Back to Journals » OncoTargets and Therapy » Volume 13
Hsa_circ_0006916 Knockdown Represses the Development of Hepatocellular Carcinoma via Modulating miR-599/SRSF2 Axis
Authors Zhu Z, Shen S, Zhao S, Wang Z
Received 12 June 2020
Accepted for publication 22 September 2020
Published 4 November 2020 Volume 2020:13 Pages 11301—11313
DOI https://doi.org/10.2147/OTT.S267471
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
This paper has been retracted.
Zhixiang Zhu, Songhe Shen, Sen Zhao, Zhixue Wang
Image Center, The First Affiliated Hospital of Henan University, Kaifeng, Henan 475001, People’s Republic of China
Correspondence: Zhixue Wang
Image Center, The First Affiliated Hospital of Henan University, 357 Ximen Street, Kaifeng, Henan 475001, People’s Republic of China
Tel +86-37122736795
Email [email protected]
Background: The aberrantly expressed circular RNAs (circRNAs) are implicated in the progression of hepatocellular carcinoma (HCC). CircRNA hsa_circ_0006916 (circ_0006916) is dysregulated in HCC, but the function and mechanism of this circRNA in HCC development remain uncertain.
Methods: Thirty paired HCC and normal tissues were collected. circ_0006916, microRNA (miR)-599 and serine/arginine rich splicing factor 2 (SRSF2) abundances were examined via quantitative reverse transcription polymerase chain reaction or Western blot. Cell viability, colony ability, migration, invasion, cell cycle and apoptosis were tested via cell counting kit-8, colony formation, wound healing analysis, transwell analysis, and flow cytometry. The interaction between miR-599 and circ_0006916 or SRSF2 was analyzed via dual-luciferase reporter and RNA immunoprecipitation analyses. The function of circ_0006916 on cell growth in vivo was analyzed via xenograft model.
Results: circ_0006916 expression was increased in HCC tissues and cell lines. circ_0006916 knockdown reduced cell viability, colony formation, migration and invasion and caused cell cycle arrest and apoptosis. miR-599 was targeted via circ_0006916, and miR-599 knockdown reversed the influence of circ_0006916 silence on HCC progression. SRSF2 was targeted via miR-599, and miR-599 overexpression suppressed cell viability, colony formation, migration and invasion and promoted cell cycle arrest and apoptosis via decreasing SRSF2. circ_0006916 could regulate SRSF2 expression via miR-599. circ_0006916 knockdown decreased HCC cell growth in the xenograft model.
Conclusion: circ_0006916 knockdown represses the progression of HCC via regulating miR-599 and SRSF2.
Keywords: hepatocellular carcinoma, circ_0006916, miR-599, SRSF2
Introduction
Hepatocellular carcinoma (HCC) is a frequent medical problem ranked as a leading cause of cancer death.1 Great improvement has gained on understanding the risk, prevention and management of HCC.2 The surgical resection is a primary option for HCC treatment, but about 70% of patients develop the recurrent cancer with poor outcomes.3 Thus, it’s necessary to find new strategies for the treatment of HCC.
The noncoding RNAs have important roles in the development, pathogenesis and treatment of HCC.4 Circular RNAs (circRNAs) are a group of highly conserved and stable noncoding RNAs without 5ʹ and 3ʹ ends, which are dysregulated and exhibit multiple functions in HCC.5 CircRNAs can communicate with mRNAs via sponging microRNAs (miRNAs) to participate in the development of HCC.6 For instance, circRNA hsa_circ_0056836 can promote HCC cell migration, proliferation and invasion via regulating miR-766-3p and Fos-related antigen 2 (FOSL2).7 Moreover, hsa_circ_0000092 contributes to the proliferation, migration, invasion and angiogenesis of HCC cells via modulating miR-338-3p and hematological and neurological expressed 1 (HN1).8 In addition, there are some circRNAs playing an anti-cancer role in HCC, such as hsa_circ_0070269 and circRNA-5692.9,10 Homer scaffolding protein 1 (HOMER1) is an important gene significantly associated with the poor survival of HCC patients.11 The circRNA hsa_circ_0006916 (circ_0006916), derived from HOMER1, can repress cell proliferation and promote apoptosis in lung cancer cells.12 Additionally, emerging evidence suggests that circ_0006916 knockdown mitigates methamphetamine (METH)-induced neuronal damage.13 More importantly, a previous report shows that circ_0006916 expression is enhanced in HCC tissues when compared to non-tumor tissues.14 Furthermore, circ_0006916 can promote HCC cell proliferation, migration and invasion via modulating miR-1322/C-X-C motif chemokine ligand 6 (CXCL6) axis.15 Nevertheless, the mechanism addressed by circ_0006916 is complex, and additional regulatory network of circ_0006916 is expected to be explored.
