Back to Journals » International Journal of General Medicine » Volume 16
Identification and Validation of Key miRNAs for Colon Cancer Based on miRNA-Gene Integration Analysis
Authors Ji Y, Gu J , Zhang H, Xu H
Received 14 September 2023
Accepted for publication 23 November 2023
Published 5 December 2023 Volume 2023:16 Pages 5703—5718
DOI https://doi.org/10.2147/IJGM.S440340
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Yi Ji,1,* Jialin Gu,1,2,* Hongqun Zhang,1,2 Houxi Xu3
1Affiliated Hospital of Integrated Traditional Chinese and Western Medicine, Nanjing University of Chinese Medicine, Nanjing, 210028, People’s Republic of China; 2The Third Clinical Medical College, Nanjing University of Chinese Medicine, Nanjing, 210046, People’s Republic of China; 3Key Laboratory of Acupuncture and Medicine Research of Ministry of Education, Nanjing University of Chinese Medicine, Nanjing, 210046, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Houxi Xu, Key Laboratory of Acupuncture and Medicine Research of Ministry of Education, Nanjing University of Chinese Medicine, Nanjing, 210046, People’s Republic of China, Email [email protected]
Background: MicroRNAs (miRNAs) plays an essential role in the pathogenesis of colon cancer. This study aims to identify and verify key miRNAs that may serve as potential biomarkers for early diagnosis and treatment of colon cancer.
Methods: Two miRNA microarray datasets (GSE53339 and GSE126093) were downloaded from the Gene Expression Omnibus (GEO) database, and the common differentially expressed miRNAs (DEMis) of both were identified by the “limma” package and the intersect function in R. Then, the miRwalk online tool was used to predict the target genes of the common DEMis and perform Gene Ontology (GO) functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on those DEMis. The miRNA-gene network was constructed using Cytoscape software to identify key miRNAs and validated using quantitative real-time PCR (qRT-PCR) and The Cancer Genome Atlas (TCGA) dataset information for experimental and external database validation, respectively. In addition, we explored the correlation between key miRNAs and infiltrating immune cells and immunotherapy biomarkers.
Results: Fourteen common DEMis were identified from the GEO database, and five targeted genes were predicted. A microRNA-gene network was subsequently constructed to identify three key miRNAs (miR-363-3p, miR-520e, and miR-330-5p). Both qRT-PCR and external database validation confirmed our findings. In addition, we found that miR-520e was significantly associated with infiltrating immune cells and established immune checkpoints.
Conclusion: Our study identified three key miRNAs that influence the development of colon cancer. In addition, further studies suggest that infiltrating immune cells may play an essential role in the pathogenesis of colon cancer. These findings assist in early diagnosis and precision-targeted therapies.
Keywords: colon cancer, GEO, miRNA, qRT-PCR, immune cell infiltration
Introduction
With nearly 2 million new cases and 1 million deaths worldwide in 2020, colorectal cancer is the third-most common cancer and the second leading cause of cancer-related deaths.1 According to data from the US Surveillance, Epidemiology and End Results program and the National Program of Cancer Registries program, the incidence of colorectal cancer among people aged 50 to 64 years and young adults under 50 years is increasing by 1% and 2% per year, respectively.2 Among them, colon cancer is nearly one-third more common than rectal cancer.1 In addition, patients with colon cancer have lower 1- and 5-year survival and a worse prognosis than patients with rectal cancer.3 Colon cancer treatment requires multimodal management, such as surgery, radiotherapy, chemotherapy, immunotherapy, and molecular targeted therapy.4 Although existing treatment strategies have achieved specific results, obstacles such as limited drug selection, drug resistance, complex anatomical localization, and post-operative recurrence rates have brought colon cancer to the forefront of the research agenda.5 The initial symptoms of colorectal cancer are atypical, and the early diagnosis rate is relatively low, with more than 50% of patients already at an advanced stage at the initial diagnosis.2 Therefore, elucidating the pathogenesis of colon cancer, identifying novel molecular targets, and developing targeted drugs are the critical measures needed to improve the treatment and survival of individuals with this aggressive disease.6
MicroRNAs (miRNAs) are a class of non-coding, small-molecule, single-stranded RNAs with a length of 20–25 nucleotides. In eukaryotic cells, miRNAs play an essential role in various physiological and pathological processes by regulating the expression of transcription factors.7 Recent studies have shown that the abnormal expression of miRNAs is closely related to tumor development, including tumor invasion, metastasis, staging, and prognosis.8–11 miRNAs can act as both tumor suppressor genes and oncogenes in tumor tissues.12 Koo et al found that a novel tumor suppressor miRNA, miR-4779, was expressed at a low level in colon cancer tissue and functions by directly targeting PAK2 and CCND3 to inhibit tumor cell growth through cell cycle arrest and apoptosis.13 Li et al demonstrated that miR-3065-3p was highly expressed in colorectal cancer tissues and promoted cancer stemness and metastasis by targeting CRLF1.14 Furthermore, another study found that miR-3651 was associated with the tumor-lymph node-metastasis stage. Inactivation of the PI3K/AKT and MAPK/ERK signaling pathways in colorectal cancer cells by downregulating miR-3651 induced growth arrest and apoptosis in cancer cells.15 MiRNAs are involved in numerous signaling pathways in colon cancer pathology and have great potential for clinical diagnosis, prognosis, and personalized, targeted therapy.16 Therefore, further studies on miRNAs will help to understand the mechanism of colon cancer and its biological functions, and provide a theoretical basis for its prevention, diagnosis, and treatment.
