords databases, while transcription factors (TFs) targeting DEGs were predicted from the ENCODE database. Finally, the top five DEGs were validated by quantitative real‐time polymerase chain reaction (qRT-PCR).
Results: A total of 112 DEGs (44 upregulated and 68 downregulated DEGs) were screened. Immune and inflammatory responses were significantly related in gene ontology analysis, and 18 signaling pathways were enriched according to Kyoto Encyclopedia of Genes and Genomes pathway analysis. The PPI network involving 85 nodes and 266 protein pairs indicated that IL1β, CXCL1, ICAM1, CCL5 and CCL4 showed higher degrees. qRT-PCR analysis of the top five DEGs revealed a decreasing trend in the VE-UHMWPE group compared with the UHMWPE group. Key microRNAs (hsa-miR-144, hsa-miR-21, and hsa-miR-221) and TFs (RELA and NFKB1) were predicted to be correlated with the pathogenesis of inflammatory osteolysis through microRNA-TF regulatory network analysis.
Conclusion: The present study helps shed light on the molecular mechanisms underlying the changes in the wear-induced inflammatory process after blending vitamin E with UHMWPE. Hub genes including IL1β, CXCL1, ICAM1, CCL5, and CCL4, key microRNAs (hsa-miR-144, hsa-miR-21, and hsa-miR-221) and TFs (RELA and NFKB1) may serve as prognostic and therapeutic targets of inflammatory osteolysis.
Keywords: inflammatory osteolysis, aseptic loosening, bioinformatics analysis, VE-UHMWPE, macrophages
Introduction
While total joint arthroplasty (TJA) is still the gold standard for the treatment of pain and dysfunction in patients with end-stage arthritis and femoral head necrosis,1,2 determining the long-term behavior of implants remains challenging in the scientific community and healthcare service systems due to polyethylene wear, with resultant osteolysis and/or aseptic loosening.3 Revision surgery is more difficult and costly than primary TJAs because of poorer prognosis and higher risk of failure and complications.2–5 Inflammatory osteolysis initiates when macrophages recognize wear debris derived from the polyethylene (PE) liners of TJAs and release a series of cytokines and chemokines that promote the recruitment of other inflammatory cells, leading to the formation of inflamed granulomas.4 Despite advances achieved in pharmacological therapies for inflammation in other skeletal diseases, there are no agents that specifically target periprosthetic inflammatory osteolysis.
A better understanding of the molecular and cellular mechanisms of inflammatory osteolysis in TJA and the development of specific therapies are urgently needed to prevent implant loosening. Attempts such as the introduction of ultra-high-molecular-weight polyethylene (UHMWPE) and cross-linking of UHMWPE using gamma irradiation (HXLPE) have been made to improve the wear resistance of implant bearing materials (acetabular components in total hip arthroplasties and tibial inserts in total knee arthroplasties) and have revealed superior durability in mid-term and long-term follow-up.4 However, the cross-linking process of polyethylene generates free radicals, reducing long-term oxidative stability and enhancing material embrittlement of HXLPE.1,6 To address this issue, the new generation of HXLPE is stabilized with vitamin E (alpha-tocopherol), a natural lipid-soluble antioxidative substance exerting potent anti-inflammatory properties on cells.7 The infusion of vitamin E into UHMWPE could donate hydrogen atoms to free radicals in polyethylene, decrease the production of reactive oxygen species (ROS) and tumor necrosis factor α (TNF-α), and interrupt the chemical reaction cascade between the macromolecules and oxygen.8 Currently, there are two methods of integrating vitamin E into UHMWPE.2 The first is to blend vitamin E with UHMWPE resin powder: after consolidation, the blend can be irradiated for cross-linking.5 However, the presence of vitamin E could reduce the cross-linking efficiency of UHMWPE and a concentration of vitamin E above 0.3% wt (mass fraction) in the irradiated surface would significantly impair wear characteristics.9,10 The second is doping the polyethylene polymers after radiation cross-linking in vitamin E, by which a maximum concentration of approximately 0.7 wt% can be reached on account of the saturation upper limit of the HXLPE.2,5 Bichara and colleagues revealed that fabricated wear particles derived from VE-UHMWPE triggered lesser cellular responses and osteolytic potentials than conventional UHMWPE in in vivo experiments,11 and a series of randomized controlled trials demonstrated lower wear rates in HXLPE/VitE cups than in HXLPE or UHMWPE cups in mid- or long-term follow-ups.1,5,7,12 However, some investigators obtained negative results for these two materials.12–14 Until now, whether the addition of vitamin E exerts a protective effect on the bearing materials of arthroplasties remained to be confirmed.
