Back to Journals » Cancer Management and Research » Volume 13
Hypomethylation-Mediated AGR2 Overexpression Facilitates Cell Proliferation, Migration, and Invasion of Lung Adenocarcinoma
Authors He J, Fu Y, Hu J, Chen J, Lou G
Received 2 February 2021
Accepted for publication 4 June 2021
Published 30 June 2021 Volume 2021:13 Pages 5177—5185
DOI https://doi.org/10.2147/CMAR.S304869
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
Junming He, Yin Fu, Jiangwei Hu, Jian Chen, Guoliang Lou
Department of Cardiothoracic Surgery, Yiwu Central Hospital, Yiwu, 322000, People’s Republic of China
Correspondence: Guoliang Lou
Department of Cardiothoracic Surgery, Yiwu Central Hospital, Yiwu, 322000, People’s Republic of China
Tel +86-13868974422
Email [email protected]
Objective: Studies have indicated that AGR2 is crucial in many cancers. However, its methylation level in lung adenocarcinoma (LUAD) is rarely known. Hence, the effect of AGR2 methylation on LUAD was explored in the study.
Methods: qRT-PCR was adopted to detect the expression of AGR2 in LUAD cells and normal lung cells. Methylation-specific PCR (MSP) was used to detect the methylation of AGR2 promoter region in different cell lines. MTT, Transwell and wound healing assays were used to verify the progression of cells in each transfection group.
Results: The expression of AGR2 was significantly up-regulated in LUAD cells relative to that in normal cells. Moreover, the expression of AGR2 was inversely modulated by DNA methylation, and the hypomethylation of CpG islands would lead to the increased expression of AGR2. Finally, overexpression and hypomethylation of AGR2 facilitated the proliferation, invasion and migration of LUAD cells.
Conclusion: These results demonstrated that hypomethylation of AGR2 promoter region promoted the expression of AGR2 in LUAD cells, thus promoting the progression of LUAD cells.
Keywords: AGR2, lung adenocarcinoma, methylation, migration, proliferation
Introduction
Lung cancer is the most frequent malignancy that is responsible for cancer-related deaths throughout the world1. Small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) are two main subtypes of lung cancer.2 Lung adenocarcinoma (LUAD) is the commonest subtype of NSCLC, accounting for 40% of all cases of lung cancer, and it comes from the glands or epithelium of the bronchi, with main clinical manifestations as dry cough, chest pain, and shortness of breath, which affects breathing, causes pain and even threatens life.3–5 In recent years, more and more therapeutic strategies have been proposed and applied to LUAD, however, due to the lack of molecular markers for early diagnosis of LUAD, the prognosis of LUAD patients after treatment is still unsatisfactory, with a 5-year survival rate of less than 20%.6–8 Therefore, understanding the molecular mechanism underlying LUAD occurrence and finding the molecular markers are vital for the diagnosis and treatment of LUAD.
Anterior gradient 2 (AGR2) is able to encode a human homologue that secrets a kind of proteins which were firstly identified in Xenopus laevis, and it is named XAG-2 according to its specific expression pattern in early development.9 Human AGR2 is mainly highly expressed in lung, stomach, colon, prostate, and small intestine tissues, and these tissues contain mucus secreting cells, mainly endocrine organs.10,11 AGR2 lies on the chromosome 7p21.3 region and is expressed by 8 exons to produce a full-length 175 amino acid protein with a molecular weight of 19.9792KD (including secretory peptides).12,13 AGR2 protein is a resident protein of endoplasmic reticulum (ER), which belongs to protein disulfide isomerase (protein-disulfide Isomerase, PDI) superfamily, and participates in the production of protein folding in the ER.14 Studies have indicated that AGR2 is a potential drug target and biomarker in breast cancer, and can promote cell proliferation.15 AGR2 is a novel target for the treatment of colorectal cancer and its high expression in colon cancer modulates cell function.16 In addition, AGR2 is highly expressed in esophageal cancer,17 pancreatic cancer,18 prostate cancer,19 and lung cancer.20,21 ARG2 is thought to promote proliferation, survival, and metastasis of tumor cells. Salmans et al demonstrated that AGR2 is a marker of breast cancer metastasis, and its overexpression in estrogen receptor-positive breast cancer is associated with poor prognosis, especially in tumors that evade anti-hormone therapy,22 suggesting that AGR2 may be involved in cell metastasis in hormone-related tumors. Taken together, AGR2 is vital in tumor therapy, and can be used as a drug target and biomarker in many cancers.