Recent studies have evaluated miRNA expression in cancer using microarray datasets or next-generation sequencing technology. The new technologies can help identify critical miRNAs related to cancer development, which are considered effective candidates for biomarkers in cancer development, thus screening for them is critical. Wu et al found that miR-155-5p is a key miRNA contributing to the aggressiveness of clear cell renal cell carcinoma by targeting PEG3.17 Li et al identified four key miRNAs for prostate cancer using bioinformatics.18 However, these studies suffer from several limitations. First, most cancer studies use a single dataset for fundamental miRNA analysis.16 This may result in inaccuracies due to differences in microarray platforms and sample specificity. Second, although some studies have used multiple microarray datasets to identify key miRNAs in cancer, they did not collect corresponding clinical cancer samples to validate them. These limitations highlight the need to use diverse datasets and validate the key miRNAs identified in the analysis of these datasets. Therefore, this study aimed to identify potential biomarkers for colon cancer and address the issues mentioned above.
Methods
miRNA Microarray Data Download
Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo) is a public database that stores high-throughput gene expression and microarray data.19 The keyword “colon cancer miRNA human” was searched in the GEO database, and two GEO datasets (GSE53339 and GSE126093) were randomly selected for download by using the GEOquery package of R software.20 Data from 32 samples (16 cancer and 16 normal tissue samples) were included. The GSE53339 dataset, published in December 2013 and based on the GPL18058 platform (Exiqon miRCURY LNA microRNA array version18), is taken from tissue samples of cancer patients, including six cancer and six adjacent tissue samples. The GSE126093 dataset, published in February 2019 and based on the GPL18058 platform (Exiqon miRCURY LNA microRNA array version 18), is also taken from tissue samples of cancer patients, including ten cancer and ten adjacent tissue samples.21 Using R software, the probe ID in the download file was converted to the miRNA SYMBOL name. All miRNA expression data were standardized and log2 transformed.
Screening for Common Differentially Expressed miRNAs (DEMis)
The GSE53339 and GSE126093 microarray datasets were analyzed using the “limma” R package (log|fold change| > 2, P-value < 0.05).22 The intersect function in R was used to identify the common DEMis among GSE53339 and GSE126093. A Venn diagram was generated by the VennDiagram R package.23
Prediction of the Target Genes of Common DEMis
MiRwalk3.0 is a freely available, comprehensive miRNA database that can provide information about predicted or experimentally confirmed miRNA-target interactions.24 MiRwalk3.0 integrates information from miRDB, TargetScan, miRTarBase, and other databases. It contains the miRNA target gene information for many species, thus providing a comprehensive technology platform for developing scientific research experiments. We used the miRwalk3.0 online tool (http://mirwalk.ummuni-heidelberg.de/0miRNA) to predict the targeted genes of the common DEMis, on the condition that the target genes could be predicted by two databases (miRDB and TargetScan) simultaneously.25,26
Gene Ontology (GO) Functional and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analyses of Common DEMis
ClusterProfiler is a GO-based tool that uses input IDs such as “groupGO” “enrich” and “enrichKEGG” to perform gene analysis and enrichment analysis.27 Additionally, clusterProfiler is used to carry out functional clustering analysis and visualize sample data, which allows complex biological information to be processed better. To determine the biological functions of the common DEMis, we used clusterProfiler to perform GO functional and KEGG pathway enrichment analyses of the target genes of the common DEMis. Screening criteria for the result of GO functional and KEGG pathway enrichment analyses were P-value < 0.05.
Construction of the miRNA-Gene Network and Identification of the Key miRNAs
Cytoscape is a software platform designed for the visualization of molecular interaction networks and biological pathways. It is commonly used to construct biological networks such as “lncRNA-miRNA-mRNA”, “protein-protein” and “miRNA-gene” interactions.28 In miRNA-gene networks, miRNA node degree can be used as a key indicator of the importance of a specific miRNA. In this study, key miRNAs were defined as node degree > 20. We used Cytoscape software to build miRNA-gene networks of common DEMis and identified key miRNAs based on node degree.
Experimental Validation of Key miRNAs
To verify the authenticity of the key miRNAs we had identified, the colon cancer tissues and colon normal tissues of fifteen colon cancer patients were selected for qRT-PCR analysis. We extracted total RNA from the tissues using TRIzol (Vazyme, Nanjing, China) reagent. Total RNA was reverse transcribed into cDNA using the miRNA 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). Expression levels of miRNA were measured using a quantitative real-time PCR system (Applied Biosystems, Foster City, CA, USA). The data was analyzed using the comparative cycle threshold (CT, 2−ΔΔCT) method. qRT-PCR program settings: pre-denaturation at 95 °C for 5 min, denaturation at 95 °C for 10 sec, annealing/extension at 60 °C for 30 sec, repeat 40 cycles, other parameters are performed according to the instrument default settings. GraphPad Prism 9.0 was used to analyze the qRT-PCR data. The reference gene for the qRT-PCR experiment was U6. The study was approved by the Ethics Committee of the Affiliated Hospital of Integrated Traditional Chinese and Western Medicine, Nanjing University of Chinese Medicine (2019LWKZ009). All patients had signed informed consent forms to allow research on the specimens. Information on the clinical characteristics of patients can be downloaded from the Supplementary Material (Table S1).
