Back to Journals » OncoTargets and Therapy » Volume 14
Identification of Critical Pathways and Potential Key Genes in Poorly Differentiated Pancreatic Adenocarcinoma
Authors Lu Y, Li D, Liu G, Xiao E , Mu S, Pan Y, Qin F, Zhai Y, Duan S , Li D, Yan G
Received 28 August 2020
Accepted for publication 17 December 2020
Published 27 January 2021 Volume 2021:14 Pages 711—723
DOI https://doi.org/10.2147/OTT.S279287
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Yuanxiang Lu,1,2,* Dongxiao Li,3,* Ge Liu,1,4 Erwei Xiao,1 Senmao Mu,1 Yujin Pan,1 Fangyuan Qin,5 Yaping Zhai,5 Shaofeng Duan,6 Deyu Li,1,2 Guoyi Yan1,4
1Department of Hepatobiliary Surgery, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou, Henan, People’s Republic of China; 2School of Clinical Medicine, Zhengzhou University, Zhengzhou, People’s Republic of China; 3Department of Gastroenterology, Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology, Wuhan, People’s Republic of China; 4School of Clinical Medicine, Henan University, Kaifeng, People’s Republic of China; 5Henan Eye Hospital, Henan Provincial People’s Hospital and People’s Hospital of Zhengzhou University, Zhengzhou, Henan, People’s Republic of China; 6School of Pharmacy, Henan University, Kaifeng, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Deyu Li; Guoyi Yan
Department of Hepatobiliary Surgery, Henan Provincial People’s Hospital, Zhengzhou University People’s Hospital, Zhengzhou 450003, People’s Republic of China
Email [email protected]; [email protected]
Introduction: The poorly differentiated pancreatic adenocarcinoma (PDAC) is an extremely lethal neoplasm without effective biomarkers for early detection and prognosis prediction, which is characteristically unresponsive to chemotherapeutic regimens. This study aims at searching for key genes which could be applied as novel prognostic biomarkers and therapeutic targets in PDAC.
Methods: Clinical samples were collected and a comprehensive differential analysis of seven PDAC samples by integrating RNA-seq data of tumor tissues and matched normal tissues from both our cohort and gene expression profiling interactive analysis (GEPIA) were performed to discover potential prognostic genes in PDAC. Pathway enrichment analysis was carried out to determine the biological function of PDAC differentially expressed genes (DEGs), and protein-protein interaction (PPI) network was constructed for functional modules analysis. Real-time PCR was performed to validate expression of hub genes.
Results: A total of 126 PDAC-specific expressed genes identified from seven PDAC samples were predominantly enriched in cell adhesion, integral component of membrane, signal transduction and chemical carcinogenesis, IL-17 signaling pathway, indicating that obtained genes might play a unique role in PDAC tumorigenesis. Furthermore, survival analysis revealed that five genes (CEACAM5, KRT6A, KRT6B, KRT7, KRT17) which exhibited high expression levels in tumor tissues were obviously correlated with the prognosis of PDAC patients and KRT7 was positively correlated with KRT6A, KRT6B, KRT17 expression. In addition, real-time PCR demonstrated that the expression level of the hub genes was consistent with RNA-seq analysis.
Discussion: The current study suggested that CEACAM5, KRT6A, KRT6B, KRT7, and KRT17 may represent novel prognostic biomarkers as well as novel therapeutic targets for poorly differentiated PDAC.
Keywords: pancreatic adenocarcinoma, biomarker, differentially expressed genes, RNA-seq, prognosis
Introduction
Pancreatic adenocarcinoma (PDAC) accounts for more than 95% of human pancreatic cancer, is a highly aggressive tumor whose mortality practically parallels incidence.1 Five-year survival of PDAC patients is no more than 5% in the world.2 By the year 2030, PDAC is predicted to be the second leading cause in tumor-related deaths.3 Less than 20% of patients can be completely resected, and most patients will get early progression even after potential curative resection, with a five-year survival rate of less than 25%.4,5 Despite advances in surgery and adjuvant chemotherapy for pancreatic cancer in recent years, the survival rate was not significantly improved. Therefore, it is urgent to explore effective targets to improve the prognosis of PDAC.