With the development of tumor studies, researchers have found that genetic variation and epigenetic modification are the two major mechanisms that promote tumor occurrence and development.23 The present studies of epigenetic mechanisms mainly focus on DNA methylation and histone modification. DNA methylation is the first discovered epigenetic phenomenon24 and refers to the transfer of methyl group to the 5ʹcarbon of cytosine with unmethylated cytosine-phosphate-guanosine (CpG) dinucleotide in DNA molecules under the catalysis of DNA methyltransferase (DNMT).25,26 Currently, DNA methylation, especially the abnormal methylation of CpG islands in gene promoter regions, has become the focus of tumor study.27 Abnormal methylation of CpG islands in gene promoter regions will lead to activation of proto-oncogenes or silencing of tumor suppressor genes, so as to change the expression of downstream key genes, thus promoting abnormal cell proliferation.28 Many studies have found that abnormal DNA methylation can promote the proliferation, invasion and metastasis of tumor cells such as colorectal cancer,29 gastric cancer,30 hepatocellular carcinoma31,32 and breast cancer.33 Sung HY et al34 found that AGR2 promoter hypomethylation and overexpression fosters migratory and invasive activities of ovarian cancer cells. However, AGR2 promoter hypomethylation in lung cancer is poorly studied.
Here, the promoter methylation level and expression of AGR2 along with their correlation in LUAD cells were investigated by bioinformatics analysis, methylation sequencing, molecular and cell experiments. In addition, the effects of hypomethylation and high expression of AGR2 on LUAD cells were investigated. Our study may offer new insights into the LUAD pathogenesis and help the discovery of new therapeutic targets.
Materials and Methods
Data Sources and Processing
The 450 k methylation data (tumor: n=475; normal: n=32) of LUAD were downloaded from TCGA database (https://portal.gdc.cancer.gov/). After the standardized treatment, the candidate methylation-driven genes were screened by “MethylMix” package. At the same time, gene expression profiles (tumor: n=522; normal: n=58) of TCGA-LUAD were downloaded from TCGA database. Differential analysis was carried out by using edgeR with the cutoff value of logFC=2 and FDR=0.05 and survival analysis was conducted by “survival” package.
Cells and Cell Culture
Human LUAD cell lines 95-D (BNCC100126), H1299 (BNCC100859), H1975 (BNCC340345), A549 (BNCC337696) and normal cell-line BEAS-2B (BNCC338205) were from BeNa Culture Collection (China). All cells were placed in RPMI-1640 (Gibco, 31800105, Life Technologies, Carlsbad) medium containing 10% fetal bovine serum (FBS) and 1% antibiotics at 37 °C with 5% CO2 for culture.
DNA Methylation-Specific PCR (MSP)
TIANamp Genomic DNA kit (Tiangen, Beijing, China) was used to extract the genomic DNA from human LUAD cell lines 95-D, H1299, H1975 and A549, as well as normal cell-line BEAS-2B according to the instructions. The genomic DNA was then treated with sodium bisulfite using the EpiTect Bisulfite Kit (Qiagen, Hilden, Germany). Two μg DNA was dissolved in 50 μL deionized water, and denatured at 50 °C for 10 min with 3 mol/L NaOH. Then, newly configured 30 mL 10 mmol/L hydroquinone and 520 μL 3.6 mol/L sodium bisulfite with pH=5.0 were added to the solution for water bath at 50 °C for 18 h. After purification by adsorption column and ethanol precipitation, DNA was dissolved in 50 μL deionized water. Subsequently, 1–2 ng sulfite transformed genomic DNA was used as a template for PCR analysis with methylation-specific primers. PCR products were loaded on 2% agarose gel electrophoresis and stained with ethidium bromide. The methylation band (M) and unmethylation band (U) appear in the CpG methylation part of MSP analysis. If there was no methylation in the CpG sequence, U appeared. However, if part of the CpG sequence was methylated, then M and U appeared at the same time. Finally, ImageJ was used to measure the density of the grayscale image (National Institutes of Health, Bethesda, MD). The primers used were listed in Supplementary Table 1.