miRNAs with ~23 nucleotides control mRNAs abundance via binding to their 3ʹUTR, thus being relevant to HCC development.16 miR-599 usually functions as a tumor suppressor via repressing the malignant growth and metastasis of human cancers, such as esophageal carcinoma and gastric cancer.17,18 More importantly, miR-599 could target Myc to restrain cell proliferation, migration and invasion in HCC.19 Serine/arginine rich splicing factor 2 (SRSF2) is highly expressed in HCC and is related to the poor prognosis of patients.20 Moreover, SRSF2 can facilitate cell proliferation and tumorigenic potential via regulating cancer-associated splice variants in HCC.21 The bioinformatic analysis predicts that circ_0006916 and SRSF2 can bind to miR-599. Hence, we hypothesize that circ_0006916 may target SRSF2 via regulating miR-599 to participate in HCC development.
In this research, we detected circ_0006916 expression in HCC tissues and cell lines. Moreover, we assessed the function of circ_0006916 on HCC development in vitro and in vivo. Additionally, we analyzed the interaction network of circ_0006916/miR-599/SRSF2.
Patients and Methods
Patients and Tissue Collection
Thirty HCC patients who underwent surgery were enrolled from the First Affiliated Hospital of Henan University. They all did not receive any other therapy before surgery. HCC tumor tissues and adjacent normal liver tissues were harvested and maintained at −80°C. All patients provided the written informed consent. This research was permitted via the ethics committee of the First Affiliated Hospital of Henan University, and conducted in line with the Helsinki Declaration.
Cell Culture
Human HCC cell lines (Huh7 and SNU-387 cells) were purchased from Procell (Wuhan, China) and grew in DMEM (Thermo Fisher, Waltham, MA, USA) plus 10% fetal bovine serum (Gibco, Gran Island, NY, USA) and 1% penicillin/streptomycin (Beyotime, Shanghai, China). The normal liver cell line (THLE-2 cells) was provided via American Type Culture Collection (Manassas, VA, USA) and cultured in BEGM (Lonza/Clonetics Corporation, Walkersville, MD, USA) without gentamycin/amphotericin and epinephrine, but with additional 5 ng/mL epidermal growth factor, 70 ng/mL phosphoethanolamine, and 10% fetal bovine serum. All cells were incubated at 37°C and 5% CO2.
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
The RNA was extracted using Trizol reagent (Thermo Fisher) following the instructions as previously reported.22 The RNA with OD260/280 ratio at 1.8~2.0 was used for cDNA generation using the specific reverse transcription kit (Fulengen, Guangzhou, China). Next, the qRT-PCR was performed using the mixture of the generated cDNA, SYBR (Solarbio, Beijing, China) and specific primers (Genscript, Nanjing, China) on an Applied Biosystems 7500 system (Thermo Fisher). The primers were listed as: circ_0006916 (sense, 5ʹ-TCAACGGGACAGATGATGAA-3ʹ; antisense, 5ʹ-TGTGTTTGGGTCAATTTGGA-3ʹ), HOMER1 (sense, 5ʹ-GGCTGCACCTTTAAATTCCC-3ʹ; antisense, 5ʹ-TCCATGTCGCTTCAAGTCCA-3ʹ), SRSF2 (sense, 5ʹ-CGGTCTCCAGATCTCGTTCG-3ʹ; antisense, 5ʹ-GGTGAGTAACCTCCGAGCAG-3ʹ), and miR-599 (sense, 5ʹ-GTTGTGTCAGTTTATCAAAC-3ʹ; antisense, 5ʹ-CTCCATATCGCACTTTAATCTCTAACT-3ʹ). U6 (sense, 5ʹ-AATTGGAACGATACAGAGAAGATTAGC-3ʹ; antisense, 5ʹ-TATGGAACGCTTCACGAATTTG-3ʹ), or GAPDH (sense, 5ʹ-GAATGGGCAGCCGTTAGGAA-3ʹ; antisense, 5ʹ-AAAAGCATCACCCGGAGGAG-3ʹ) was used as a reference. Relative RNA abundance was calculated by 2−ΔΔCt method.23
Detection of circRNA Stability
RNase R (a 3ʹ to 5ʹ exonuclease) and Actinomycin D (a transcription inhibitor) were applied to analyze the stability of circ_0006916 as previous reports.24,25 For RNase R assay, the extracted RNA was treated with 3 U/μg of RNase R (GeneSeed, Guangzhou, China) for 30 min at 37°C, and then the abundances of circ_0006916 and the linear HOMER1 were detected via qRT-PCR. For Actinomycin D assay, cells were stimulated via 2 μg/mL of Actinomycin D (Amyjet, Wuhan, China) for 24 h, and then the RNA was isolated for the detection of circ_0006916 and the linear HOMER1 expression. The untreated group was regarded as the mock group.