Given that macrophages are the first immune cells encountering wear particles, we attempted to analyze the genomic mechanisms of human macrophages after exposure to VE-UHMWPE and conventional UHMWPE particles. Notably, Terkawi and his colleagues originally submitted the GSE104589 dataset to the Gene Expression Omnibus (GEO) database. They screened correlated gene expression in human macrophages exposed to VE-UHMWPE and UHMWPE particles, demonstrating the involvement of IL-27 in the macrophage response to VE-UHMWPE particles and its regulatory role in osteolysis.15 However, they only investigated the function of one transcriptome gene (IL-27),15 and there were still multiple genes that could play vital roles in these processes. Therefore, to identify the differentially expressed genes (DEGs) and correlated pathways in macrophages after exposure to VE-UHMWPE and UHMWPE, and provide a more comprehensive view of the genetic mechanisms in inflammatory osteolysis, in the present study these microarray data were retrieved from the GEO database and reanalyzed by using various bioinformatics technologies and in vitro experiments. Furthermore, the present study constructed a transcription factor (TF)‐DEG network and a microRNA (miRNA)‐DEG network. The associations between the TFs and their targets were predicted by using the Encyclopedia of DNA Elements (ENCODE), while the regulatory associations between miRNAs and their targets were extracted from the TarBase, miRTarBase and miRecords databases. The integration of these two networks was then performed to establish a TF‐microRNA coregulatory network from which key microRNAs and TFs can be identified. Our analysis provided insight into genomics at the cellular level and explored the potential mechanism of inflammatory osteolysis.
Materials and Methods
High-Throughput Gene Expression Data
The high-throughput gene expression profile was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/), under the accession number GSE104589. The GSE104589 dataset, based on GPL16791 (a platform for Illumina HiSeq 2500, Homo sapiens) contained 9 samples, including 3 samples from macrophages exposed to VE-UHMWPE particles, and 3 samples from macrophages exposed to UHMWPE and 3 control samples from macrophages cultured alone, which were all selected for deep analysis in this study.
Data Preprocessing and Identification of DEGs
Based on the annotation information in the platform, the probe sets were transformed into the corresponding gene symbol. The mean value of multiple probe sets was calculated if they corresponded to the same gene symbol. The data were normalized using quantile normalization with Illumina16 in R software (Version 3.6.2) and DEGs were identified using the limma package (Version 3.42.2) in R software. Individual p-values were calculated and converted to adjusted p-values (adj. p. val) for comparisons by false discovery rate correction of the Benjamini and Hochberg test. The cutoff point of adj. p. val < 0.05 and |log2fold change (FC)| >2 were used to select the DEGs. Then, the heatmap and volcano plot of the DEGs were generated using the ggplot2 package in R software.
Functional and Pathway Enrichment Analyses of DEGs
The up- and downregulated DEGs were uploaded to an online biological information database, the Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.8 Beta (https://david‐d.ncifcrf.gov/),17,18 for further analysis, including gene ontology (GO) and Kyoto Encyclopedia of Gene and Genome (KEGG) pathways, which were visualized in the R ggplot2 package. The significance criterion was set at p-val < 0.05.
Protein‐protein Interaction Network Construction and Module Analysis
The PPI network of DEGs was predicted using the multiple protein online tool in the STRING database,19 a search tool for the Retrieval of Interacting Genes database (version 11.0, http://string-db.org) and visualized in Cytoscape software (Version 3.6.2).20,21 The most significant modules in the PPI network were identified and visualized using Molecular Complex Detection (MCODE), which is a tool in Cytoscape to select densely connected areas in the acquired network. The selection criteria were as follows: MCODE scores >5, node score cutoff = 0.2, degree cutoff = 2, max depth = 100 and k‐score = 2. The biological process analysis of the top 20 genes was performed and visualized using the Biological Networks Gene Ontology tool (BiNGO) (version 3.0.3) plugin in Cytoscape.22
Hub Gene Selection
The hub genes (top 10) in the PPI network were identified by Maximal Clique Centrality (MCC), Degree, and Maximum Neighborhood Component (MNC) from the cytoHubba plugin in Cytoscape.
TF‐DEG Network and miRNA‑DEG Network Construction
The targeted TFs of DEGs were predicted using the ENCODE database in NetworkAnalyst (https://www.networkanalyst.ca/faces/home.xhtml), a visual analytics platform for comprehensive gene expression profiling and meta‐analysis. Meanwhile, the microRNA‐DEG pairs were identified using the TarBase, miRTarBase and miRecords databases. The generated pairs of TF‐DEG and miRNA‐DEG were retained for further analysis.
TF‐miRNA Integrated Network Construction
The integrated network of TF-miRNA was constructed according to the TF‐DEG and miRNA‐DEG networks. The coregulated DEGs targeted by miRNAs and TFs were first selected, and subsequent miRNAs and TFs were extracted. Eventually, the integrated regulatory network was constructed and visualized in Cytoscape software (Version 3.6.2).
Preparation of Polyethylene Particles and Macrophage Culture
The materials were prepared from a single batch of medical-grade GUR 1050 resin (Ticona, Bayport, TX, USA). The VE-UHMWPE and UHMWPE particles were first fabricated using the method presented by Oral.23 The human monocyte cell line THP-1 (ATCC; American Type Culture Collection, USA) was seeded in a 6-well plate at a density of 5×105 cells/well and cultured with RPMI-1640 medium (BSigma Aldrich, St. Louis, MO, USA) supplemented with 10% inactivated fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine. Cells were incubated in a humidified atmosphere with 5% CO2 environment at 37°C. The cells were differentiated into macrophages by using 125 ng/well phorbol 12-myristate 13-acetate (PMA) (Sigma Aldrich, St. Louis, MO, USA) for 48 h. Then the cells were cultured with free-endotoxin VE-UHMWPE or UHMWPE particles at a ratio of 1:4000 μm3 for 24 h using an inverted cell culture technique that allows particles to directly interact with macrophages. Control macrophages were cultured alone.
Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT–PCR)
Purified 0.5 mg DNA-free RNA samples were reverse transcribed using a One Step PrimeScript® miRNA cDNA Synthesis Kit (Takara, Japan, D350A), and the amplification reaction was conducted using a SYBR® Premix Ex TaqTM II kit (Takara, Japan, DRR820A). Gene expression of each target was calculated using the ΔΔCT method after normalization to the expression of GAPDH housekeeping genes.
ELISA
Secreted IL-1β, IL-1, IL6 and TNF-α levels in the supernatant of macrophages in different groups were detected using quantitative ELISA kits (R&D Systems Inc. MN, USA) according to each manufacturer’s instruction.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism Software, version 7.0 (GraphPad Software Inc., USA). One-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison procedure was used to compare the gene expression and levels of inflammatory factors. The results are presented as the means ± standard errors (SEM), and a p-value < 0.05 was considered statistically significant.
Results
Data Normalization
Data normalization and cross‐comparability were evaluated via the box plot visualization of the gene expression of all samples and principal component analysis (PCA) for confirming biological variability between different samples. Figure 1 demonstrates that the black lines were almost in the same position after data normalization, indicating an excellent degree of standardization, which can ensure the accuracy of subsequent data processing. Figure 2A shows that all samples involving UHMWPE, VE-UHMWPE, and no particles were grouped separately, indicating globally distinct expression profiles.
 |
Figure 1 The distribution of expression of all samples before and after normalization. |
Identification of DEGs
The high-throughput sequencing data from the GSE104589 dataset were analyzed to select DEGs according to the predefined criteria. In total, 112 DEGs were detected between the VE-UHMWPE group and the UHMWPE group, including 44 upregulated and 68 downregulated DEGs. In addition, a volcano plot and a heatmap of all DEGs were generated using the R ggplot2 package (Figure 2B and C). Meanwhile, 376 DEGs were identified between the UHMWPE group and the control group, including 182 upregulated and 194 downregulated DEGs, and the top 50 genes are listed in Supplementary Table 1.
GO Function and KEGG Enrichment Analysis of DEGs
GO enrichment analysis of 112 DEGs between the VE-UHMWPE group and the UHMWPE group was performed to identify the most relevant biological processes (BPs), molecular functions (MFs), and cellular components (CCs). The top ten enriched terms in BP, CC, and MF are presented in Figure 3A–C. Additionally, based on KEGG pathway analysis, the DEGs were significantly enriched in 18 signaling pathways, such as the chemokine signaling pathway, cytokine–cytokine receptor interaction, rheumatoid arthritis, herpes simplex infection and influenza A, measles, and legionellosis (Figure 3D). The top 20 terms associated with 112 DEGs, enriched in BP, CC and MF are presented in Table 1. GO enrichment analysis and related pathways of 376 DEGs between the UHMWPE group and the control group are presented in Supplementary Figure 1 and Supplementary Table 2. Based on KEGG pathway analysis, the DEGs were significantly enriched in 30 signaling pathways, such as the Toll-like receptor signaling and phagosome pathways.
 |
Table 1 Top 20 Biological Processes of DEGs |
PPI Network Construction and Module Analysis
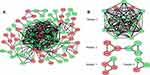
The interactions between the proteins expressed from DEGs, which consisted of 85 nodes and 266 edges (Figure 4A), were constructed from the STRING database and visualized using Cytoscape. In addition, 5 of the top genes with relatively high connectivity degrees (≥20) were IL1β (degree = 23), CXCL1 (degree = 22), ICAM1 (degree = 21), CCL5 (degree = 20), and CCL4 (degree = 20). Four significant modules (Figure 4B) were obtained by module analysis in the PPI network using MCODE from Cytoscape based on the aforementioned criteria. The crucial nodes with a high MCODE score in these four modules were IL1RN, C3AR1, C5AR1, ICAM1, HCAR2, IDO1, CCL5, CXCL9, CXCL1, CCL20, STAT1, CCL4, CCL3, CXCL5, PTGS2, IL1B, and CCL1. The biological process analysis of the top 20 genes was performed and visualized using BiNGO in Cytoscape, which is shown in Figure 5.
Hub Gene Selection
Hub genes were selected by CytoHubba. The top 10 hub genes, which were selected based on the 3 most commonly used classification methods in cytoHubba, are presented in Table 2. By overlapping the first 13 genes, 7 central genes (IL1β, CXCL1, CCL5, CCL4, CCL20, CXCL9 and C3AR1) were consequently identified as presented in Figure 6A. After being marked on selected modules, we observed that the hub genes were downregulated and gathered in module 1 (Figure 6B).