Survival Analysis of Key miRNAs
The Cancer Genome Atlas (TCGA) is a widely used public database, jointly established by the National Cancer Institute and the National Human Genome Research Institute.29 It contains relevant data on various cancers, such as miRNA expression, mRNA expression, lncRNA expression, and clinical data. Therefore, the colon cancer dataset in the TCGA database was used to verify whether the key miRNAs are related to survival outcomes. Forest plots and Kaplan-Meier plots were constructed using the survival package of R software. A P-value < 0.05 was considered statistically significant.
Assessment of Immune Cell Infiltration in Colon Cancer
CIBERSORT (https://cibersortx.stanford.edu/) was used to evaluate immune cell infiltration of colon cancer in the TCGA database.30 CIBERSORT is a tool for the deconvolution of the human immune cell subtype expression matrix based on linear support vector regression principles. CIBERSORT provides a set of gene expression signatures for 22 immune cell subtypes by default. The “pheatmap” package and the “ggplot2” package were used to create a boxplot of immune cell infiltration.31
Correlation Analysis of Key miRNAs with Infiltrating Immune Cells and Immunotherapy Biomarkers
Correlations between key mRNAs and infiltrating immune cells were explored using Spearman’s method, and the results were visualized using the “ggplot2” package.31 Furthermore, given immunotherapy’s potential as a treatment for colon cancer, this study also examined the associations between essential miRNAs and crucial immune checkpoints, including PD-L1, CTLA-4, PDCD1, LAG-3, HAVCR2, TIGIT, ICOS, IDO1, and BTLA. A P-value < 0.05 was considered statistically significant.
Results
Screening for Common DEMis
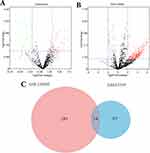
We used the “limma” R package to analyze the GSE53339 and GSE126093 miRNA microarray datasets and obtained the DEMis. We screened 99 miRNAs from the GSE53339 microarray dataset and 307 miRNAs from the GSE126093 microarray dataset. Figure 1A and B show the DEMis of the GSE53339 and GSE126093 microarray datasets, respectively. As shown in the figure, the red dots represent up-regulated DEMis, and the green dots represent down-regulated DEMis. The intersect function in R was used to identify the common DEMis among GSE53339 and GSE126093. The following 14 common DEMis were identified: miR-548b-5p, miR-363-3p, miR-145-3p, miR-18a-3p, miR-4677-5p, miR-520e, miR-5583-5p, miR-521, miRPlus-C1066, miR-431-5p, miR-548n, miR-938, miR-330-5p, and miR-4776-3p. As shown in Figure 1C, the red area represents the DEMis number of GSE53339, the blue area represents the DEMis number of GSE126093, and the purple area in the middle indicates the number of common DEMis.
Prediction of the Target Genes of Common DEMis Using GO Functional and KEGG Pathway Enrichment Analyses
Utilizing the miRwalk 3.0 online resource, we predicted target genes for common differentially expressed miRNAs (DEMiRs) identified by both miRDB and TargetScan databases. The target genes of the following five miRNAs were predicted: miR-18a-3p, miR-330-5p, miR-363-3p, miR-431-5p, and miR-520e. To study the biological functions and metabolic pathways of these miRNAs, clusterProfiler software was employed to analyze the miRNA target genes. GO functional analysis revealed the following miRNA-associated terms: (i) “biological processes” involving the regulation of oxidative stress-induced neuronal death, peptidyl-lysine trimethylation, and tube morphogenesis; (ii) “cell components” comprising the ruffle, leading-edge, and basal plasma membranes; and (iii) “molecular functions” involving phosphatidylinositol, gamma-catenin, and beta-catenin binding properties (Figure 2). KEGG pathway enrichment analysis showed that the target genes of miRNAs were mainly involved in ECM-receptor interaction and the PI3K/Akt, Ras, and TGF-β signaling pathways (Figure 3).
 |
Figure 3 KEGG pathway enrichment analysis of the common DEMis. The horizontal axis represents the number of genes, the vertical axis represents the name of the KEGG pathway. |
Construction of the miRNA-Gene Network and Identification of Key miRNAs
The miRNA-gene network was constructed using Cytoscape software (Figure 4), and the node degrees of the common DEMis were determined. Supplementary Table S2 shows the node degree information for the common DEMis. Three of the miRNAs had very high node degrees (miR-363-3p, miR-520e, and miR-330-5p), and they were therefore considered to be highly associated with the development of colon cancer.