Aberrant gene expression is a common theme of human disease, corresponding mRNA play pivotal roles in the occurrence and progress of tumor-related disease.6 The emergence of high-throughput sequencing technology allows investigators to detect a complete data of the mutational and transcriptional landscape of most tumors.7 To date, numerous investigators have performed comprehensive next-generation sequencing analysis on PDAC samples, aiming to determine novel biological markers whose mutations are involved in tumor formation. The RNA-seq has been considered as a promising tool to filtrate the hub genetic markers of PDAC progression.7 Bioinformatic analysis of the RNA-seq data and transcriptomes has found a group of altered genes in PDAC, which dramatically promoted our understanding of the biological processes of tumorigenesis as well as remedy in human tissue samples.8 It is demonstrated that SMAD3 take an active part in inducing epithelial-mesenchymal transition (EMT) and was considered as an effective biomarker indicating poor prognosis.9 Some study revealed that THBS1 enriched in the phosphoinositide 3-kinase-Akt pathway could promote the development and progression of PDAC.10 Some scholars6,11 have considered KRAS, TP53, DPC4, and SMAD4 as tumor driver genes that may result tumor recurrence and antitumor immunity in PDAC, meanwhile, targeted drugs against these pathways can be developed.12,13 However, despite enormous progress in understanding the tumor initiation and progression of PDAC has made in gene level, there exist no widely recognized prognostic biomarkers with high sensitivity and specificity which could predict prognosis precisely and can be applied for postoperative individualized treatment. At present, RNA-seq technology combined with bioinformatics analysis enable it a promising way to comprehensively explore the aberrations of mRNA expression across the formation and development of PDAC. Investigation into these genes could provide valuable information that can help to devise the optimal treatment for patients and even predict disease relapse.
In the present study, we carried out a complicated differential analysis of PDAC and paired normal samples by integrating RNA-seq data both from our cohort aiming to elucidate the differentially expressed genes (DEGs) and explore the potential biofunctions by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. Furthermore, the protein-protein interaction (PPI) network was constructed, and five hub genes were identified from it. Furthermore, survival analysis revealed that the hub genes were obviously correlated with the prognosis of PDAC. The present study may provide new insight into the understanding of PDAC pathogenesis and the identified hub genes may serve as potential targets for diagnosis and treatment.
Materials and Methods
Sample Preparation and RNA-seq Analysis
Seven patients underwent radical resection of the primary tumor was included in this study and all of them were confirmed by postoperative pathology as poorly differentiated adenocarcinoma of the pancreas. PDAC tissues and their adjacent normal tissues were collected for transcriptome sequencing analysis and were stored in RNA later solution (Ambion, USA) immediately after surgery and stored at –80°C prior to RNA extraction. This study has been approved by the Ethics Committee of Henan Provincial People’s Hospital, and all patients have signed informed consent before the surgery.
Total RNA of each specimen was isolated using TRIzol reagent (the RNeasy Mini Kit; Qiagen, Germany) following the manufacturer’s instructions. An Agilent 2100 RNA Nano 6000 Assay Kit (Agilent Technologies, CA, USA) was used to qualify the integrity and concentration of total RNA. Later, all the specimens were sent to BGI (The Beijing Genomics Institute) Corporation (Wuhan, China) for further RNA-seq detection and analysis via BGISEQ-500 sequencer.
Identification of DEGs
DEGs between PDAC and non-PDAC samples from the generated data (RNA-seq dataset) were screened using R scripts, only the data with |log2FC| ≥ 1 and Q-value ≤0.05 were left for as DEGs for further investigation.
GO Analysis, Enrichment, KEGG Pathway Analyses
To uncover the functional roles of DEGs, we determined the transcriptional profiles acquired using RNA-seq and performed GO and KEGG enrichment analyses. Based on the DR.TOM system of BGI, we analyzed the functional enrichment of DEGs, concentrating on the pathways interrelated with these genes that were enriched in the terms biological process, molecular function and cellular component. The KEGG pathway analysis was applied to determine the significant pathways of the DEGs. The threshold of the hypergeometric distribution test for default enrichment results was 0.05.