Vector and Oligonucleotide Construct and Cell Transfection
SRSF2 overexpression vector (pc-SRSF2) was constructed via inserting the full-length sequence of SRSF2 into pcDNA3.1 vector (Thermo Fisher) by endonuclease sites Nhe I and Xho I, with the empty vector as negative control (pc-NC). The siRNA for circ_0006916 (si-circ_0006916, 5ʹ-UUCACAUAGGGAACAACCUAU-3ʹ), negative control of siRNA (si-NC) (5ʹ-AAGACAUUGUGUGUCCGCCTT-3ʹ), miR-599 mimic (5ʹ-GUUGUGUCAGUUUAUCAAAC-3ʹ), negative control of mimic (miRNA NC, 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ), miR-599 inhibitor (5ʹ-GUUUGAUAAACUGACACAAC-3ʹ), and negative control of inhibitor (inhibitor NC, 5ʹ-UGAGCUGCAUAGAGUAGUGAUUA-3ʹ) were formed via GenePharma (Shanghai, China). Huh7 and SNU-387 cells were transfected with the constructed vectors or oligonucleotides by Lipofectamine 3000 (Thermo Fisher) for 24 h.
Cell Counting Kit-8 (CCK8)
Cell viability was analyzed via CCK8. 1 × 104 Huh7 and SNU-387 cells were added into 96-well plates, and grown for 72 h. Next, 10 μL of CCK8 solution (Glpbio, Montclair, CA, USA) was added, and incubated with cells for 2 h. The absorbance was examined through a microplate reader (Bio-Rad, Hercules, CA, USA) with a wavelength of 450 nm. Relative cell viability was normalized to the control group.
Colony Formation Analysis
1 × 103 Huh7 and SNU-387 cells were seeded into the 6-well plates, and then cultured for 10 days. Subsequently, cells were fixed and dyed with 0.5% crystal violet (Beyotime). The colony formation was captured and the numbers were counted.
Wound Healing Analysis
The mobility of Huh7 and SNU-387 cells was assessed via wound healing analysis. 2 × 105 Huh7 and SNU-387 cells were added to 12-well plates, and incubated until reaching ~90% confluence. A straight wound was caused via a 10-μL sterile pipette tip. Next, cells were cultured for 24 h, and the width of wound was measured under a microscope (Nikon, Tokyo, Japan).
Transwell Analysis
The migrated and invasive abilities of cells were analyzed via transwell analysis using transwell chamber (BD, Franklin Lakes, NJ, USA). 5 × 105 Huh7 and SNU-387 cells in serum-free medium were added into the upper chamber precoated with 100 μL of Matrigel (Solarbio) for invasion assay, and 1 × 105 cells in non-serum medium were added to the upper chambers without Matrigel for migration assay. 600 μL of medium with 10% serum was added into the lower chambers. Following culture for 24 h, cells were dyed with 0.5% crystal violet, followed by observation under a microscope (magnification ×100) with 3 randomly selected fields.
Flow Cytometry
Cell cycle distribution and apoptosis were investigated via flow cytometry. For cell cycle detection, Huh7 and SNU-387 cells were incubated in 12-well plates for 72 h, and then cells were fixed with 70% ethanol, and interacted with 1 mg/mL of RNase and 50 μg/mL of propidium iodide (PI) for 30 min. 1 × 104 Huh7 and SNU-387 cells were examined via a flow cytometer (Agilent, Hangzhou, China), and the proportion of cells at G0/G1, S or G2/M phase was tested via ModFit software (Verity Software House, Topsham, ME, USA).
Cell apoptosis was tested with Annexin V-FITC apoptosis kit (Thermo Fisher). 2 × 105 Huh7 and SNU-387 cells were added in 6-well plates and cultured for 72 h. Next, cells were incubated with Annexin V-FITC binding buffer, and then dyed with Annexin V-FITC and PI, followed via the detection with a flow cytometer. The apoptotic rate was displayed as a percentage of cells in the upper and lower right quadrants.