5-Aza-2ʹ-Deoxycytidine (5-AzadC) Treatment
To demethylate the methylated CpG sites, 95-D and H1299 cells were treated with different doses of 5-aza-dC (0, 5, 10 µM) for 3 d.
Cell Transfection
The oe-NC and oe-AGR2 plasmids were designed and built by Sangon Biotech (Shanghai) Co., Ltd. The plasmids were transfected instantaneously into 95-D and H1299 cell lines using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions, and cells were cultured in the corresponding medium with 5% CO2 at 37 °C. All cells should be cultured in medium for at least 24 h before transfection and washed with phosphate buffer solution (PBS, pH 7.4) prior to transient transfection.
qRT-PCR
Trizol (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from tissues and cells. The ratio of 260/230 and 260/280 was detected by nanodrop 2000 micro-ultraviolet spectrophotometer (1011U, nanodrop, USA) and the concentration and purity of extracted total RNA were determined. mRNA was reversely transcribed into cDNA by TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix kit (AT311-02, TransGen Biotech Co., Ltd., Beijing), and the cDNA was diluted to 50 ng/μL. THUNDERBIRD SYBR qPCR Mix (TOYOBO) was used for mRNA detection. The reaction amplification system was 25 μL. ABI7500 quantitative PCR instrument (7500, ABI, USA) was used to perform real-time fluorescence quantitative PCR. β-actin was used as internal reference for mRNA and each group was measured in triplicate. Primer sequences shown in Supplementary Table 2 were all purchased from Sangon Biotech Co., Ltd. (Shanghai, China). The difference in relative expression was calculated by 2−ΔΔCt method.
Western Blotting
Total proteins were isolated with radioimmunoprecipitation assay (RIPA) lysis (Invitrogen; Thermo Fisher Scientific, Inc.). Bicinchoninic acid (BCA) protein assay kit (Beyotime Institute of Biotechnology, Haimen, China) was implemented to detect protein concentration. The total proteins were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and were then transferred onto polyvinylidene fluoride (PVDF) membrane (Amersham, USA). The PVDF membrane was blocked with 5% skim milk for 1 h. The confining liquid was poured. Next, the membrane was incubated overnight at 4 °C with primary antibodies AGR2 (Abcam, UK) and GAPDH (Abcam, UK). Subsequently, the membrane was rinsed 3 times with PBST (PBS containing 0.1% Tween-20) buffer (10 min/time), and was incubated with horseradish peroxidase-labeled secondary antibody goat anti-rabbit IgG H&L (HRP) (ab6721, Abcam, UK) at room temperature for 1 h. The enhanced chemiluminescence (ECL; Solarbio, Beijing, China) was added for development, and gel imaging software was employed for analysis.
Cell Proliferation Assay
Cell proliferation was detected by MTT assay. When the cell growth density reached about 80% after transfection, cells were washed with PBS twice and digested with 0.25% trypsin to obtain the single-cell suspension. After cell counting, cells in (3–6) ×103 cells/well were inoculated in 96-well plates with a volume of 0.2 mL/well. Each group was made in sextuplicate. Cells were cultured in an incubator and at 24 h, 48 h, 72 h, 96 h and 120 h, respectively, the medium was changed to medium containing 10% MTT solution (5 g/L) (GD-Y1317, Guduo Biotechnology Co., LTD., Shanghai, China), and cells were cultured for 4 h. The supernatant was discarded and 100 μL dimethyl sulphoxide (D5879-100 mL, Sigma, USA) was added to each well. The mixture was placed on a shaker for 10 min to fully dissolve the formazan crystals produced by living cells. The absorbance values of each well at 450 nm were detected by the microplate reader (Nanjing Detielab Co., LTD.). Each experiment was repeated three times, and the cell activity curves were plotted using the time point as the abscissa.