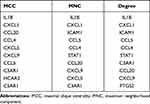 |
Table 2 List of the Top 10 Hub Genes Selected by MCC, MNC and Degree Methods in cytoHubba |
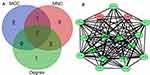 |
Figure 6 Identification of the hub genes. (A) Hub genes were identified by the overlap of the three methods in CytoHubba; (B) the selected hub genes were all included in the most significant module. |
Construction of the TF‐DEG Network Analysis
According to TF binding site data and genetic coordinate position information provided in ENCODE, a potential regulatory network between DEGs and TFs was constructed to analyze the functional roles of selected DEGs. A total of 73 associations between 22 TFs and 13 DEGs were predicted. As shown, KLF16 regulated 5 DEGs (eg, C5AR1 and CCL20), TFDP1 regulated 5 DEGs (eg, IL2RA and THBS1), and NFIC regulated 4 DEGs (eg, CCL3 and CCL44) (Figure 7).
 |
Figure 7 The network of TF-DEG was obtained from the ENCODE database. Abbreviations ENCODE, Encyclopedia of DNA Elements; TF, transcription factor; DEG, differentially expressed gene. |
Construction of miRNA‐DEG Network Analysis
The microRNA‐DEG pairs were identified through network analysis of 20 DEGs using the TarBase, miRTarBase and miRecords databases. Ultimately, a total of 64 associations between 25 microRNAs and only 18 DEGs were identified, and then the network was visualized in Cytoscape (Figure 8). THBS1 regulated 8 interacting microRNAs, hsa-let-7b-5p, hsa-let-7f-5p, hsa-mir-17-5p, hsa-mir-1-3p, hsa-mir-155-5p, hsa-mir-410-3p, hsa-mir-4302 and hsa-mir-2467-3p. A hub microRNA, hsa-mir-335-5p, was predicted to interact with five DEGs, including PTGS2, ICAMI1, CCL20, C5AR1 and CXCL9.
 |
Figure 8 The network of DEG-miRNA was obtained from the tarbase, miRTarBase, and miRecords databases. Abbreviations: DEG, differentially expressed gene; miRNA, microRNA. |
TF‐miRNA Interaction Network
The TF-microRNA interaction network was constructed through network analysis of 20 DEGs in Cytoscape (Figure 9), which included 16 DEGs, 28 TFs, and 13 microRNAs, with 70 associations between the TFs and DEGs and 19 associations between the miRNAs and DEGs. We separately analyzed the degree of 16 DEGs in the TF‐DEG network and the microRNA‐DEG network (Table 3). We found that RELA and NFKB1 regulated six interacting DEGs. Simultaneously, hsa-miR-144, hsa-miR-21, and hsa-miR-221 regulated three interacting DEGs.
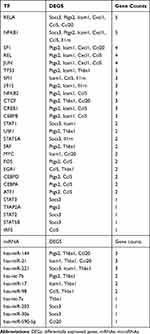 |
Table 3 Co-DEGs Regulated by TF and miRNAs |
 |
Figure 9 Integrative regulatory network of TF-DEG-miRNA. Abbreviations: TF, transcription factor; DEG, differentially expressed gene; miRNA, microRNA. |
Assessment of Polyethylene Particles, qRT-PCR and ELISA
As visualized with a transmission electron microscope, VE-UHMWPE particles were engulfed by macrophages (Figure 10A and B). To further validate the transcriptional RNA-seq data, the expression of 5 hub genes (IL1β, CXCL1, ICAM1, CCL5, and CCL4) with high connectivity degrees (≥20) was examined by qRT-PCR (Figure 10C). Notably, the results of the gene expression examined by qRT-PCR and high-throughput sequencing were highly correlated. Moreover, ELISA revealed a significant increase in the levels of inflammatory factors including IL-1β, IL-2, IL-6 and TNF-α in the supernatant of the UHMWPE group in comparison with the control groups, while the levels of these inflammatory factors were decreased in the VE-UHMWPE group in comparison with the UHMWPE groups (Figure 10D).
Discussion
Bearing in mind that wear particles continuously released from the surfaces of implant-bearing materials could result in local infiltration and activation of inflammatory cells, an ideal approach to minimize periprosthetic osteolysis at the site of implantation should be emphasized to avoid an inflammatory response. As the major source of inflammatory mediators associated with impaired bone metabolism and increased activity of resorptive osteoclasts, macrophages can colonize the site of implantation and initiate an inflammatory cascade.24 Several studies have focused on the characterization of the immune response of macrophages to implant particles in order to understand the molecular mechanisms of inflammatory osteolysis.25–27 Terkawi revealed that the differentiated gene profile of macrophages stimulated by conventional UHMWPE particles mainly involves common gene expression signatures for inflammation and rheumatoid arthritis.15 By using informatics analysis of the gene profile of macrophages stimulated by UHMWPE and VE-UHMWPE particles, it was revealed that VE-UHMWPE triggered reduced biological activity and inflammatory responses in comparison with UHMWPE. A total of 112 DEGs were identified between the two groups, which may aid in advancing our knowledge on regulatory factors with respect to aseptic loosening of orthopedic implants.