Dual-Luciferase Reporter and RNA Immunoprecipitation (RIP) Analyses
The targets of circ_0006916 were predicted via CircInteractome (http://circinteractome.nia.nih.gov/),26 and the targets of miR-599 were predicted by starBase (http://starbase.sysu.edu.cn/).27 The dual-luciferase reporter and RIP analyses were carried out to explore the target correlation of miR-599 and circ_0006916 or SRSF2. In brief, the wild-type (circ_0006916 WT or SRSF2-3ʹUTR WT) or mutant luciferase reporter vectors (circ_0006916 MUT or SRSF2-3ʹUTR MUT) were generated via inserting the corresponding sequence containing wild-type or mutant miR-599 complementary sites in luciferase gene downstream in PGL3-Control vectors (Promega, Madison, WI, USA) via endonuclease site Xba I. These constructed vectors, control vectors and miR-599 mimic or miRNA NC were transfected into Huh7 and SNU-387 cells for 24 h. The luciferase activity was measured via a dual-luciferase analysis kit (Promega) and GloMax 20/20 Luminometer (Promega).
RIP analysis was conducted with a Magna RIP kit (Sigma, St. Louis, MO, USA). 1 × 107 Huh7 and SNU-387 cells were lysed and interacted with magnetic beads conjugated via anti-Ago2 for 4 h. Next, The RNA enriched on the beads was extracted and used for the detection of circ_0006916, miR-599 and SRSF2 levels. IgG was employed as a negative control, and input functioned as a positive control.
Western Blot
Cells or tissues were lysed in RIPA buffer (Solarbio) for the collection of total protein, and the protein concentration was examined via a BCA kit (Solarbio). The protein samples (30 μg) were loaded on SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). Five percent bovine serum albumin (Solarbio) was used to block the membranes. Next, the membranes were incubated with Abcam antibody (Cambridge, MA, USA) against SRSF2 (ab28428, 1:3000 dilution) overnight and the IgG conjugated via HRP (ab205718, 1:10,000 dilution) for 2 h. GAPDH (ab22555, 1:5000 dilution) functioned as a loading reference. Following exposing to ECL reagent (Abcam), the visualized blots were tested via Image Lab software (Bio-Rad).
Xenograft Experiment
The lentiviral vectors carried shRNA for circ_0006916 (sh-circ_0006916) or negative control (sh-NC) were generated via GenePharma, and transfected into SNU-387 cells. Male BALB/c nude mice (5-week-old) were provided via Charles River (Beijing, China), and arbitrarily divided into sh-circ_0006916 and sh-NC groups (n=6/group). In sh-circ_0006916 group, mice were subcutaneously injected with 5 × 106 SNU-387 cells with stable transfection of sh-circ_0006916. In sh-NC group, mice were subcutaneously injected with 5 × 106 SNU-387 cells with stable transfection of sh-NC. The tumor size was detected every 7 days, and volume was calculated via 0.5 × length × width2. After 28 days, mice were euthanized via 5% isoflurane. Tumor tissues were harvested and weighed. Next, the expression of circ_0006916, miR-599 and SRSF2 in tumor tissues was detected via qRT-PCR or Western blot. This experiment was approved via the Animal Ethical Committee of the First Affiliated Hospital of Henan University, and performed in line with the National Institutes of Health.
Statistical Analysis
Four replicates were conducted in vitro, and the experiment was repeated 3 times, unless otherwise indicated. The data were reported as mean ± SD, and compared via Student’s t-test or ANOVA with Tukey’s test via GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA). The linear correlation of gene expression in HCC tissues was assessed via Pearson test. The difference was considered as significant at P<0.05.
Results
circ_0006916 Expression is Enhanced in HCC
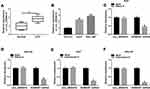
To detect the expression change of circ_0006916 in HCC, 30 paired tumor and normal tissues were collected. As exhibited in Figure 1A, circ_0006916 abundance was higher in HCC tissues than normal samples. Furthermore, circ_0006916 level was evidently elevated in HCC cell lines (Huh7 and SNU-387 cells) compared with normal liver cell line (THLE-2 cells) (Figure 1B). To test the stability of circ_0006916, circ_0006916 and corresponding linear HOMER1 levels were detected in cells after treatment of RNase R or Actinomycin D. As displayed in Figure 1C–F, circ_0006916 was more resistant to RNase R and Actinomycin D than the linear HOMER1. These results indicated that circ_0006916 was stably expressed in HCC, and the increased expression of circ_0006916 might be associated with HCC development.