Protein-protein Interaction (PPI) Network Analysis and Identification of Hub Genes
Search tool for the retrieval of interacting genes (STRING) database is online tool supplied to illustrating the PPI information. To further explore the potential interplay among those DEGs, we mapped the DEGs to STRING, and only interactions that enjoyed a minimum required combined score >0.4 were set as significant. Subsequently, the plug-in MCODE (version 1.5, http://apps.cytoscape.org/apps/mcode) was carried out to identify significant modules of the constructed network. Furthermore, the key genes were identified based on the PPI network by using cytoHubba (version 0.1, apps.cytoscape.org/apps/cytohubba), which is another plug-in of Cytoscape.
Survival Analysis and Correlation of DEGs
To study prognostic implications of hub genes, online server PROGgeneV2 was employed to conduct a survival analysis of data from TCGA (https://cancergenome.nih.gov/). In the public available TCGA datasets, PDAC samples were dichotomized into hub gene high- and low-expression groups by the median transcript per million (TPM) value as a cutoff, respectively. Cox regression analysis was carried out to unravel the correlation between hub gene expression and prognosis in PDAC and the log rank test was used for the hypothesis tests. The genes interrelated with OS were carried out with further analysis, incorporating Pearson correlation analysis as well as analysis of expression levels between tumor and normal tissues via GEPIA Online tools. A p-value of <0.05 is considered statistically significant.
Quantitative Reverse Transcriptional PCR (qRT-PCR)
Total RNA of the seven patients’ specimens were isolated using the E.Z.N.A. Total RNA Kit I (Invitrogen) following standard procedures. Complementary DNA (cDNA) was synthesized subsequently by using the Prime Script RT Reagent Kit (Takara Bio, Japan). Real-time PCR was performed on the ABI 7500 Touch RealTime PCR Detection System (ABI. USA) to determine the mRNA expression level of screened hub genes according to the comparative Ct method. GAPDH was set as a normalization control. The sequences of primers are presented in Supplementary material.
Results
DEGs Associated with PDAC
Paired differential gene expression analysis by RNA-seq revealed that 126 genes were found to be differentially expressed in seven paired PDAC samples from our cohort. Among the DEGs, 82 were upregulated genes and 44 were downregulated genes (Supplementary material). Heatmaps of the distributions of DEGs are presented in Figure 1.
 |
Figure 1 Heat map of the differentially expressed genes in seven PDAC samples. |
GO Enrichment Analyses of DEGs
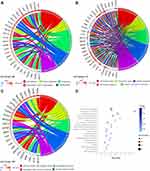
To further elaborate the function of DEGs in human PDAC, GO annotations and KEGG pathway enrichment analysis was carried out. GO analysis determined 51 terms significantly enriched, incorporating terms correlated with molecular function (BP), biological process (CC) and cellular component (MF) (Figure 2A–C). Changes in BP were significantly enriched in cell adhesion, blood coagulation, proteolysis, keratinization, extracellular matrix organization. For CC, the DEGs were mainly concerned with extracellular region, extracellular space, integral component of membrane, plasma membrane, extracellular exosome. Moreover, for category MF, the main functions of proteins encoded by the DEGs were calcium ion binding, peptidase activity, metallopeptidase activity, extracellular matrix structural constituent, structural molecule activity.
KEGG Pathway Analysis
The pathway terms associated with the obtained DEGs were tested for significance using Fisher’s exact test (p<0.05). Pathway terms with prominent enrichment were figured out and DEGs were opt for the pathway category analysis. Interestingly, most of the enriched pathways were correlated to the signal transduction and specific types of cancer-related pathways, including chemical carcinogenesis, IL-17 signaling pathway. In addition, the DEGs were also enriched in protein digestion and absorption, drug metabolism-cytochrome p450 and metabolism of xenobiotics by cytochrome p450 (Figure 2D).