Integrated analysis of differential gene expression indicated that the top 10 DEGs (IL1β, CXCL1, CCL20, CCL4, CXCL5, CXCL9, CCL5, C3AR1, HCAR2 and C5AR1) in the PPI network were the hub genes affecting the inflammatory response of macrophages to VE-UHMWPE, all showing a decreasing trend compared to the UHMWPE group. We found that most of these DEGs were chemokines (IL1β, CXCL1, CCL20, CCL4, CXCL5, CXCL9 and CCL5) and three (C3AR1, HCAR2 and C5AR1) were inflammation-related receptors. These findings agreed with the fact that proinflammatory mediators including SPP1, CCL2, CCL3, CCL5 and CCL4 were highly expressed in macrophages cultured with UHMWPE in comparison with the blank control.15 Importantly, large numbers of DEGs identified between the UHMWPE group and the VE-UHMWPE group also serve regulatory roles in osteoclastogenesis and bone resorption.15,28 Some inflammatory diseases are associated with increased bone resorption and fracture rates because inflammatory cytokines produced by innate and adaptive immune cells could not only facilitate inflammation but also activate bone degeneration and inhibit bone formation.29 The degree of inflammation is related to the extent of local and systemic bone loss.30 For instance, chemokines including CCL3, CCL4, CCL5, CXCL9, CCL5 and CCL20 are involved in the remodeling of bone tissue through mediating the development and function of osteoblasts and osteoclasts.15,24,31,32 CXCL1 could contribute to osteolysis and facilitate metastasis of prostate cancer and osteosarcoma,33,34 while several published results confirmed the role of IL1β inducing bone resorption and osteoporosis.29 C5AR1 signaling in osteoblasts plays a detrimental role in bone regeneration by interacting with TLR2 and stimulating CXCL10.28 The expression of HCAR2 in peripheral blood mononuclear cells was significantly lower in nonunion patients than in those with healed long bone fractures, thus acting as a valuable biomarker for nonunion diagnosis.35 Tian and his colleagues demonstrated that C3AR1 might participate in the osteocyte apoptosis induced by myeloma cells.36 The expression of the top 5 hub genes (IL1β, CXCL1, ICAM1, CCL5, and CCL4) with connectivity degrees ≥20 showed a consistent trend by qRT-PCR assay and high-throughput sequencing. Moreover, ELISA revealed that UHMWPE particles stimulated a significant increase in the levels of IL-1β, IL-2, IL-6 and TNF-α, which was consistent with a study by Terkawi et al4 and diffusion of vitamin E to UHMWPE could reduce the secretion of these inflammatory factors.
We then used a variety of bioinformatics analysis methods to analyze these DEGs to obtain a comprehensive understanding of these genes. As a result, GO terms of 112 DEGs for biological process categories included positive regulation of the signal transduction process, inflammatory response and immune response. Furthermore, the KEGG pathway enrichment analysis revealed that these DEGs were predominantly associated with the chemokine signaling pathway and cytokine–cytokine receptor interactions. The DEGs in the chemokine signaling pathway were CXCL1, CCL1, CCL3, CXCL5, FGR, CXCL9, STAT1, CCL5, CCL4, CCL7, CCL22, CCL20 and RAP1B, while the DEGs in the cytokine–cytokine receptor interaction pathway were CCL1, CXCL1, CCL3, IL2RA, CXCL5, CXCL9, CCL5, CCL4, CCL7, CCL22, CCL20, IL1B and IL3RA. Nineteen DEGs were involved in the inflammatory response and fourteen were related to the immune response. Our bioinformatics analysis demonstrated the involvement of the rheumatoid arthritis pathway and measles pathway of DEGs in the response of macrophages to VE-UHMWPE and UHMWPE particles. Of significance, the cytokine–cytokine receptor interaction pathway, which was reported to be the most significantly enriched in patients with rheumatoid arthritis and measles, was the major pathway involved in the response of macrophages to UHMWPE particles.37 Many genes that were downregulated in macrophages encountering VE-UHMWPE particles in comparison with the UHMWPE group such as IL-1α, IL-27, CCL3, CCL20, SOCS3 and CCL5 were abundantly expressed in rheumatoid synovial tissues and played substantial roles in the pathogenesis of rheumatoid arthritis.38,39 There is overwhelming evidence that macrophages serve a prominent role in the pathogenesis of rheumatoid arthritis.38 They are the most abundant cell type in the rheumatoid synovium and can release an array of inflammatory cytokines resulting in the recruitment of other inflammatory cells, T-cell polarization, fibroblast activation and osteoclast differentiation.40 Analogously, the exposure of macrophages to UHMWPE particles may lead to a sustained local inflammatory response associated with chronic synovitis, joint pain, swelling and bone loss. These results presented an important implication that VE-UHMWPE particles initiated fewer rheumatoid arthritis-like immune responses around the implant than UHMWPE particles, hindering the development of inflammatory osteolysis.