Verification of Key miRNAs
We selected colon cancer and matched paracancerous tissues from 15 patients for qRT-PCR analysis to verify the authenticity of the key miRNAs we identified. The results showed that miR-330-5p and miR-520e were significantly upregulated in colon cancer tissues, while miR-363-3p was downregulated, with all differences being statistically significant (P-value < 0.05) (Figure 5). For detailed information regarding qRT-PCR primers of miRNA, please refer to the Supplementary Material (Table S3).
 |
Figure 5 Comparison of relative expression of miRNAs between the colon cancer tissues and the paracancerous tissues. *P < 0.05 and **P < 0.01 indicate that the data are statistically significant. |
Survival Analysis of Key miRNAs
According to the median expression of miRNA, patients were divided into high-expression and low-expression groups. Then, the Kaplan–Meier diagram of miRNA was drawn using the survival package of R software. The results showed that the expression of two miRNAs (miR-363-3p and miR-520e) was related to the prognosis of patients (Figure 6A–C). Subsequent univariate analysis identified a prognostic association with miR-363-3p expression (Figure 6D); however, the multivariate analysis did not corroborate this finding (Table S4). Future studies with larger, well-characterized cohorts may provide more evidence.
 |
Figure 6 The Kaplan-Meier plots (A–C) and univariate analysis of forest plots (D) for the three key miRNAs signature in colon cancer patients. |
Immune Cell Infiltration Results
CIBERSORT software was used to evaluate the differences in immune infiltration between 22 immune cell types between colon cancer tissues and normal controls. Figure 7A shows the proportions of immune cells in colon cancer tissues and normal controls. Compared with normal control tissues, naive B cells, plasma cells, and M2 macrophages were more infiltrated in colon cancer tissues, while M0 macrophages, M1 macrophages, follicular B helper T cells, and activated mast cells were less infiltrated (Figure 7B). The relevant heatmaps of 22 immune cells are shown in Figure 8. M2 macrophages and activated mast cells showed the strongest negative correlation (correlation coefficient = −0.47), while M1 macrophages and resting dendritic cells showed the strongest positive correlation (correlation coefficient = 0.4).
Correlation of Key miRNAs with Infiltrating Immune Cells and Immunotherapy Biomarkers
Correlation analysis of miR-363-3p, miR-330-5p, and miR-520e with infiltrating immune cells showed that miR-520e was significantly associated with infiltrating immune cells. miR-520e was positively correlated with activated CD4 memory T cells (P-value < 0.001; r = 0.24) (Figure 9A–C). Furthermore, a significant association was observed between these pivotal miRNAs and established immune checkpoints (Figure 9D).
Discussion
Colon cancer is a common malignant tumor of the digestive system that severely threatens human health and life expectancy.32 More than half of colon cancer patients have disease progression due to a lack of timely diagnosis. Furthermore, approximately 60% of patients will eventually develop metastatic recurrence, and their 5-year survival rate is less than 15%.33,34 However, the current treatment strategy for colon cancer has begun a revolutionary shift from surgery, radiation, and chemotherapy to immunotherapy and molecular targeted therapy. Nonetheless, drug resistance is still a problem in 50% of colon cancer patients.35 Therefore, the discovery of new therapeutic targets remains a top priority. Fortunately, with the rapid development of molecular biotechnology, details regarding the pathogenesis and therapeutic targets of colon cancer have been identified at the gene level. As a result, miRNA homeostasis has become one of the most popular research hotspots in the biomedical field, with miRNA disorders being implicated in almost all types of diseases, including tumors.36,37
In the pathological process of tumors, miRNAs play a dual role as both tumor promoters and suppressors. Abnormal miRNA expression is closely related to tumor carcinogenesis, malignancy, and drug resistance. Lin and his team found that up-regulation of miR-181a-5p promoted apoptosis and enhanced the sensitivity of breast cancer cells to chemotherapeutic agents.38 Another study showed that overexpression of miRNA-93-5p could increase the migration and invasiveness of breast cancer cells by targeting FGD5.39 Moreover, Li et al demonstrated that serum miR-486-5p was significantly overexpressed in patients with cervical cancer and was responsible for stimulating the proliferation, migration, and invasion of cervical cancer cells by inhibiting PTEN expression and activating the carcinogenic PI3K/Akt pathway.40 There is ample evidence that miRNAs are involved in numerous signaling pathways in colon cancer and have great potential for clinical diagnosis, prognosis, and personalized targeted therapy.16 Therefore, it is necessary to identify and validate key miRNAs in colon cancer to explore their potential research value in early diagnosis and precise targeting therapy.
Our study integrated two previously generated miRNA microarray datasets to identify 14 common DEMis. The genes targeted by the common DEMis in both miRDB and TargetSca databases were predicted using the miRwalk3.0 online tool. Five miRNAs (miR-18a-3p, miR-330-5p, miR-363-3p, miR-431-5p, and miR-520e), which targeted genes, were subjected to GO functional and KEGG pathway enrichment analyses. GO functional analysis revealed the following miRNA-associated terms: (i) “biological processes” involving the regulation of oxidative stress-induced neuronal death, peptidyl-lysine trimethylation, and tube morphogenesis; (ii) “cell components” comprising the ruffle, leading-edge and basal plasma membranes; and (iii) “molecular functions” involving phosphatidylinositol, gamma-catenin, and beta-catenin binding properties. KEGG pathway enrichment analysis showed that miRNAs were mainly involved in extracellular matrix (ECM)-receptor interaction as well as the PI3K/Akt, Ras, and TGF-β signaling pathways. The PI3K/Akt intracellular signaling pathway is primarily responsible for cell growth and glucose metabolism, and its activation is closely related to the metastasis of various cancers.41 Research has shown that in the colon cancer cell line HT-29, the PI3K/Akt pathway evades apoptosis and promotes cell proliferation through Akt activation.42 The exact pathway also can up-regulate matrix metalloproteinases and increase the invasiveness of colon cancer.43 These results are not only consistent with our findings, but also demonstrate the importance of these miRNAs.