Protein-protein Interaction (PPI) Network Analysis and Functional Module Analysis
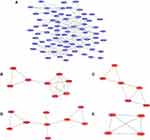
This network consists of convinced interactions from curated databases and those that were experimentally determined; involved interactions analysis with regards to gene neighborhood, gene fusions and gene co-occurrence; and text-mining, co-expression, respectively. The direct or indirect function interaction of proteins was visualized with the help of String database (Figure 3A). Furthermore, clustering analysis carried out by Cytoscape plug-in MCODE identified four functional modules from the whole network (Figure 3B–E), and the subsequently pathway enrichment analyses for genes of the four modules which could help to comprehend the potential biofunctions showed that these modules were mostly involved in tumorigenesis, cell adhesion and energy metabolism incorporating cell adhesion molecules (CAMs), pathways in cancer, cell cycle, PPAR signaling pathway, estrogen signaling pathway, cholesterol metabolism (Table 1).
 |
Table 1 KEGG Enrichment Analysis of Top Four Modules Identified from PPI Network |
Survival Curves, Expression Levels and Correlation Analysis
CEACAM5, KRT6A, KRT6B, KRT7, KRT17 were suggested to have prognostic relevance with PDAC, patients with high expression of the five genes may have a worse outcome in survival condition (Figure 4). We further explored the relationship between mRNA expression level and human tumor samples. Expression levels of CEACAM5, KRT6A, KRT6B, KRT7, KRT17 are displayed in Figure 5. CEACAM5, KRT6A, KRT6B, KRT7, KRT17 demonstrate high expression levels in PDAC tissues. And then, we carried out Pearson correlation analyses among the expressed genes revealed in Figure 6. It was demonstrated that CEACAM5 was positively correlated with KRT7 (p=1.7e−08, R=0.3); KRT7 was also positively correlated with KRT6A, KRT6B, KRT17: KRT6A (p-value=0, R=0.43); KRT6B (p-value=2.7e−15, R=0.41); KRT17 (p-value=0, R=0.57).
 |
Figure 4 Prognostic survival analysis of CEACAM5, KRT6A, KRT6B, KRT7, KRT17 in PDAC were analyzed by the online server PROGgeneV2 tool, data of survival analysis is from TCGA (https://cancergenome.nih.gov/). |
 |
Figure 5 Expression analysis of a CEACAM5, KRT6A, KRT6B, KRT7, KRT17 in normal and tumor pancreatic tissues. |
Expression Validation
To further validate the reliability of the RNA-seq data, we validated the expression of hub genes in cancers and noncancer samples using qRT-PCR. As a result, the relative expression level of CEACAM5, KRT6A, KRT6B, KRT7, KRT17 in PDAC samples were significantly higher compared to nonPDAC samples (p<0.05). This result was consistent with the RNA-seq analysis (Figure 7).
 |
Figure 7 Validation of hub genes expression in PDAC and non-PDAC clinical samples using qRT-PCR. The relative expression level of the five hub genes. |
Discussion
Tumorigenesis is sophisticated and usually accompanied by multiple genetic alterations at the transcription level.14 The high mortality rate of pancreatic cancer has made it a worldwide public health problem. Many clinicians are not able to make an early diagnosis, this might be caused by lack of identification of specific biomarkers, either in genes or proteins.15 Fortunately, the emergence of RNA-seq technology has provided a possibility for exploring mechanisms underlying tumor formation at the mRNA level as well as identification of effective molecular therapeutic targets. The combination of bioinformatics and high-throughput sequencing have produced a vast amount of gene expression profiles, which could perform statistical analysis in disease occurrence, progression, and recurrence, serving an important role in tumor analysis.
In the current study, 126 DEGs were identified based on transcriptome analysis. GO and KEGG enrichment analyses revealed that the DEGs were predominately associated with cell adhesion, integral component of membrane, signal transduction and chemical carcinogenesis, IL-17 signaling pathway. Furthermore, we performed an integrative analysis of potential prognostic association between PDAC and DEGs by PROGgeneV2. CEACAM5, KRT6A, KRT6B, KRT7, KRT17 have potential prognostic value for PDAC patients. The PPI network and module analysis were carried out to further explore functional sub-networks and CEACAM5, KRT6A, KRT6B, KRT7, KRT17 represented core hub genes in the network. Moreover, these genes are not only correlated with PDAC, but also to the signal transduction pathway.