Wound Healing and Transwell Invasion Assays
The transfected cells were digested with trypsin and seeded in a 6-well plate at a density of 5×106 cells/well until cells were adherent to the wall. Subsequently, 10 μL sterile pipette tip was used to scratch the single layer of cells, and the scraped cells were removed by washing with sterile PBS. The medium was replaced with serum-free medium for further culture. The scratch distance was observed and photographed after 0 h and 24 h, respectively. Wound healing present (%)=(0 h cell distance - 24 h cell distance)/0 h cell distance×100%.
Matrigel (356234, BD Company, USA) was dissolved overnight at 4 °C, diluted with serum-free medium at 1:3, and added to the upper chamber of Transwell chamber at 50 μL/well, and balanced in the incubator for 30 min. Cell density was diluted to 1×105 cells/mL by serum-free medium in the upper chamber, and the medium containing 15% FBS was added to the lower chamber. After incubation at 37 °C for 48 h, the cells that did not pass through the membrane were removed with a cotton swab, and the cells in the lower chamber were stained with 0.1% crystal violet. The number of invading cells in each group was used as an indicator to evaluate invasion ability. Four random fields were selected under the microscope to count and photograph the invading cells. The average number of cells in each field was counted.
Statistics Analysis
All data were analyzed using GraphPad Prism 6.0 (La Jolla, CA). Each experiment was repeated 3 times, including 3 biological replicates and 3 technical replicates. The results were expressed as mean ± standard deviation, and the comparison between two groups was analyzed by t-test. P<0.05 indicated statistically significant difference, while P<0.01 indicated extremely significant difference.
Results
AGR2 is Hypomethylated and Highly Expressed in LUAD
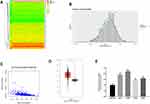
Methylation data of LUAD were downloaded from TCGA database for MethylMix analysis to screen out 78 methylation-driven genes (Figure 1A), among which AGR2 was significantly hypomethylated in tumor tissues (Figure 1B), and the methylation level of AGR2 was inversely related with its expression level (Figure 1C). Expression analysis of LUAD and normal tissues in the TCGA database showed that the expression level of AGR2 in tumor tissues was greatly up-regulated (Figure 1D). Additionally, the expression of AGR2 in LUAD cell lines 95-D, H1299, H1975, A549 and in the normal cell-line BEAS-2B were detected by qRT-PCR, and it was found that the expression of AGR2 in LUAD cell lines was higher than that in the normal cell line (Figure 1E). It could be concluded that AGR2 was hypomethylated and highly expressed in LUAD.
Methylation of AGR2 in LUAD Cells and Normal Cells
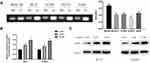
High expression of AGR2 in LUAD may be caused by the hypomethylation of its DNA promoter region through bioinformatics analysis. Therefore, we further explored the methylation of AGR2 in LUAD cells through in vitro experiments. In AGR2 is Hypomethylated and Highly Expressed in LUAD, the mRNA expression level of AGR2 in the normal cell line and four LUAD cell lines were measured by qRT-PCR. It was discovered that the mRNA expression of AGR2 in LUAD cells, especially in 95-D and H1299 cells, was significantly higher than that in normal cells, so 95-D and H1299 cells were selected for subsequent functional experiments (Figure 1E). Then, DNA in LUAD cells and normal cells was treated with bisulfite and the CpG island methylation of AGR2 promoter was determined by MSP. It was exhibited that the methylation level of AGR2 in 4 LUAD cell lines was lower than that in normal cells (P<0.05, Figure 2A), which further supported our previous finding that there was a negative correlation between methylation and expression of AGR2. Later, we treated 95-D and H1299 cells with 5 µM and 10 µM methyltransferase inhibitor 5-AzadC for 3 d, and detected the mRNA expression of AGR2 after treatment. It was observed that the mRNA expression level of AGR2 was significantly increased after treatment in a dose-dependent manner. Hence, 10 µM 5-AzadC was selected in the following assays (P<0.01) (Figure 2B). The similar results were observed in Western blotting (Figure 2C). The result suggested that inhibition of AGR2 promoter methylation promoted the expression of AGR2 in LUAD cells. These results further demonstrated that the methylation level and expression level of AGR2 were negatively correlated in LUAD, and the high expression of AGR2 was caused by the hypomethylation of the AGR2 promoter region.