We subsequently constructed the miRNA-gene network and identified three colon cancer-related miRNAs (miR-330-5p, miR-363-3p, and miR-520e). miR-330-5p is the product of the miR-330 gene, and abnormal expression of miR-330-5p is closely related to the development of various cancers, including cell proliferation, migration, invasion, apoptosis, and epithelial-mesenchymal transition. However, the exact role of miR-330-5p in cancer remains controversial.44–47 Deng et al found that miR-330-5p regulates proliferation, apoptosis, and invasion of colorectal cancer cells by targeting SND1.48 However, in some cancers (such as thyroid, liver, and cervical cancer), miR-330-5p can promote both tumorigenesis and anti-tumorigenesis. Gao and his team found that miR-330-5p can target the downstream tumor promoter TLR4 and inhibit its expression, thereby inhibiting the progression of thyroid cancer.49 Moreover, Jing et al found that miR-330-5p promotes thyroid cancer by inhibiting the expression of SRF gene tumor formation.50 The miR-520e gene is located at locus 19q13.4 on the chromosome and is one of the critical members of the miR-520 family. The abnormal expression of miR-520e is closely related to the occurrence and development of various cancers, such as breast, liver, and colorectal cancer. A study by Yi et al found that miR-520e expression was significantly increased in breast cancer tissues and promoted proliferation and migration of breast cancer cells in vitro.51 Tian et al found that miR-520e could target the EphA2 gene and inhibit its expression, thereby suppressing the development of liver cancer.52 MiR-363-3p belongs to the miR-363 family. The abnormal expression of miR-363-3p is also closely related to the occurrence and development of various cancers. miR-363-3p can act as a tumor suppressor to inhibit the proliferation, metastasis, and invasion of various cancers.53–55 A study has shown that miR-363-3p could target the SphK2 gene and inhibit its expression, thereby inhibiting the development of colorectal cancer.56 Research indicates that miR-363-3p targets the SphK2 gene, suppressing its expression, which in turn may impede colorectal cancer progression.56 While miR-363-3p is implicated as an tumor suppressor gene, analysis of the TCGA database reveals that its elevated expression correlates with poorer outcomes. This paradox is also reflected in other datasets.57,58 In addition, these key miRNAs did not show significance in multivariate analyses, despite the fact that these miRNAs are strongly associated with tumor progression and prognosis. To resolve these inconsistencies, further stratified analysis is required to uncover variation across demographics such as race, gender, and age.
After that, we verified the reliability of the three key miRNAs by qRT-PCR to reveal the pathogenesis of colon cancer and identify new therapeutic targets. The results showed that miR-330-5p, miR-363-3p, and miR-520e were significantly different between colon cancer and normal tissues (P < 0.05). Previous study has shown a strong relationship between miR-330-5p and miR-520e with colon cancer pathology. We searched the three key miRNAs in the human microRNA-mRNA interactome database. The results showed that miR-330-5p and miR-363-3p had been verified in human samples, and this information also confirms the authenticity of our results.59 The results of each study showed that these miRNAs are key miRNAs of colon cancer and play important roles in the development of colon cancer. This study also allowed us to recognize the limitations of small sample sizes and the differences between patients with colon cancer. Therefore, we will demonstrate the association between these key miRNAs and colon cancer by expanding the clinical sample size in subsequent studies.
Finally, we used CIBERSORT to assess the differential expression of immune cell infiltration patterns in colon cancer. Increased infiltration of naive B cells, plasma cells, and M2 macrophages and decreased infiltration of M0 macrophages, M1 macrophages, follicular B helper T cells, and activated mast cell infiltration may be related to the pathogenesis and progression of colon cancer. A recent study showed that colon cancer patients had a higher proportion of M2 macrophages than normal controls.60 Therefore, M2 macrophages may be involved in the pathogenesis of colon cancer. An earlier study showed that PLD4 promotes the antitumor effect of M1 macrophages in colon cancer cells.61 Nonetheless, the roles of naive B cells, plasma cells, follicular B helper T cells, and activated mast cells in colon cancer remain unclear. Based on our correlation analysis of key miRNAs and infiltrating immune cells, we found that only miR-520e was significantly associated with infiltrating immune cells. Therefore, we speculate that these miRNAs may not be involved in immune response pathways. Notably, significant associations were also observed between these key miRNAs and established immune checkpoints, suggesting their utility as potential prognostic biomarkers and therapeutic targets in immunotherapy for colon cancer. Current study posits that miRNAs offer novel insights into the molecular regulatory mechanisms governing immune checkpoints, which play a critical role in tumor evasion and the establishment of the tumor microenvironment.62–64
In summary, the innovative nature of our study stems from (i) using an integrated approach instead of a single miRNA microarray dataset, (ii) constructing a miRNA-gene network and using the node degree to identify key miRNAs, and (iii) performing qRT-PCR verification on the key miRNAs. The combination of the above methods can effectively screen more reliable biomarkers, thereby helping to improve the diagnosis, treatment, and, ultimately, the prognosis of colon cancer patients.