CEACAM5 is a key member of the glycosylphosphatidylinositol (GPI)-linked immunoglobulin (Ig) superfamily, attached to the plasma membrane of cell.16 The CEACAM5 gene, encoding the most extensively studied tumor markers-the carcinoembryonic antigen (CEA), is notorious for it is vital role in the clinical work as a tumor marker for several malignancies.17 The encoded protein as an intercellular adhesion molecule, plays an irreplaceable role in several process incorporating cell-cell recognition, proliferation, differentiation and cancer invasion and metastasis.18 Furthermore, it is indispensable in forming tissue architecture and neovascularization. Expression of CEACAM5 has been revealed to have significant correlation with the PDAC patients’ lymph node status.19 It has been conclusively demonstrated that the overexpression of CEACAM5 will contribute to pancreatic cancer progression, particularly invasion and metastasis.20 Knockdown of the CEACAM5 gene could effectively inhibit tumor proliferation of tumor-bearing mouse.21 Distant metastasis is an important sign of a poor prognosis in PDAC patients. Expression of CEACAM5 in PDAC tissues is higher than that in normal tissues, which may contribute to its invasion and it is consistent with previous studies. Our study found a significant difference of OS between overexpression of CEACAM5 and the rest, this may be a conceivable marker in predicting poor prognosis of patients with PDAC.
The keratins are the typical intermediate filament proteins of epithelia, revealing a remarkable degree of molecular diversity.22 As part structural basis of the epithelial, keratins play pivotal roles in the mechanical stability and integrity of epithelial cells and tissues, involving intracellular signaling pathways.23 Keratins are abundantly expressed in hair and skin cells, its aberrant expression in other tissues may cause pathological process, involved in the progression of various tumors.24 It has been extensively explored that the role of keratins in cancer development was related with epithelial-derived tumors such as bladder cancer,25 lung cancer, colorectal cancer, urothelial cancer, and gastric cancer.26 Here in this work, we focused on the roles of KRTA/6B, KRT7, KRT17 in PDAC.
KRT6 belongs to the type II intermediate filament protein, including three isoforms (KRT6A, KRT6B, and KRT6C).27 KRT6A could be detected in ductal myoepithelial cells in human mammary duct.28 KRT6A/KRT6B proteins have been unraveled to play roles in various cancers. Slusser-Nore et al found that the expression of KRT6 is regulated by the activation of the ERK1/2 pathway in human urothelial cells.29 It is reported that the interaction of KRT6 with notch1 could aggravate the invasion of renal cell carcinoma.30 However, the role of KRT in PDAC was unknown. Our RNA sequencing data demonstrated that mKRT6A/mKRT6B was hyperexpressed in PDAC. Based on the survival analysis, we found for the first time that patients with higher mKRT6A/mKRT6B expression showed shorter OS period. Thus, further studies are needed to investigate the function in detail and it may be the potential biomarker and novel therapeutic target for pancreatic cancer. In addition, we found that KRT6A/B is positively correlated with KRT7/KRT17 in tumor tissues; moreover, they were almost absent in the adjacent tissue counterparts.
Our recognition of KRT7 (keratin 7) in PDAC is concordant with previous findings. Actually, KRT7 is an intermediate filament protein, which is selectively expressed in simple epithelia.31 In the normal pancreas, only minimal expression of it could be detected in the duct, but are absent from acinar and endocrine cells,32 is considered as a characteristic marker for ductal cells of the hepato-pancreatic ductal tree.33 It was demonstrated that KRT7 is significantly overexpressed in pancreatic cancer tissues, the coding protein was routinely detected by immunohistochemistry analysis for the diagnosis of PDAC.34 KRT7 also participated in the context of differentiation of pancreatic cancer cells and it was investigated that KRT7 promoter is available for targeting PDAC cells either in vitro or in vivo. The AdK7Luc virus invasion assays further demonstrated that by incorporating the KRT7 promoter fragment in the adenovirus could increase cancer specificity.32 Studies have shown that overexpression of KRT7 in pancreatic cancer was markedly associated with liver metastasis and it was recommended as marker in distinguishing pancreatic and ampulla of Vater adenocarcinomas.35 KRT7 has been extensively explored in cancer research, nevertheless, no studies have linked the expression of KRT7 to poor prognosis in pancreatic cancers. Our survival analysis showed for the first time that patients with high expression of KRT7 mRNA enjoyed significantly shorter OS than that of the low expression one, and the gene was positively correlated with KRT6A, KRT6B, and KRT17, respectively.