Promoter Hypomethylation and Overexpression of AGR2 Promote the Progression of LUAD Cells
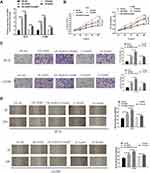
To investigate the effect of AGR2 promoter hypomethylation and AGR2 overexpression on the growth of LUAD, we overexpressed AGR2 and treated LUAD cells with methyltransferase inhibitor. The results manifested that AGR2 expression was notably increased in two LUAD cell lines in AGR2 overexpressing and inhibitor group. 5-AzadC could further increase AGR2 expression in cells compared with that in AGR2 overexpression group (oe-AGR2) (Figure 3A). From the results of MTT (Figure 3B), Transwell (Figure 3C) and wound healing (Figure 3D) assays, the proliferation, invasion and migration capacities were obviously increased in two LUAD cells with AGR2 overexpression or methyltransferase inhibitor. 5-AzadC enhanced proliferation, migration and invasion of AGR2-overexpressing cells compared with oe-AGR2 group, as shown in Figure 3B and D. These results revealed that overexpression and hypomethylation of AGR2 could stimulate the proliferation, migration and invasion of LUAD cells.
Discussion
A number of studies showed that DNA methylation is closely related to the occurrence and development of LUAD. Studies have reported that four of these five genes HOXA9, TAL1, ATP8A2, ENG and SPARCL1 have highly methylated CpG sites in their promoters, which are important in the diagnosis and treatment of LUAD.35 In addition, STXBP6 is differentially expressed in LUAD, and has been identified as a novel biomarker in LUAD. The possible role of hypermethylation of STXBP6 in the poor clinical prognosis of lung cancer patients provides a basis for the genetic etiology of LUAD.36 In contrast to hypermethylation, increased expression of S100A15 and reduced DNA methylation of its gene promoter have also been proved to be associated with high metastatic potential and poor prognosis of LUAD.37 In this study, we demonstrated that the AGR2 promoter region was hypomethylated and AGR2 was highly expressed in LUAD cells through bioinformatics analysis and molecular experiments, which was consistent with studies of AGR2 promoter methylation in other cancers. Sung HY et al found that the promoter of AGR2 is hypomethylated and AGR2 is highly expressed in ovarian cancer cells.34 These results indicate that the hypomethylation of AGR2 may play a role in promoting the occurrence and development of cancer. Our results and previous studies indicate that in a variety of cancers, including LUAD, the AGR2 promoter region is hypomethylated and AGR2 is highly expressed, while the methylation level is negatively correlated with the expression level.
To explore whether AGR2 promoter hypomethylation could modulate the expression of AGR2 and affect the growth of LUAD cells, methylation sequencing, cell functional experiments combined with different doses of DNA methyltransferase inhibitor were performed to find that hypomethylation promoted AGR2 expression, and the expression of AGR2 in cells treated with 5-AzadC was markedly increased in a dose-dependent manner. In addition, hypomethylation and overexpression of AGR2 promoted the proliferation and migration of LUAD cells. 5-AzadC enhanced proliferation, migration and invasion of AGR2-overexpressing cells compared with oe-AGR2 group. Xiu et al found that AGR2 as an oncogene promotes the proliferation and metastasis of breast cancer cells.38 Xue et al demonstrated that AGR2 is associated with poor prognosis and promotes cell proliferation in NSCLC.39 Our results further confirmed the role of AGR2 in promoting cancer and verified that both hypomethylation and overexpression of AGR2 promoted LUAD.