Conclusions
In this study, we explored the key miRNAs that influence the development of colon cancer from the perspective of molecular biology using bioinformatics methods. We found that three key miRNAs (miR-330-5p, miR-363-3p, and miR-520e) may be highly associated with the pathogenesis of colon cancer. We then verified the reliability of these three key miRNAs by qRT-PCR. The results of survival analysis also confirmed that the key miRNAs were related to the prognosis of patients. Finally, we believe that these three miRNAs are key miRNAs of colon cancer and play important roles in the development of colon cancer. Collectively, these findings will improve our understanding of colon cancer’s pathogenesis and the molecular mechanisms involved.
Abbreviations
DEMis, Differentially expressed miRNAs; ECM, Extracellular matrix; GEO, Gene Expression Omnibus; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA, MicroRNA; qRT-PCR, Quantitative Real-Time Polymerase Chain Reaction; TCGA, The Cancer Genome Atlas.
Data Sharing Statement
The datasets generated and/or analyzed during the current study are available in the GEO database repository: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53339 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126093.
Ethics Approval and Consent to Participate
The research was approved by the Affiliated Hospital of Integrated Traditional Chinese and Western Medicine, Nanjing University of Chinese Medicine (2019LWKZ009). All the procedures were followed in accordance with the Declaration of Helsinki under the Ethics approval and consent to participate heading.
Author Contributions
Yi Ji: Investigation, Writing - Original Draft and Funding acquisition; Jialin Gu: Visualization and Writing - Review & Editing; Hongqun Zhang: Supervision; Houxi Xu: Conceptualization, Methodology and Funding acquisition. All authors contributed to data analysis, drafting, or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82004288) and Traditional Chinese and Western Medicine Clinical Medicine Brand Construction Project of Jiangsu Higher Education Institutions (Phase II).
Disclosure
The authors declare that they have no competing interests.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca A Cancer J Clinicians. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. Ca A Cancer J Clinicians. 2020;70(3):145–164. doi:10.3322/caac.21601
3. Araghi M, Arnold M, Rutherford MJ, et al. Colon and rectal cancer survival in seven high-income countries 2010-2014: variation by age and stage at diagnosis (the ICBP SURVMARK-2 project). Gut. 2021;70(1):114–126. doi:10.1136/gutjnl-2020-320625
4. Yoshino T, Argilés G, Oki E, et al. Pan-Asian adapted ESMO Clinical Practice Guidelines for the diagnosis treatment and follow-up of patients with localised colon cancer. Ann Oncol. 2021;32(12):1496–1510. doi:10.1016/j.annonc.2021.08.1752
5. Pastille E, Frede A, McSorley HJ, et al. Intestinal helminth infection drives carcinogenesis in colitis-associated colon cancer. PLoS Pathog. 2017;13(9):e1006649. doi:10.1371/journal.ppat.1006649
6. Piawah S, Venook AP. Targeted therapy for colorectal cancer metastases: a review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer. 2019;125(23):4139–4147. doi:10.1002/cncr.32163
7. Bartel DP. Metazoan MicroRNAs. Cell. 2018;173(1):20–51. doi:10.1016/j.cell.2018.03.006
8. Gao G, Guo X, Gu W, Lu Y, Chen Z. miRNA-142-3p functions as a potential tumor suppressor directly targeting FAM83D in the development of ovarian cancer. Aging (Albany NY). 2022;14(8):3387–3399. doi:10.18632/aging.203998
9. Wang Y, Hu S, Zhang H, et al. MiRNA-186-5p exerts an anticancer role in breast cancer by downregulating CXCL13. J Healthc Eng. 2022;2022:4891889. doi:10.1155/2022/4891889
10. Yang X, Liu R. Long non-coding RNA HCG18 promotes gastric cancer progression by regulating miRNA-146a-5p/tumor necrosis factor receptor-associated factor 6 axis. Bioengineered. 2022;13(3):6781–6793. doi:10.1080/21655979.2022.2034565
11. Yu Z, Liu Y, Li Y, et al. miRNA-338-3p inhibits glioma cell proliferation and progression by targeting MYT1L. Brain Res Bull. 2022;179:1–12. doi:10.1016/j.brainresbull.2021.11.016