circ_0006916 Knockdown Inhibits HCC Development in vitro
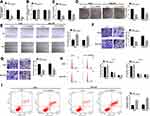
To analyze the function of circ_0006916 in HCC development, circ_0006916 expression was knocked down in Huh7 and SNU-387 cells using the siRNA targeting circ_0006916 (si-circ_0006916). The knockdown efficacy of si-circ_0006916 is confirmed in Figure 2A. Meanwhile, the introduction of si-circ_0006916 did not affect the abundance of the linear HOMER1 (Figure 2B). The results of CCK8 and colony formation analyses displayed that circ_0006916 silence markedly decreased cell viability and colony ability (Figure 2C and D). Furthermore, the data of wound healing and transwell analyses displayed that circ_0006916 silence obviously inhibited cell migration and invasion (Figure 2E–G). Additionally, circ_0006916 interference caused cell cycle arrest at G0/G1 phase, revealed by the increased proportion of cells at G0/G1 phase and the reduced proportion of cells at S phase (Figure 2H). Besides, circ_0006916 silence evidently promoted cell apoptosis in Huh7 and SNU-387 cells (Figure 2I). Taken together, circ_0006916 knockdown repressed HCC cell progression.
miR-599 is Targeted via circ_0006916
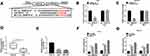
To explore the regulatory network mediated via circ_0006916, the targeted miRNA was searched. miR-599 was one predicted target of circ_0006916. To confirm this prediction, we constructed the luciferase reporter vectors circ_0006916 WT and circ_0006916 MUT, and the targeted sequence is shown in Figure 3A. The data of dual-luciferase reporter analysis showed that miR-599 addition evidently decreased the luciferase activity of circ_0006916 WT, while it did not alter the luciferase activity of circ_0006916 MUT (Figure 3B and C). Furthermore, miR-599 expression was lower in HCC tissues than normal tissues (n = 30) (Figure 3D). And miR-599 expression was negatively associated with circ_0006916 level in HCC tissues (r = −0.6014, P = 0.0004) (Supplementary Figure 1A). In addition, miR-599 level was significantly decreased in Huh7 and SNU-387 cells in comparison to THLE-2 cells (Figure 3E). Besides, there were amount of circ_0006916 and miR-599 enriched via Ago2 RIP (Figure 3F and G). These results suggested that miR-599 was targeted via circ_0006916.
miR-599 Knockdown Reverses the Influence of circ_0006916 Silence on HCC Development in vitro
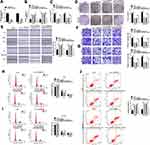
To explore whether miR-599 was required for circ_0006916-mediated HCC development, Huh7 and SNU-387 cells were transfected with si-NC, si-circ_0006916, si-circ_0006916 + inhibitor NC or miR-599 inhibitor. The efficacy of miR-599 inhibitor is identified in Figure 4A. Moreover, miR-599 expression was evidently up-regulated via circ_0006916 knockdown, which was weakened via transfection of miR-599 inhibitor (Figure 4B). Moreover, miR-599 knockdown attenuated silence of circ_0006916-mediated inhibition of cell viability and colony ability (Figure 4C and D). In addition, miR-599 knockdown reversed interference of circ_0006916-mediated suppression of migration and invasion (Figure 4E–G). Besides, miR-599 down-regulation mitigated knockdown of circ_0006916-induced cycle arrest and apoptosis (Figure 4H–J). These results indicated that circ_0006916 silence repressed HCC cell progression via regulating miR-599.
SRSF2 is Targeted via miR-599
To further analyze the regulatory network involved in this study, the target of miR-599 was searched. SRSF2 was a candidate target, and the luciferase reporter vectors SRSF2-3ʹUTR WT and SRSF2-3ʹUTR MUT were constructed (Figure 5A). The data of dual-luciferase reporter analysis displayed that miR-599 overexpression remarkably reduced the luciferase activity of SRSF2-3ʹUTR WT, while it showed little impact on the luciferase activity of SRSF2-3ʹUTR MUT (Figure 5B and C). Moreover, RIP assay showed that miR-599 and SRSF2 could be enriched in the same complex via Ago2 RIP (Figure 5D and E). Additionally, SRSF2 protein abundance was evidently elevated in HCC tissues and cells when compared to the corresponding normal tissues or cells (Figure 5F and G). And SRSF2 abundance was negatively correlated with miR-599 level in HCC tissues (r = −0.4771, P = 0.0077) (Supplementary Figure 1B). Besides, the influence of miR-599 on SRSF2 expression was investigated. The efficacy of SRSF2 overexpression vector is validated in Figure 5H. Furthermore, miR-599 overexpression evidently reduced SRSF2 protein expression, and this effect was attenuated via introduction of SRSF2 overexpression vector (Figure 5I). These results suggested that SRSF2 was targeted via miR-599.
