Back to Journals » OncoTargets and Therapy » Volume 13
HOPX Is an Epigenetically Inactivated Tumor Suppressor and Overexpression of HOPX Induce Apoptosis and Cell Cycle Arrest in Breast Cancer
Authors You Q, Geng Y, Ye H, Zhu G, Gao X, Zhu H
Received 18 February 2020
Accepted for publication 23 May 2020
Published 23 June 2020 Volume 2020:13 Pages 5955—5965
DOI https://doi.org/10.2147/OTT.S250404
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Qinghua You, Yuanyuan Geng, Huiying Ye, Guixiang Zhu, Xiaofang Gao, Hongbo Zhu
Department of Pathology, Shanghai Pudong Hospital, Shanghai 201399, People’s Republic of China
Correspondence: Xiaofang Gao; Hongbo Zhu
Department of Pathology, Shanghai Pudong Hospital, 2800 Gongwei Road, Huinan Yown, Pudong New District, Shanghai 201399, People’s Republic of China
Email [email protected]; [email protected]
Background: Evidence has been shown that abnormal DNA methylation plays a vital role in the progression of breast cancer via silencing of gene expression. The results of bisulfite sequencing showed that the methylation status of HOPX in breast cancer tissues was higher than that in normal breast cancer tissues, but little known about the biological functions of HOPX in breast cancer.
Methods: A total of 13 paired breast cancer and adjacent noncancerous tissues were subjected to bisulfite sequencing. Meanwhile, the methylation levels of cg218995965 and cg24862548 in breast cancer cells were detected by methylation-specific PCR (MSP). Flow cytometry, wound healing and transwell invasion assays were used to detect the apoptosis, migration and invasion in breast cancer cells. In addition, the expressions of HOPX, p21, cyclin D1 and CDK4 in cells were detected with Western blot assay.
Results: Bisulfite sequencing indicated that the CpG sites (cg218995965 and cg24862548) in the HOPX promoter region showed significantly higher methylation in breast cancer tissues. In addition, methylation-specific PCR revealed that HOPX was significantly hypermethylated in breast cancer cell lines MDA-MB-468 and MCF-7. Furthermore, overexpression of HOPX significantly inhibited the proliferation of MDA-MB-468 and MCF-7 cells via inducing the apoptosis. Moreover, upregulation of HOPX markedly inhibited the migration and invasion abilities of MDA-MB-468 cells. Meanwhile, overexpression of HOPX obviously induced cell cycle arrest in MDA-MB-468 cells via upregulation of p21, and downregulation of cyclin D1 and CDK4. Additionally, overexpression of HOPX suppressed tumor growth of breast cancer in vivo.
Conclusion: Our data showed that HOPX, a tumor suppressor, is epigenetically silenced in breast cancer. Overexpression of HOPX could suppress the progression of breast cancer, and thus indicating that it might serve as a potential target for the treatment of patients with breast cancer.
Keywords: breast cancer, DNA methylation, HOPX, apoptosis
Introduction
Breast cancer was the most common cancer in women all over the world.1 During 2012–2016, worldwide breast cancer incidence rate slightly rose (by 0.3% per year).1 Breast cancer is characterized by the continuous growth of malignant mammary gland cells, and generally classified into estrogen receptor-negative (MDA-MB-468) or estrogen receptor-positive (MCF-7) subtypes.2–4 Evidence has been shown that ductal carcinoma in situ, inflammatory breast cancer, invasive ductal carcinoma, and metastatic breast cancer are the main types of breast cancer.5 Recent years, surgical resection, radiotherapy and chemotherapy are the main treatment options for patients with breast cancer.6 However, the prognosis and survival rate in advanced-stage patients remain unsatisfactory.7 Therefore, it is urgently needed to explore novel treatment options for breast cancer.
Previous studies indicated that epigenetic modifications are participated in various biological processes.8,9 In addition, epigenetic reprogrammings have been shown to be involved in various human diseases, including breast cancer.10 DNA methylation, histone modification, and chromatin remodeling are the major types of epigenetic modifications.11 Among of these, DNA methylation at cytosine guanine (CpG) sites is a major form of epigenetic modification.12 DNA methylation process is the addition of the methyl group at the carbon 5-position of cytosine within a CpG dinucleotide.13 It has been shown that DNA methylation located in a gene promoter normally acts to inhibit gene transcription.14 Yari et al indicated that patients with breast cancer were reported to have higher levels of DNA methylation compared to normal individual.15 In addition, some researches indicated a marked relationship between the methylation level of gene and susceptibility to breast cancer.15,16 Moreover, the promoter methylation is closely associated with the gene expression, and some DNA methylation markers have been proven to have prognostic value.17
In this study, differentially methylated CpG sites were screened using Illumina Infinium Methylation EPIC BeadChip between breast cancer tissues and adjacent normal tissues. Our data found that the CpG sites (cg218995965 and cg24862548) in the HOPX promoter region showed significantly higher methylation in breast cancer tissues than that in the adjacent tissues. Therefore, the aim of this study was to investigate the role of HOPX in breast cancer, along with the potential mechanisms, thus offering therapeutic strategy for the treatment of breast cancer.
Materials and Methods
Patients’ Samples
A total of 13 paired breast cancer and adjacent noncancerous tissues were obtained from patients with breast cancer who underwent breast surgery in the Shanghai Pudong Hospital between from June 2017 to April 2019 with written informed consent in accordance with the Declaration of Helsinki were obtained from all the patients. None of these patients were subjected to chemotherapy or radiotherapy before surgery. This study was approved by the Institutional Ethical Committee of Shanghai Pudong Hospital.