TFs and microRNAs have been demonstrated to be important regulatory factors in the development of inflammatory osteolysis.41,42 Thus, we constructed a TF-microRNA interaction regulatory network to show the potential interactions among TFs, DEGs, and microRNAs under wear debris-induced inflammatory conditions. As shown in the network, RELA was connected to 6 target genes (SOCS3, PTGS2, ICAM1, CXCL1, CCL5, CCL20) for TF nodes; NFKB1 was also connected to 6 target genes (SOCS3, PTGS2, ICAM1, CXCL1, CCL5, IL1RN). These results suggested their importance in the development of inflammatory osteolysis. RELA, also known as p65, is a REL-associated protein involved in NF-kappaB (NF-κB) heterodimer formation, nuclear translocation, and activation.43 The phosphorylation and acetylation of RELA are pivotal post-translational modifications required for NF-κB activation.43 NFKB1 is a 105 kD protein that is considered a REL-specific transcription inhibitor. After being processed by the 26S proteasome, NFKB1 can produce a 50 kD protein acting as the DNA binding subunit of the NF-κB protein complex.44 NF-κB is an essential transcription factor complex involved in multiple cellular processes.45 As shown in the GO terms, the target genes of RELA and NFKB1 were enriched predominantly with respect to terms such as protein binding, inflammatory response, immune response, chemotaxis, chemokine-mediating signaling pathway, chemokine activity, etc. Thus, RELA and NFKB1 may be new drug targets for regulating inflammatory osteolysis in orthopedic implants. Additionally, the most connected microRNA node identified was hsa-mir-335-5p, which targeted five DEGs (PTGS2, ICAMI1, CCL20, C5AR1, and CXCL9). A series of studies have demonstrated that an increased level of miR-335-5p could facilitate bone formation, which may also be related to the decreased potential of osteoclastogenesis.46,47 Meanwhile, microRNA-21 could act as a functional factor in the pathogenesis of particle-induced osteolysis.48 Although there is no direct evidence to prove the role of hsa-miR-144 and hsa-miR-221 in the pathogenesis of inflammatory osteolysis until now, the target genes of these microRNAs such as ICAM1 and CCL20 are considered candidate genetic markers associated with the inflammatory process.
The current study provided several advances in the field. At present, a systematic bioinformatics analysis on the different performances of VE-UHMWPE and UHMWPE in the pathological process of osteolysis is still lacking. We performed a rigorous informatics analysis based on multiple databases and tools and provided results on this topic as comprehensively as possible, proposing multiple key genes, transcription factors, and miRNAs that may play important roles in the process of reduction of inflammatory osteolysis by VE-UHMWPE. These findings could provide research directions and molecular targets for further basic and clinical trials. However, it should be admitted that the current investigation is only fundamental research. Although multiple hub genes, key miRNAs and TFs were selected, the detailed functions of these molecules in inflammatory osteolysis should be further identified by in vivo and in vitro studies. Currently, whether the addition of vitamin E to UHMWPE could decrease the incidence of aseptic loosening of orthopedic implants has not been verified by randomized controlled trials. Analyses of protein expression and changes in osteoclastogenesis in the VE-UHMWPE and UHMWPE groups are warranted to further explore the roles of these materials in the pathogenesis of inflammatory osteolysis of orthopedic implants.
Conclusion
Combined with various informatics analyses and in vitro experiments, IL1β, CXCL1, ICAM1, CCL5, and CCL4 may serve as prognostic and therapeutic targets of inflammatory osteolysis in orthopedic devices. Additionally, the TF-microRNA interaction regulatory network revealed that key microRNAs (hsa-miR-144, hsa-miR-21, and hsa-miR-221) and TFs (RELA and NFKB1) were important regulators at the transcriptional level in this process. Further experiments are required to probe the functions of these genes in the pathogenesis of inflammatory osteolysis of orthopedic implants.
Data Sharing Statement
All data generated or analysed during this study are included in this published article.
Ethics Approval
This investigation involved the THP-1 cell lines that were purchased commercially from American Type Culture Collection in the USA and no further approval was required.
Funding
This work was supported by the China Scholarship Council (CSC) (grant number 201808080126), the Young Taishan Scholars Program of Shandong Province (grant number tsqn201909183), the Academic promotion programme of Shandong First Medical University (grant number 2020RC008) and the Natural Science Foundation of Shandong Province (grant number ZR201911090016).
Disclosure
The authors declare there are no conflicts of interest regarding the publication of this paper.
References
1. Massier J, Van Erp J, Snijders TE, Gast A. A vitamin E blended highly cross-linked polyethylene acetabular cup results in less wear: 6-year results of a randomized controlled trial in 199 patients. Acta Orthop. 2020;91(6):705–710. doi:10.1080/17453674.2020.1807220
2. Rochcongar G, Remazeilles M, Bourroux E, et al. Reduced wear in vitamin E-infused highly cross-linked polyethylene cups: 5-year results of a randomized controlled trial. Acta Orthop. 2020:1–5. doi:10.1080/17453674.2020.1852785.