In summary, our results indicated that AGR2 played an oncogenic role in LUAD, and the hypomethylation of AGR2 promoted the invasion and migration of LUAD cells. These conclusions offer a better understanding of the function of AGR2 methylation in LUAD and provide a foundation for seeking new targeted therapeutic approaches for LUAD.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Giovino GA. Epidemiology of tobacco use in the United States. Oncogene. 2002;21:7326–7340. doi:10.1038/sj.onc.1205808
2. Nasim F, Sabath BF, Eapen GA. Lung Cancer. Med Clin North Am. 2019;103:463–473. doi:10.1016/j.mcna.2018.12.006
3. Liu Z, Xu L. UBE2S promotes the proliferation and survival of human lung adenocarcinoma cells. BMB Rep. 2018;51:642–647. doi:10.5483/BMBRep.2018.51.12.138
4. Imielinski M, Berger A, Hammerman P, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi:10.1016/j.cell.2012.08.029
5. McGuire S. World Cancer Report 2014. Adv Nutr. 2016;7:418–419. doi:10.3945/an.116.012211
6. Alberg AJ, Brock MV, Ford JG, Samet JM, Spivack SD. Epidemiology of lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143:e1S–e29S. doi:10.1378/chest.12-2345
7. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi:10.1002/ijc.29210
8. Wood SL, Pernemalm M, Crosbie PA, Whetton AD. Molecular histology of lung cancer: from targets to treatments. Cancer Treat Rev. 2015;41:361–375. doi:10.1016/j.ctrv.2015.02.008
9. Brychtova V, Vojtesek B, Hrstka R. Anterior gradient 2: a novel player in tumor cell biology. Cancer Lett. 2011;304:1–7. doi:10.1016/j.canlet.2010.12.023
10. Shih LJ, Lu Y-F, Chen Y-H, et al. Characterization of the agr2 gene, a homologue of X. laevis anterior gradient 2, from the zebrafish, Danio rerio. Gene Expr Patterns. 2007;7:452–460. doi:10.1016/j.modgep.2006.11.003
11. Thompson DA, Weigel RJ. hAG-2, the human homologue of the Xenopus laevis cement gland gene XAG-2, is coexpressed with estrogen receptor in breast cancer cell lines. Biochem Biophys Res Commun. 1998;251:111–116. doi:10.1006/bbrc.1998.9440
12. Fletcher GC, Patel S, Tyson K, et al. hAG-2 and hAG-3, human homologues of genes involved in differentiation, are associated with oestrogen receptor-positive breast tumours and interact with metastasis gene C4.4a and dystroglycan. Br J Cancer. 2003;88:579–585. doi:10.1038/sj.bjc.6600740
13. Myung JK, Frischer T, Afjehi-Sadat L, Pollak A, Lubec G. Mass spectrometrical analysis of the processed metastasis-inducing anterior gradient protein 2 homolog reveals 100% sequence coverage. Amino Acids. 2008;35:485–494. doi:10.1007/s00726-007-0551-y
14. Fessart D, de Barbeyrac C, Boutin I, et al. Extracellular AGR2 triggers lung tumour cell proliferation through repression of p21(CIP1). Biochim Biophys Acta Mol Cell Res. 2021;1868:118920. doi:10.1016/j.bbamcr.2020.118920
15. Salmans ML, Zhao F, Andersen B. The estrogen-regulated anterior gradient 2 (AGR2) protein in breast cancer: a potential drug target and biomarker. Breast Cancer Res. 2013;15(2):204. doi:10.1186/bcr3408
16. Gao H, Xu X, Chen B, et al. Anterior gradient 2: a new target to treat colorectal cancer. Med Hypotheses. 2013;80:706–708. doi:10.1016/j.mehy.2013.02.015
17. DiMaio MA, Kwok S, Montgomery KD, Lowe AW, Pai RK. Immunohistochemical panel for distinguishing esophageal adenocarcinoma from squamous cell carcinoma: a combination of p63, cytokeratin 5/6, MUC5AC, and anterior gradient homolog 2 allows optimal subtyping. Hum Pathol. 2012;43(11):1799–1807. doi:10.1016/j.humpath.2012.03.019
18. Chen R, Pan S, Duan X, et al. Elevated level of anterior gradient-2 in pancreatic juice from patients with pre-malignant pancreatic neoplasia. Mol Cancer. 2010;9:149. doi:10.1186/1476-4598-9-149
19. Zhang Y, Forootan SS, Liu D, et al. Increased expression of anterior gradient-2 is significantly associated with poor survival of prostate cancer patients. Prostate Cancer Prostatic Dis. 2007;10:293–300. doi:10.1038/sj.pcan.4500960