KRT17 belongs to type I intermediate filament, which is mainly involved in formation of basal epithelial cells. It was striking, however, that KRT17 was routinely regenerated and highly expressed in human tissue samples.36 Mature pancreatic duct epithelium cell express no or little KRT17, nevertheless, it was determined that the KRT17 mRNA level was eminently upregulated in PDAC samples, comparing with normally differentiated pancreatic tissues, and an identical trend was detected in neoplastic cells, compared with normal pancreatic cells.37 Furthermore, immunohistochemistry analysis revealed that KRT17 was overexpressed in tumor tissues compared with adjacent noncancerous tissues.38 Due to it is function as structural constituents of the tissue, KRT17 has been reported to play a key role as a multifunctional promoter and oncogene to accelerate proliferation, metastasis, invasion of malignancies.39 In addition, Chen et al discovered that inhibition of the proliferation of PANC‑1 human pancreatic cancer cells could be replicated by silencing of KRT17 through lentivirus‑mediated short hairpin RNA.40 And it was found that upregulation of KRT17 promoted PDAC cell proliferation, invasion, and metastasis through EMT.40 In addition, some studies raised concerns that overexpression of KRT17 mRNA was associated with a poor outcome for PDAC patients.41 In our study, RNA-seq analysis demonstrated that mRNA expression of KRT17 was markedly increased in cancer tissues compared to adjacent normal tissue. And the bioinformatics analysis predicted that overexpression is associated with tumor metastasis and poor prognosis, indicating that KRT17 could serve as a potential prognostic biomarker and a therapeutic target for PDAC.
Taken together, we identified 126 DEGs in patients with PDAC by employing RNA-seq method, and defined the potential novel therapeutic targets as well as evaluating their prognostic values in PDAC patients by bioinformatic analysis. GO-term and KEGG-pathway analysis were enriched in cellular component, cellular process related GO terms, and signal transduction as well as specific types of cancers related KEGG pathways. CEACAM5, KRT6A, KRT6B, KRT7, KRT17 may serve as novel biomarkers of poor survival in PDAC, and CEACAM5, KRT6A/B, KRT17 may even play a carcinogenic role which might provide effective anti-cancer target in PDAC therapy. Of course, our study has limitation, since TCGA database does not rovide sufficient information for us to identify poorly differentiated PDAC from the non-PDAC ones, the survival analysis of the recognized hub genes was carried out using samples of all pancreatic cancer subtypes. Thus, although this result contributes to the understanding of the biological relevance of the hub genes, further studies are needed to verify the function of hub genes.
Conclusion
In this work, 126 DEGs were identified between poorly differentiated pancreatic adenocarcinoma and the matched normal tissues. Among the DEGs, five genes were considered as hub genes which were related to many pathways of tumor progress and leading to poor prognosis. The result of this study may provide novel biomarkers for PDAC which is valuable for further investigation.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Ethics Approval and Consent to Participate
Approval was obtained from the ethics committee of People’s Hospital of Zhengzhou University. The procedures used in this study adhere to the tenets of the Declaration of Helsinki. Informed consent was obtained from all individual participants included in the study.
Acknowledgments
The authors are grateful for the support of Postdoctoral Research Project in Henan Province and Medical science and technology research project of Henan Provincial Department of Health (201403177, 201701021).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi:10.3322/caac.21208
2. Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi:10.1038/nature14169
3. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi:10.1158/0008-5472.can-14-0155
4. Garrido-Laguna I, Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12(6):319–334. doi:10.1038/nrclinonc.2015.53
5. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi:10.1016/S0140-6736(16)00141-0
6. Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271(5247):350–353. doi:10.1126/science.271.5247.350
7. Shang M, Zhang L, Chen X, Zheng S. Identification of hub genes and regulators associated with pancreatic ductal adenocarcinoma based on integrated gene expression profile analysis. Discov Med. 2019;28(153):159–172.
8. Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A, Cicenas J. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer. Cancers (Basel). 2017;9:5. doi:10.3390/cancers9050042
9. Yamazaki K, Masugi Y, Effendi K, et al. Upregulated SMAD3 promotes epithelial-mesenchymal transition and predicts poor prognosis in pancreatic ductal adenocarcinoma. Lab Invest. 2014;94(6):683–691. doi:10.1038/labinvest.2014.53
10. Zheng B, Peng J, Mollayup A, et al. Construction of a prognostic prediction system for pancreatic ductal adenocarcinoma to investigate the key prognostic genes. Mol Med Rep. 2018;17(1):216–224. doi:10.3892/mmr.2017.7850
11. Xu JZ, Wang WQ, Zhang WH, et al. The loss of SMAD4/DPC4 expression associated with a strongly activated hedgehog signaling pathway predicts poor prognosis in resected pancreatic cancer. J Cancer. 2019;10(17):4123–4131. doi:10.7150/jca.30883
12. Huijberts S, van Geel R, van Brummelen EMJ, et al. Phase I study of lapatinib plus trametinib in patients with KRAS-mutant colorectal, non-small cell lung, and pancreatic cancer. Cancer Chemother Pharmacol. 2020;85(5):917–930. doi:10.1007/s00280-020-04066-4
13. Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2020;17(3):153–168. doi:10.1038/s41575-019-0245-4
14. Hallajzadeh J, Maleki Dana P, Mobini M, et al. Targeting of oncogenic signaling pathways by berberine for treatment of colorectal cancer. Med Oncol. 2020;37(6):49. doi:10.1007/s12032-020-01367-9
15. Moore A, Donahue T. Pancreatic cancer. JAMA. 2019;322(14):1426. doi:10.1001/jama.2019.14699
16. Hammarstrom S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9(2):67–81. doi:10.1006/scbi.1998.0119
17. Kelleher M, Singh R, O’Driscoll CM, Melgar S. Carcinoembryonic antigen (CEACAM) family members and inflammatory bowel disease. Cytokine Growth Factor Rev. 2019;47:21–31. doi:10.1016/j.cytogfr.2019.05.008
18. Beauchemin N, Draber P, Dveksler G, et al. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp Cell Res. 1999;252(2):243–249. doi:10.1006/excr.1999.4610
19. Blumenthal RD, Hansen HJ, Goldenberg DM. Inhibition of adhesion, invasion, and metastasis by antibodies targeting CEACAM6 (NCA-90) and CEACAM5 (carcinoembryonic antigen). Cancer Res. 2005;65(19):8809–8817. doi:10.1158/0008-5472.can-05-0420
20. Sato K, Mori R, Hiroshima Y, et al. RT-PCR of peritoneal washings predicts peritoneal pancreatic cancer recurrence. J Surg Res. 2018;226:122–130. doi:10.1016/j.jss.2017.11.009
21. Johnson B, Mahadevan D. Emerging role and targeting of carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) in human malignancies. Clin Cancer Drugs. 2015;2(2):100–111. doi:10.2174/2212697x02666150602215823
22. Chu PG, Weiss LM. Keratin expression in human tissues and neoplasms. Histopathology. 2002;40(5):403–439. doi:10.1046/j.1365-2559.2002.01387.x
23. Depianto D, Kerns ML, Dlugosz AA, Coulombe PA. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat Genet. 2010;42(10):910–914. doi:10.1038/ng.665
24. Chen PJ, Hsu JD, Han CP. Anti-cytokeratin CAM 5.2 is specific for K8 and to a lesser extent for the closely related K7, but shows no reactivity with K18 or K19: comment on: “keratin expression in endocrine organs and their neoplasms” Endocr Pathol. 2009 Spring; 20(1):1–10. Endocr Pathol. 2010;21(1):69–70. doi:10.1007/s12022-010-9108-9
25. Masuda N, Murakami K, Kita Y, et al. Trp53 mutation in Krt5-expressing basal cells facilitates the development of basal squamous-like invasive bladder cancer in the chemical carcinogenesis of mouse bladder. Am J Pathol. 2020;190(8):1752–1762. doi:10.1016/j.ajpath.2020.04.005