3. Khan M, Osman K, Green G, Haddad FS. The epidemiology of failure in total knee arthroplasty: avoiding your next revision. Bone Joint J. 2016;98-B:105–112. doi:10.1302/0301-620X.98B1.36293
4. Terkawi MA, Kadoya K, Takahashi D, et al. Identification of IL-27 as potent regulator of inflammatory osteolysis associated with vitamin E-blended ultra-high molecular weight polyethylene debris of orthopedic implants. Acta Biomater. 2019;89:242–251. doi:10.1016/j.actbio.2019.03.028
5. Busch A, Jäger M, Klebingat S, et al. Vitamin E-blended highly cross-linked polyethylene liners in total hip arthroplasty: a randomized, multicenter trial using virtual CAD-based wear analysis at 5-year follow-up. Arch Orthop Trauma Surg. 2020;140:1859–1866. doi:10.1007/s00402-020-03358-x
6. McKellop H, Shen FW, Lu B, et al. Effect of sterilization method and other modifications on the wear resistance of acetabular cups made of ultra-high molecular weight polyethylene. A hip-simulator study. J Bone Joint Surg Am. 2000;82:1708–1725. doi:10.2106/00004623-200012000-00004
7. van Erp J, Massier J, Halma JJ, et al. 2-year results of an RCT of 2 uncemented isoelastic monoblock acetabular components: lower wear rate with vitamin E blended highly cross-linked polyethylene compared to ultra-high molecular weight polyethylene. Acta Orthop. 2020;91:254–259. doi:10.1080/17453674.2020.1730073
8. Bracco P, Oral E. Vitamin E-stabilized UHMWPE for total joint implants: a review. Clin Orthop Relat Res. 2011;469:2286–2293. doi:10.1007/s11999-010-1717-6
9. Oral E, Godleski BC, Malhi AS, Muratoglu OK. The effects of high dose irradiation on the cross-linking of vitamin E-blended ultrahigh molecular weight polyethylene. Biomaterials. 2008;29:3557–3560. doi:10.1016/j.biomaterials.2008.05.004
10. Rowell SL, Oral E, Muratoglu OK. Comparative oxidative stability of α-tocopherol blended and diffused UHMWPEs at 3 years of real-time aging. J Orthop Res. 2011;29(5):773–780. doi:10.1002/jor.21288
11. Bichara DA, Malchau E, Sillesen NH, et al. Vitamin E-diffused highly cross-linked UHMWPE particles induce less osteolysis compared to highly cross-linked virgin UHMWPE particles in vivo. J Arthroplasty. 2014;29:232–237. doi:10.1016/j.arth.2014.03.044
12. Busch A, Jäger M, Wegner A, Haversath M. Vitamin E-blended versus conventional polyethylene liners in prostheses: prospective, randomized trial with 3-year follow-up. Orthopade. 2020;49:1077–1085. doi:10.1007/s00132-019-03830-6
13. Nebergall AK, Greene ME, Laursen MB, et al. Vitamin E diffused highly cross-linked polyethylene in total hip arthroplasty at five years: a randomised controlled trial using radiostereometric analysis. Bone Joint J. 2017;99-B:577–584. doi:10.1302/0301-620X.99B5.37521
14. Galea VP, Rojanasopondist P, Laursen M, et al. Evaluation of vitamin E-diffused highly crosslinked polyethylene wear and porous titanium-coated shell stability: a seven-year randomized control trial using radiostereometric analysis. Bone JOINT J. 2019;101-B:760–767. doi:10.1302/0301-620X.101B7.BJJ-2019-0268.R1
15. Terkawi MA, Hamasaki M, Takahashi D, et al. Transcriptional profile of human macrophages stimulated by ultra-high molecular weight polyethylene particulate debris of orthopedic implants uncovers a common gene expression signature of rheumatoid arthritis. Acta Biomater. 2018;65:417–425. doi:10.1016/j.actbio.2017.11.001
16. Cobos EJ, Nickerson CA, Gao F, et al. Mechanistic Differences in Neuropathic Pain Modalities Revealed by Correlating Behavior with Global Expression Profiling. Cell Rep. 2018;22:1301–1312. doi:10.1016/j.celrep.2018.01.006
17. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi:10.1038/nprot.2008.211
18. Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi:10.1093/nar/gkn923
19. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi:10.1093/nar/gky1131
20. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi:10.1101/gr.1239303
21. Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 2014;47:8–13. doi:10.1002/0471250953.bi0813s47
22. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi:10.1093/bioinformatics/bti551
23. Oral E, Greenbaum ES, Malhi AS, et al. Characterization of irradiated blends of alpha-tocopherol and UHMWPE. Biomaterials. 2005;26:6657–6663. doi:10.1016/j.biomaterials.2005.04.026
24. Mbalaviele G, Novack DV, Schett G, Teitelbaum SL. Inflammatory osteolysis: a conspiracy against bone. J Clin Invest. 2017;127:2030–2039. doi:10.1172/JCI93356
25. Goodman SB. Wear particles, periprosthetic osteolysis and the immune system. Biomaterials. 2007;28:5044–5048. doi:10.1016/j.biomaterials.2007.06.035
26. Garrigues GE, Cho DR, Rubash HE, et al. Gene expression clustering using self-organizing maps: analysis of the macrophage response to particulate biomaterials. Biomaterials. 2005;26:2933–2945. doi:10.1016/j.biomaterials.2004.06.034
27. Kandahari AM, Yang X, Laroche KA, et al. A review of UHMWPE wear-induced osteolysis: the role for early detection of the immune response. Bone Res. 2016;4:16014. doi:10.1038/boneres.2016.14
28. Modinger Y, Rapp A, Pazmandi J, et al. C5aR1 interacts with TLR2 in osteoblasts and stimulates the osteoclast-inducing chemokine CXCL10. J Cell Mol Med. 2018;22:6002–6014. doi:10.1111/jcmm.13873
29. Ruscitti P, Cipriani P, Carubbi F, et al. The role of IL-1beta in the bone loss during rheumatic diseases. Mediators Inflamm. 2015;2015:782382. doi:10.1155/2015/782382
30. Redlich K, Smolen JS. Inflammatory bone loss: pathogenesis and therapeutic intervention. Nat Rev Drug Discov. 2012;11:234–250.