miR-599 Inhibits HCC Development via Targeting SRSF2
To analyze the function of miR-599 and whether it required SRSF2, Huh7 and SNU-387 cells were transfected with miRNA NC, miR-599 mimic, miR-599 mimic + pc-NC or pc-SRSF2. Moreover, miR-599 overexpression suppressed cell viability and colony ability in Huh7 and SNU-387 cells, which was mitigated via SRSF2 up-regulation (Figure 6A and B). In addition, miR-599 up-regulation evidently reduced the abilities of migration and invasion, which was alleviated via SRSF2 overexpression (Figure 6C–E). Furthermore, miR-599 overexpression facilitated cell cycle arrest at G0/G1 phase and apoptosis, and this effect was weakened via SRSF2 overexpression (Figure 6F–H). These data suggested that miR-599 repressed HCC cell progression via regulating SRSF2.
circ_0006916 Knockdown Decreases SRSF2 Expression via miR-599
To explore how and whether circ_0006916 could regulate SRSF2, Huh7 and SNU-387 cells were transfected with si-NC, si-circ_0006916, si-circ_0006916 + pc-NC or pc-SRSF2. As displayed in Figure 7A, SRSF2 abundance was evidently decreased via circ_0006916 knockdown, which was restored via transfection of SRSF2 overexpression vector. Moreover, Huh7 and SNU-387 cells were transfected with si-NC, si-circ_0006916, si-circ_0006916 + inhibitor NC or miR-599 inhibitor. Results showed that miR-599 knockdown mitigated the suppressive effect of circ_0006916 silence on SRSF2 expression in Huh7 and SNU-387 cells (Figure 7B). These results showed that circ_0006916 could regulate SRSF2 expression via modulating miR-599.
circ_0006916 Silence Decreases Xenograft Tumor Growth
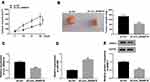
To explore the function of circ_0006916 in HCC development in vivo, SNU-387 cells with stable transfection of sh-NC or sh-circ_0006916 were inoculated into mice to establish the xenograft model. The mice were divided into sh-NC or sh-circ_0006916 group (n = 6). After 28 days, the tumor volume and weight were evidently reduced in sh-circ_0006916 group compared with sh-NC group (Figure 8A and B). Moreover, circ_0006916, miR-599 and SRSF2 abundances were detected in the tumor tissues. As displayed in Figure 8C–E, circ_0006916 and SRSF2 abundances were evidently reduced and miR-599 expression was enhanced in sh-circ_0006916 group in comparison to sh-NC group. These results indicated circ_0006916 down-regulation inhibited HCC development in vivo.
Discussion
HCC is the major liver cancer with high incidence and mortality.28 Moreover, the overall survival of HCC patients is lower than 18%.29 CircRNAs are implicated in the progression of liver diseases including HCC via regulating various cell events.30 A previous study suggested that circ_0006916 played an important role in HCC development.15 In this study, we found that circ_0006916 could repress HCC development. Moreover, we were the first time to validate it was associated with circ_0006916/miR-599/SRSF2 network in HCC.
In this research, we found circ_0006916 abundance was elevated in HCC, which was also consistent with former reports.14,15 Moreover, by treatment of RNase R and Actinomycin D, we found that circ_0006916 was stably expressed in HCC cells. To explore the potential role of circ_0006916, we performed the loss-of-function experiments, and found that circ_0006916 interference restrained HCC development via reducing cell viability, colony ability, migration and invasion and increasing cycle arrest and apoptosis, which was also like that in a previous report.15 Nevertheless, the role of circ_0006916 in HCC was opposite to that in lung cancer as a previous report.12 We hypothesized that the differences of circ_0006916 expression and function between the two cancers might be caused via the different tumor microenvironment. Furthermore, to explore the function of circ_0006916 in vivo, we established the xenograft model, and validated the anti-HCC role of circ_0006916 interference.