26. Chen W, Zhang W, Wu R, Cai Y, Xue X, Cheng J. Identification of biomarkers associated with histological grade and prognosis of gastric cancer by co-expression network analysis. Oncol Lett. 2019;18(5):5499–5507. doi:10.3892/ol.2019.10869
27. Yang B, Zhang W, Zhang M, Wang X, Peng S, Zhang R. KRT6A promotes EMT and cancer stem cell transformation in lung adenocarcinoma. Technol Cancer Res Treat. 2020;19:1533033820921248. doi:10.1177/1533033820921248
28. Grimm SL, Bu W, Longley MA, Roop DR, Li Y, Rosen JM. Keratin 6 is not essential for mammary gland development. Breast Cancer Res. 2006;8(3):R29. doi:10.1186/bcr1504
29. Slusser-Nore A, Garrett SH, Zhou XD, Sens DA, Sens MA, Somji S. The expression of keratin 6 is regulated by the activation of the ERK1/2 pathway in arsenite transformed human urothelial cells. Toxicol Appl Pharmacol. 2017;331:41–53. doi:10.1016/j.taap.2017.05.007
30. Hu J, Zhang LC, Song X, Lu JR, Jin Z. KRT6 interacting with notch1 contributes to progression of renal cell carcinoma, and aliskiren inhibits renal carcinoma cell lines proliferation in vitro. Int J Clin Exp Pathol. 2015;8(8):9182–9188.
31. Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31(1):11–24. doi:10.1016/0092-8674(82)90400-7
32. Pujal J, Huch M, Jose A, et al. Keratin 7 promoter selectively targets transgene expression to normal and neoplastic pancreatic ductal cells in vitro and in vivo. FASEB J. 2009;23(5):1366–1375. doi:10.1096/fj.08-115576
33. Regitnig P, Spuller E, Denk H. Insulinoma of the pancreas with insular-ductular differentiation in its liver metastasis–indication of a common stem-cell origin of the exocrine and endocrine components. Virchows Arch. 2001;438(6):624–628. doi:10.1007/s004280100406
34. Bournet B, Pointreau A, Souque A, et al. Gene expression signature of advanced pancreatic ductal adenocarcinoma using low density array on endoscopic ultrasound-guided fine needle aspiration samples. Pancreatology. 2012;12(1):27–34. doi:10.1016/j.pan.2011.12.003
35. Tot T. Adenocarcinomas metastatic to the liver: the value of cytokeratins 20 and 7 in the search for unknown primary tumors. Cancer. 1999;85(1):171–177. doi:10.1002/(SICI)1097-0142(19990101)85:1<171::AID-CNCR24>3.0.CO;2-V
36. Moll R. Cytokeratins as markers of differentiation in the diagnosis of epithelial tumors. Subcell Biochem. 1998;31:205–262.
37. Cheng L, Zhao Y, Tang M, Luo Z, Wang X. Knockdown of ISOC1 suppresses cell proliferation in pancreatic cancer in vitro. Oncol Lett. 2019;17(5):4263–4270. doi:10.3892/ol.2019.10082
38. Goldstein NS, Bassi D. Cytokeratins 7, 17, and 20 reactivity in pancreatic and ampulla of vater adenocarcinomas. Percentage of positivity and distribution is affected by the cut-point threshold. Am J Clin Pathol. 2001;115(5):695–702. doi:10.1309/1ncm-46qx-3b5t-7xhr
39. Liu Z, Yu S, Ye S, et al. Keratin 17 activates AKT signalling and induces epithelial-mesenchymal transition in oesophageal squamous cell carcinoma. J Proteomics. 2020;211:103557. doi:10.1016/j.jprot.2019.103557
40. Chen P, Shen Z, Fang X, et al. Silencing of keratin 17 by lentivirus-mediated short hairpin RNA inhibits the proliferation of PANC-1 human pancreatic cancer cells. Oncol Lett. 2020;19(5):3531–3541. doi:10.3892/ol.2020.11469
41. Roa-Pena L, Leiton CV, Babu S, et al. Keratin 17 identifies the most lethal molecular subtype of pancreatic cancer. Sci Rep. 2019;9(1):11239. doi:10.1038/s41598-019-47519-4
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.