31. Liu Z, Liang W, Kang D, et al. Increased Osteoblastic Cxcl9 Contributes to the Uncoupled Bone Formation and Resorption in Postmenopausal Osteoporosis. Clin Interv Aging. 2020;15:1201–1212. doi:10.2147/CIA.S254885
32. Sucur A, Jajic Z, Artukovic M, et al. Chemokine signals are crucial for enhanced homing and differentiation of circulating osteoclast progenitor cells. Arthritis Res Ther. 2017;19:142. doi:10.1186/s13075-017-1337-6
33. Chao CC, Lee CW, Chang TM, et al. CXCL1/CXCR2 Paracrine Axis Contributes to Lung Metastasis in Osteosarcoma. Cancers (Basel). 2020;12:459. doi:10.3390/cancers12020459
34. Hardaway AL, Herroon MK, Rajagurubandara E, Podgorski I. Marrow adipocyte-derived CXCL1 and CXCL2 contribute to osteolysis in metastatic prostate cancer. Clin Exp Metastasis. 2015;32:353–368. doi:10.1007/s10585-015-9714-5
35. Wu J, Liu L, Hu H, et al. Bioinformatic analysis and experimental identification of blood biomarkers for chronic nonunion. J Orthop Surg Res. 2020;15:208. doi:10.1186/s13018-020-01735-1
36. Tian H. Identification of candidate genes for myeloma-induced osteocyte death based on microarray data. J Orthop Surg Res. 2016;11:81. doi:10.1186/s13018-016-0411-0
37. Liu G, Jiang Y, Chen X, et al. Measles contributes to rheumatoid arthritis: evidence from pathway and network analyses of genome-wide association studies. PLoS One. 2013;8:e75951. doi:10.1371/journal.pone.0075951
38. Cobelli N, Scharf B, Crisi GM, et al. Mediators of the inflammatory response to joint replacement devices. Nat Rev Rheumatol. 2011;7:600–608. doi:10.1038/nrrheum.2011.128
39. Aarts J, Roeleveld DM, Helsen MM, et al. Systemic overexpression of interleukin-22 induces the negative immune-regulator SOCS3 and potently reduces experimental arthritis in mice. Rheumatology (Oxford).2021;(60) Issue 4:1974–1983. doi:10.1093/rheumatology/keaa589
40. Santegoets KC, van Bon L, van den Berg WB, et al. Toll-like receptors in rheumatic diseases: are we paying a high price for our defense against bugs? FEBS Lett. 2011;585:3660–3666. doi:10.1016/j.febslet.2011.04.028
41. Lin TH, Pajarinen J, Lu L, et al. NF-kappaB as a Therapeutic Target in Inflammatory-Associated Bone Diseases. Adv Protein Chem Struct Biol. 2017;107:117–154.
42. Li W, Wang X, Chang L, Wang F. MiR-377 inhibits wear particle-induced osteolysis via targeting RANKL. Cell Biol Int. 2019;43:658–668. doi:10.1002/cbin.11143
43. Nolan GP, Ghosh S, Liou HC, et al. DNA binding and I kappa B inhibition of the cloned p65 subunit of NF-kappa B, a rel-related polypeptide. Cell. 1991;64:961–969.
44. Meyer R, Hatada EN, Hohmann HP, et al. Cloning of the DNA-binding subunit of human nuclear factor kappa B: the level of its mRNA is strongly regulated by phorbol ester or tumor necrosis factor alpha. Proc Natl Acad Sci U S A. 1991;88:966–970. doi:10.1073/pnas.88.3.966
45. Hussen BM, Azimi T, Hidayat HJ, et al. NF-KappaB interacting LncRNA: review of its roles in neoplastic and non-neoplastic conditions. Biomed Pharmacother. 2021;139:111604. doi:10.1016/j.biopha.2021.111604
46. Zhang L, Tang Y, Zhu X, et al. Overexpression of MiR-335-5p Promotes Bone Formation and Regeneration in Mice. J Bone Miner Res. 2017;32:2466–2475. doi:10.1002/jbmr.3230
47. Sui L, Wang M, Han Q, et al. A novel Lipidoid-MicroRNA formulation promotes calvarial bone regeneration. Biomaterials. 2018;177:88–97. doi:10.1016/j.biomaterials.2018.05.038
48. Zhou Y, Liu Y, Cheng L. miR-21 expression is related to particle-induced osteolysis pathogenesis. J Orthop Res. 2012;30:1837–1842. doi:10.1002/jor.22128
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.