The regulatory network of circRNA/miRNA/mRNA is a major mechanism for circRNA in a competing endogenous RNA (ceRNA) manner.31 A previous study has confirmed the circ_0006916/miR-1322/CXCL6 regulatory network in HCC.15 Our study wanted to explore an additional network. Here we used dual-luciferase reporter and RIP analyses to identify miR-599 was targeted via circ_0006916. The decreased miR-599 was detected in HCC tissues and cells, indicating the decreased miR-599 might be associated with the malignancy of HCC. Moreover, we found that miR-599 overexpression using the miR-599 mimic repressed HCC development via decreasing cell viability, colony formation, migration and invasion. These results implied that miR-599 played a suppressive role in HCC, which was like that in a previous report.19 This is also consistent with the role of miR-599 in other cancers, including esophageal carcinoma, gastric cancer, breast cancer, glioma and anaplastic thyroid cancer.32–36 In addition, we found that miR-599 knockdown attenuated the effect of circ_0006916 silence on HCC development, indicating circ_0006916 could regulate HCC development via controlling miR-599.
Next, we identified miR-599 could target SRSF2. Here we found that SRSF2 expression was enhanced in HCC, which was also in agreement with a previous study.20 Luo et al reported that SRSF2 could contribute to HCC cell proliferation.21 Furthermore, Ma et al showed that SRSF2 down-regulation repressed cell proliferation and caused G1 phase arrest in HCC.37 These reports suggested SRSF2 played a carcinogenic role in HCC. Moreover, we found that SRSF2 overexpression attenuated the suppressive effect of miR-599 on HCC development. In addition, our results showed that circ_0006916 could regulate SRSF2 via competitively binding to miR-599. Hence, we concluded that circ_0006916 could regulate HCC development via modulating miR-599/SRSF2 axis.
In conclusion, circ_0006916 silence repressed HCC development, possibly via regulating miR-599 and SRSF2. This research proposed a novel insight into the pathogenesis of HCC, and indicated circ_0006916 could be used as a target for the treatment of HCC.
Highlights
- circ_0006916 abundance is elevated in hepatocellular carcinoma.
- circ_0006916 knockdown restrains cell viability, colony formation, migration and invasion.
- circ_0006916 knockdown facilitates cell apoptosis.
- circ_0006916 could modulate SRSF2 via sponging miR-599.
- circ_0006916 knockdown decreases xenograft tumor growth.
Ethics Approval
This work was approved by Henan Province Medical Science and Technology (LHGJ20190515)
Disclosure
The authors declare that they have no financial or non-financial conflicts of interest for this work.
References
1. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi:10.1016/S0140-6736(18)30010-2
2. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi:10.1038/s41575-019-0186-y
3. Tabrizian P, Jibara G, Shrager B, Schwartz M, Roayaie S. Recurrence of hepatocellular cancer after resection: patterns, treatments, and prognosis. Ann Surg. 2015;261(5):947–955. doi:10.1097/SLA.0000000000000710
4. Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67(3):603–618. doi:10.1016/j.jhep.2017.04.009
5. Qiu L, Xu H, Ji M, et al. Circular RNAs in hepatocellular carcinoma: biomarkers, functions and mechanisms. Life Sci. 2019;231:116660. doi:10.1016/j.lfs.2019.116660
6. Sun X, Ge X, Xu Z, Chen D. Identification of circular RNA-microRNA-messenger RNA regulatory network in hepatocellular carcinoma by integrated analysis. J Gastroenterol Hepatol. 2020;35(1):157–164. doi:10.1111/jgh.14762
7. Li Z, Liu Y, Yan J, et al. Circular RNA hsa_circ_0056836 functions an oncogenic gene in hepatocellular carcinoma through modulating miR-766-3p/FOSL2 axis. Aging (Albany NY). 2020;12(3):2485–2497. doi:10.18632/aging.102756
8. Pu J, Wang J, Li W, et al. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J Cell Mol Med. 2020. doi:10.1111/jcmm.15010
9. Su X, Su J, He H, Zhan Y, Liu H. Hsa_circ_0070269 inhibits hepatocellular carcinoma progression through modulating miR-182/NPTX1 axis. Biomed Pharmacother. 2019;120:109497. doi:10.1016/j.biopha.2019.109497
10. Liu Z, Yu Y, Huang Z, et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019;10(12):900. doi:10.1038/s41419-019-2089-9
11. Jiang L, Zhao L, Bi J, et al. Glycolysis gene expression profilings screen for prognostic risk signature of hepatocellular carcinoma. Aging (Albany NY). 2019;11(23):10861–10882.
12. Dai X, Zhang N, Cheng Y, et al. RNA-binding protein trinucleotide repeat-containing 6A regulates the formation of circular RNA circ0006916, with important functions in lung cancer cells. Carcinogenesis. 2018;39(8):981–992. doi:10.1093/carcin/bgy061
13. Li J, Sun Q, Zhu S, et al. Knockdown of circHomer1 ameliorates METH-induced neuronal injury through inhibiting Bbc3 expression. Neurosci Lett. 2020;732:135050. doi:10.1016/j.neulet.2020.135050
14. Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38(1):98. doi:10.1186/s13046-019-1041-2
15. Zhao M, Dong G, Meng Q, Lin S, Li X. Circ-HOMER1 enhances the inhibition of miR-1322 on CXCL6 to regulate the growth and aggressiveness of hepatocellular carcinoma cells. J Cell Biochem. 2020.