12. Segal M, Slack FJ. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin Drug Discov. 2020;15(9):987–992. doi:10.1080/17460441.2020.1765770
13. Koo KH, Kwon H. MicroRNA miR-4779 suppresses tumor growth by inducing apoptosis and cell cycle arrest through direct targeting of PAK2 and CCND3. Cell Death Dis. 2018;9(2):77. doi:10.1038/s41419-017-0100-x
14. Li Y, Xun J, Wang B, et al. miR-3065-3p promotes stemness and metastasis by targeting CRLF1 in colorectal cancer. J Transl Med. 2021;19(1):429. doi:10.1186/s12967-021-03102-y
15. Li C, Ding D, Gao Y, Li Y. MicroRNA‑3651 promotes colorectal cancer cell proliferation through directly repressing T‑box transcription factor 1. Int J Mol Med. 2020;45(3):956–966. doi:10.3892/ijmm.2020.4458
16. Cekaite L, Eide PW, Lind GE, Skotheim RI, Lothe RA. MicroRNAs as growth regulators, their function and biomarker status in colorectal cancer. Oncotarget. 2016;7(6):6476–6505. doi:10.18632/oncotarget.6390
17. Wu H, Wu H, Sun P, Zhu D, Ma M, Fan W. miR-155-5p promotes cell proliferation and migration of clear cell renal cell carcinoma by targeting PEG3. Urol Int. 2021;105(9–10):906–915. doi:10.1159/000514416
18. Li D, Hao X, Song Y. Identification of the key MicroRNAs and the miRNA-mRNA regulatory pathways in prostate cancer by bioinformatics methods. Biomed Res Int. 2018;2018:6204128. doi:10.1155/2018/6204128
19. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–5. doi:10.1093/nar/gks1193
20. Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi:10.1093/bioinformatics/btm254
21. Chen Z, Ren R, Wan D, et al. Hsa_circ_101555 functions as a competing endogenous RNA of miR-597-5p to promote colorectal cancer progression. Oncogene. 2019;38(32):6017–6034. doi:10.1038/s41388-019-0857-8
22. Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20(18):3705–3706. doi:10.1093/bioinformatics/bth449
23. Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinf. 2011;12:35. doi:10.1186/1471-2105-12-35
24. Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One. 2018;13(10):e0206239. doi:10.1371/journal.pone.0206239
25. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. doi:10.7554/eLife.05005
26. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–d131. doi:10.1093/nar/gkz757
27. Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2(3):100141. doi:10.1016/j.xinn.2021.100141
28. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi:10.1093/bioinformatics/btq675
29. Tomczak K, Czerwińska P, Wiznerowicz M. The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19(1a):A68–77. doi:10.5114/wo.2014.47136
30. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259. doi:10.1007/978-1-4939-7493-1_12
31. Wickham H. ggplot2. Wiley Interdiscip Rev. 2011;3(2):180–185. doi:10.1002/wics.147
32. Chakrabarti S, Peterson CY, Sriram D, Mahipal A. Early stage colon cancer: current treatment standards, evolving paradigms, and future directions. World J Gastrointest Oncol. 2020;12(8):808–832. doi:10.4251/wjgo.v12.i8.808
33. Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14(2):89–103. doi:10.5114/pg.2018.81072
34. Littlejohns P, Tamber S, Ranson P, Campbell B. Treatment for liver metastases from colorectal cancer. Lancet Oncol. 2005;6(2):73. doi:10.1016/s1470-2045(05)01729-8
35. Hu T, Li Z, Gao CY, Cho CH. Mechanisms of drug resistance in colon cancer and its therapeutic strategies. World J Gastroenterol. 2016;22(30):6876–6889. doi:10.3748/wjg.v22.i30.6876
36. Kadkhoda S, Ghafouri-Fard S. The importance of miRNA-630 in human diseases with an especial focus on cancers. Cancer Cell Int. 2022;22(1):105. doi:10.1186/s12935-022-02531-z
37. Huang L, Zhang L, Chen X. Updated review of advances in microRNAs and complex diseases: experimental results, databases, webservers and data fusion. Brief Bioinform. 2022;23(6). doi:10.1093/bib/bbac397
38. Lin J, Chen X, Sun M, et al. Upregulation of microRNA-181a-5p increases the sensitivity of HS578T breast cancer cells to cisplatin by inducing vitamin D receptor-mediated cell autophagy. Oncol Lett. 2021;21(4):247. doi:10.3892/ol.2021.12508
39. Zhou D, Bai J, Hu Y, et al. miR-93-5p promotes invasion and migration of triple negative breast cancer cells by targeting FGD5. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2020;36(9):794–801.