20. Alavi M, Mah V, Maresh EL, et al. High expression of AGR2 in lung cancer is predictive of poor survival. BMC Cancer. 2015;15:655. doi:10.1186/s12885-015-1658-2
21. Luu TT. Overexpression of AGR2 Is associated with drug resistance in mutant non-small cell lung cancers. Anticancer Res. 2020;40:1855–1866. doi:10.21873/anticanres.14139
22. Ong HS, Ji T, Wang LZ, Yu ZW, Zhang CP. Dermatofibrosarcoma protuberans on the right neck with superior vena cava syndrome: case report and literature review. Int J Oral Maxillofac Surg. 2013;42:707–710. doi:10.1016/j.ijom.2013.02.014
23. Wu C, Morris JR. Genes, genetics, and epigenetics: a correspondence. Science (New York, N Y). 2001;293:1103–1105. doi:10.1126/science.293.5532.1103
24. Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948;175:315–332. doi:10.1016/S0021-9258(18)57261-6
25. Morgan AE, Davies TJ, Mc Auley MT. The role of DNA methylation in ageing and cancer. Proc Nutr Soc. 2018;77:412–422. doi:10.1017/s0029665118000150
26. Holčáková J. Effect of DNA Methylation on the Development of Cancer. Klinicka Onkologie Casopis Ceske a Slovenske Onkologicke Spolecnosti. 2018;31:41–45. doi:10.14735/amko20182S41
27. Mersakova S, Nachajova M, Szepe P, Kasajova PS, Halasova E. DNA methylation and detection of cervical cancer and precancerous lesions using molecular methods. Tumor Biol. 2016;37(1):23–27. doi:10.1007/s13277-015-4197-1
28. Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi:10.1016/b978-0-12-380866-0.60002-2
29. Liu X, Chen X, Zeng K, et al. DNA-methylation-mediated silencing of miR-486-5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/β-catenin signal pathways. Cell Death Dis. 2018;9:1037. doi:10.1038/s41419-018-1105-9
30. Xin L, Liu L, Liu C, et al. DNA-methylation-mediated silencing of miR-7-5p promotes gastric cancer stem cell invasion via increasing Smo and Hes1. J Cell Physiol. 2020;235:2643–2654. doi:10.1002/jcp.29168
31. Cai C, Xie X, Zhou J, et al. Identification of TAF1, SAT1, and ARHGEF9 as DNA methylation biomarkers for hepatocellular carcinoma. J Cell Physiol. 2020;235:611–618. doi:10.1002/jcp.28999
32. Fu Y, Feng M-X, Yu J, et al. DNA methylation-mediated silencing of matricellular protein dermatopontin promotes hepatocellular carcinoma metastasis by α3β1 integrin-Rho GTPase signaling. Oncotarget. 2014;5:6701–6715. doi:10.18632/oncotarget.2239
33. Yao R, Jiang H, Ma Y, et al. PRMT7 induces epithelial-to-mesenchymal transition and promotes metastasis in breast cancer. Cancer Res. 2014;74:5656–5667. doi:10.1158/0008-5472.Can-14-0800
34. Sung HY, Choi EN, Lyu D, et al. Aberrant hypomethylation-mediated AGR2 overexpression induces an aggressive phenotype in ovarian cancer cells. Oncol Rep. 2014;32:815–820. doi:10.3892/or.2014.3243
35. Yan H, Guan Q, He J, et al. Individualized analysis reveals CpG sites with methylation aberrations in almost all lung adenocarcinoma tissues. J Transl Med. 2017;15:26. doi:10.1186/s12967-017-1122-y
36. Lenka G. Identification of Methylation-Driven, Differentially Expressed STXBP6 as a Novel Biomarker in Lung Adenocarcinoma. Sci Rep. 2017;7:42573. doi:10.1038/srep42573
37. Chen YC, Lin M-C, Hsiao -C-C, et al. Increased S100A15 expression and decreased DNA methylation of its gene promoter are involved in high metastasis potential and poor outcome of lung adenocarcinoma. Oncotarget. 2017;8:45710–45724. doi:10.18632/oncotarget.17391
38. Xiu B, Chi Y, Liu L, et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer. 2019;18:187. doi:10.1186/s12943-019-1115-y
39. Xue X, Fei X, Hou W, et al. miR-342-3p suppresses cell proliferation and migration by targeting AGR2 in non-small cell lung cancer. Cancer Lett. 2018;412:170–178. doi:10.1016/j.canlet.2017.10.024
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.