16. Xu X, Tao Y, Shan L, et al. The role of microRNAs in hepatocellular carcinoma. J Cancer. 2018;9(19):3557–3569.
17. Ba Y, Liu Y, Li C, Zhu Y, Xing W. HIPK3 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-599/c-MYC axis. Onco Targets Ther. 2020;13:1967–1978. doi:10.2147/OTT.S217087
18. Zhang XH, Li BF, Ding J, et al. LncRNA DANCR-miR-758-3p-PAX6 molecular network regulates apoptosis and autophagy of breast cancer cells. Cancer Manag Res. 2020;12:4073–4084. doi:10.2147/CMAR.S254069
19. Tian J, Hu X, Gao W, et al. Identification a novel tumor-suppressive hsa-miR-599 regulates cells proliferation, migration and invasion by targeting oncogenic MYC in hepatocellular carcinoma. Am J Transl Res. 2016;8(6):2575–2584.
20. Wang P, Guo L, Li K, et al. Serine/arginine rich splicing factor 2 expression and clinic pathological features indicating a prognostic factor in human hepatocellular carcinoma patients. Cancer Biomark. 2018;21(3):681–687. doi:10.3233/CBM-170770
21. Luo C, Cheng Y, Liu Y, et al. SRSF2 regulates alternative splicing to drive hepatocellular carcinoma development. Cancer Res. 2017;77(5):1168–1178.
22. Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc. 2006;1(2):581–585. doi:10.1038/nprot.2006.83
23. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
24. Peng QS, Cheng YN, Zhang WB, Fan H, Mao QH, Xu P. circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit hippo signaling pathway. Cell Death Dis. 2020;11(2):112. doi:10.1038/s41419-020-2273-y
25. Jia Q, Ye L, Xu S, et al. Circular RNA 0007255 regulates the progression of breast cancer through miR-335-5p/SIX2 axis. Thorac Cancer. 2020;11(3):619–630. doi:10.1111/1759-7714.13306
26. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13(1):34–42. doi:10.1080/15476286.2015.1128065
27. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–97. doi:10.1093/nar/gkt1248
28. Ryerson AB, Eheman CR, Altekruse SF, et al. Annual report to the nation on the status of cancer, 1975–2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122(9):1312–1337. doi:10.1002/cncr.29936
29. Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462. doi:10.1056/NEJMra1713263
30. Chien Y, Tsai PH, Lai YH, et al. CircularRNA as novel biomarkers in liver diseases. J Chin Med Assoc. 2020;83(1):15–17.
31. Wu J, Liu S, Xiang Y, Qu X, Xie Y, Zhang X. Bioinformatic analysis of circular RNA-associated ceRNA network associated with hepatocellular carcinoma. Biomed Res Int. 2019;2019:8308694. doi:10.1155/2019/8308694
32. Wang C, Tang D, Wang H, et al. Circular RNA hsa_circ_0030018 acts as a sponge of miR-599 to aggravate esophageal carcinoma progression by regulating ENAH expression. J Cell Biochem. 2019.
33. Cheng Y, Su Y, Wang S, et al. Identification of circRNA-lncRNA-miRNA-mRNA competitive endogenous RNA network as novel prognostic markers for acute myeloid leukemia. Genes (Basel). 2020;11(8). doi:10.3390/genes11080868.
34. Wang Y, Sui Y, Zhu Q, Sui X. Hsa-miR-599 suppresses the migration and invasion by targeting BRD4 in breast cancer. Oncol Lett. 2017;14(3):3455–3462. doi:10.3892/ol.2017.6651
35. Jiang Y, Wang X, Zhang J, Lai R. MicroRNA-599 suppresses glioma progression by targeting RAB27B. Oncol Lett. 2018;16(1):1243–1252.
36. Bi JW, Zou YL, Qian JT, Chen WB. MiR-599 serves a suppressive role in anaplastic thyroid cancer by activating the T-cell intracellular antigen. Exp Ther Med. 2019;18(4):2413–2420.
37. Ma K, He Y, Zhang H, et al. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J Biol Chem. 2012;287(8):5639–5649. doi:10.1074/jbc.M111.291229
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.