40. Li C, Zheng X, Li W, Bai F, Lyu J, Meng QH. Serum miR-486-5p as a diagnostic marker in cervical cancer: with investigation of potential mechanisms. BMC Cancer. 2018;18(1):61. doi:10.1186/s12885-017-3753-z
41. Noorolyai S, Shajari N, Baghbani E, Sadreddini S, Baradaran B. The relation between PI3K/AKT signalling pathway and cancer. Gene. 2019;698:120–128. doi:10.1016/j.gene.2019.02.076
42. Yang F, Gao JY, Chen H, Du ZH, Zhang XQ, Gao W. Targeted inhibition of the phosphoinositide 3-kinase impairs cell proliferation, survival, and invasion in colon cancer. Onco Targets Ther. 2017;10:4413–4422. doi:10.2147/ott.S145601
43. Jin Y, Chen W, Yang H, et al. Scutellaria barbata D. Don inhibits migration and invasion of colorectal cancer cells via suppression of PI3K/AKT and TGF-β/Smad signaling pathways. Exp Ther Med. 2017;14(6):5527–5534. doi:10.3892/etm.2017.5242
44. Tréhoux S, Lahdaoui F, Delpu Y, et al. Micro-RNAs miR-29a and miR-330-5p function as tumor suppressors by targeting the MUC1 mucin in pancreatic cancer cells. Biochim Biophys Acta. 2015;1853(10 Pt A):2392–2403. doi:10.1016/j.bbamcr.2015.05.033
45. Cui LH, Xu HR, Yang W, Yu LJ. lncRNA PCAT6 promotes non-small cell lung cancer cell proliferation, migration and invasion through regulating miR-330-5p. Onco Targets Ther. 2018;11:7715–7724. doi:10.2147/ott.S178597
46. Qin L, Sun X, Zhou F, Liu C. CircLRP6 contributes to prostate cancer growth and metastasis by binding to miR-330-5p to up-regulate NRBP1. World J Surg Oncol. 2021;19(1):184. doi:10.1186/s12957-021-02287-2
47. Karimi L, Jaberi M, Asadi M, et al. Significance of microRNA-330-5p/TYMS expression axis in the pathogenesis of colorectal tumorigenesis. J Gastrointest Cancer. 2022;53(4):965–970. doi:10.1007/s12029-021-00695-x
48. Deng J, Liu S, Zhao L, et al. SND1 acts as a functional target of miR-330-5p involved in modulating the proliferation, apoptosis and invasion of colorectal cancer cells. Biochem Biophys Res Commun. 2022;615:116–122. doi:10.1016/j.bbrc.2022.05.045
49. Gao Y, Wang F, Zhang L, et al. LINC00311 promotes cancer stem-like properties by targeting miR-330-5p/TLR4 pathway in human papillary thyroid cancer. Cancer Med. 2020;9(4):1515–1528. doi:10.1002/cam4.2815
50. Jing QB, Tong HX, Tang WJ, Tian SD. Clinical significance and potential regulatory mechanisms of serum response factor in 1118 cases of thyroid cancer based on gene chip and RNA-sequencing data. Med Sci Monit. 2020;26:e919302. doi:10.12659/msm.919302
51. Yi M, Li M, Long X, et al. miR-520e regulates cell proliferation, apoptosis and migration in breast cancer. Oncol Lett. 2016;12(5):3543–3548. doi:10.3892/ol.2016.5085
52. Tian JH, Liu WD, Zhang ZY, et al. Influence of miR-520e-mediated MAPK signalling pathway on HBV replication and regulation of hepatocellular carcinoma cells via targeting EphA2. J Viral Hepat. 2019;26(4):496–505. doi:10.1111/jvh.13048
53. Dong S, Xue S, Sun Y, et al. MicroRNA-363-3p downregulation in papillary thyroid cancer inhibits tumor progression by targeting NOB1. J Investig Med. 2021;69(1):66–74. doi:10.1136/jim-2020-001562
54. Guo Q, Dong L, Zhang C, Liu D, Peng P. MicroRNA-363-3p, negatively regulated by long non-coding RNA small nucleolar RNA host gene 5, inhibits tumor progression by targeting Aurora kinase A in colorectal cancer. Bioengineered. 2022;13(3):5357–5372. doi:10.1080/21655979.2021.2018972
55. Song B, Yan J, Liu C, Zhou H, Zheng Y. Tumor suppressor role of miR-363-3p in gastric cancer. Med Sci Monit. 2015;21:4074–4080. doi:10.12659/msm.896556
56. Dong J, Geng J, Tan W. MiR-363-3p suppresses tumor growth and metastasis of colorectal cancer via targeting SphK2. Biomed Pharmacother. 2018;105:922–931. doi:10.1016/j.biopha.2018.06.052
57. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
58. Chandrashekar DS, Karthikeyan SK, Korla PK, et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia. 2022;25:18–27. doi:10.1016/j.neo.2022.01.001
59. Plotnikova O, Baranova A, Skoblov M. Comprehensive analysis of human microRNA-mRNA interactome. Front Genet. 2019;10:933. doi:10.3389/fgene.2019.00933
60. Lan J, Sun L, Xu F, et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019;79(1):146–158. doi:10.1158/0008-5472.Can-18-0014
61. Gao L, Zhou Y, Zhou SX, et al. PLD4 promotes M1 macrophages to perform antitumor effects in colon cancer cells. Oncol Rep. 2017;37(1):408–416. doi:10.3892/or.2016.5216
62. Kipkeeva F, Muzaffarova T, Korotaeva A, et al. The features of immune checkpoint gene regulation by microRNA in cancer. Int J Mol Sci. 2022;23(16):9324. doi:10.3390/ijms23169324
63. Wang Q, Lin W, Tang X, et al. The roles of microRNAs in regulating the expression of PD-1/PD-L1 immune checkpoint. Int J Mol Sci. 2017;18(12):2540. doi:10.3390/ijms18122540
64. Skafi N, Fayyad-Kazan M, Badran B. Immunomodulatory role for MicroRNAs: regulation of PD-1/PD-L1 and CTLA-4 immune checkpoints expression. Gene. 2020;754:144888. doi:10.1016/j.gene.2020.144888
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.