Microarray Data and Discovery of Differential Methylation Position
DNA was extracted and purified using a Genomic DNA Extraction Kit (TaKaRa, Dalian, China). A bisulfite conversion reaction was employed through the EZ-96 DNA Methylation Kit (Zymo Research, Irvine, CA, USA). Samples were assayed on the Illumina Infinium Methylation EPIC BeadChip (Illumina Inc., San Diego, CA). EPIC experiments were performed following bisulphite conversion according to the manufacturer’s protocol.18 β-value is the estimate of methylation level (β = methylated intensity/(methylated intensity + unmethylated intensity + 100)). After that, the differential methylation position (DMP) was screened using champ package. An adj.p.value < 0.05 or |delta beta| ≥ 0.2 was set as criteria.
DNA Methylation Analysis by Quantitative Methylation Specific PCR (qMSP)
Genomic DNA was extracted and purified using a Genomic DNA Extraction Kit (TaKaRa, Dalian, China). A bisulfite conversion reaction was employed through the EZ-96 DNA Methylation Kit (Zymo Research, Irvine, CA, USA). The unmethylated primers of cg24852548 were 5ʹ-GAGGTTGTAGAGTGGTAGAGTGG-3ʹ (Forward) and 5ʹ- ACAAATCACAAAAATAAATTCACC-3ʹ (Reverse). The methylated primers of cg24852548 were 5ʹ-GAGGTTGTAGAGCGGTAGAGTGG-3ʹ (Forward) and 5ʹ- AAATCGCGAAAATAAATTCGC-3ʹ (Reverse). The unmethylated primers of cg21899596 were 5ʹ-GGTGAGGGTTTGTGGAATTATT-3ʹ (Forward) and 5ʹ- AACCTCCCTCCCTAAACTAAACA-3ʹ (Reverse). The methylated primers of cg21899596 were 5ʹ-GTGAGGGTTCGCGGAATTA-3ʹ (Forward) and 5ʹ- ACCTCCCTCCCTAAACTAAACG-3ʹ (Reverse). The PCR reaction conditions were as follows: 98°C for 4 min, 40 cycles of 98°C for 30 s, 56°C for 30 s, and 72°C for 30 s, and then 72°C for 10 min. After that, agarose gel electrophoresis was performed. Finally, the blots were visualized by ultraviolet (UV) illumination.
Cell Culture
Human normal breast cell line MCF-10A, human breast cancer cell lines MDA-MB-468, MCF-7, SKBR3, MDA-MB-231 and BT474 and 293T cell line were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA). All these cell lines were maintained in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS) and 100 U/mL antibiotics (penicillin–streptomycin) at 37°C with 5% CO2.
Lentivirus Production and Exogenous HOPX Overexpression
Lentiviral constructs of HOPX overexpression (lenti-HOPX) was obtained from GenePharma (Shanghai, China). After that, lenti-HOPX plasmids were transfected into 293T cells. After 72 h of incubation, cell supernatants were collected, and used for transfection (24 h) of MCF-7 and MDA-MB-468 cells with NC and lenti-HOPX supernatants respectively. Subsequently, cells were incubated with puromycin (2.5 μg/mL) to select stable HOPX-overexpression cells for another 48 h. RT-qPCR assay was used to verify the expression of HOPX in cells.
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA samples were extracted from cultured cells using TRIzol reagent according to the manufacturer’s instructions. For reverse transcription of mRNA, a PrimeScript RT reagent Kit (Takara Bio Inc. Shiga, Japan) was used to synthesize cDNA. After that, qPCR amplification was performed using the SYBR Premix Ex Taq II kit (Takara) on a Light Cycler 480 II Real-Time PCR system (Roche Diagnostics, Basel, Switzerland). The PCR primers were as follows: HOPX, forward: 5ʹ-CCTGGAGTACAACTTCAACAAGG-3ʹ; reverse: 5ʹ-CTGCTTAAACCATTTCTGGGTC-3ʹ. β-actin, forward: 5ʹ-GTCCACCGCAAATGCTTCTA-3ʹ; reverse: 5ʹ-TGCTGTCACCTTCACCGTTC-3ʹ. β-actin was used as the internal control for normalizing HOPX expression. Data were analyzed using the 2–ΔΔCT method.
Cell Proliferation Assay
Cell Counting Kit-8 (CCK-8, Dojindo) was used to determine the cell viability. MCF-7 and MDA-MB-468 cells (10,000 cells/well) were seed onto 96-well plates overnight at 37°C. After that, cells were infected with lenti-HOPX for 24, 48 and 72 h. Then, 10 μL CCK-8 reagent was added into each well followed by incubation at 37°C for 2 h. Subsequently, the absorbance was evaluated at 450 nm using a microplate Reader.
Flow Cytometric Analysis
Apoptosis in MCF-7 and MDA-MB-468 cells were carried out by an Annexin V-FITC/PI Apoptosis Detection Kit (Sigma-Aldrich, St. Louis, USA). Cells were washed three times with pre-cold PBS, and then resuspended in binding buffer. After that, cells were stained with 5 μL of propidium iodide (PI) and Annexin V-FITC at room temperature for 15 min. Subsequently, a FACS Calibur flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) was used to measure the percentage of apoptotic cells.
For analysis of cell cycle, cells were washed twice with pre-cold PBS, and then resuspended with 70% ethanol overnight at 4°C. After that, cells were stained with 1 mg/mL of PI/RNase Staining Buffer (BD Biosciences) in the dark for 30 min. Subsequently, a FACS Calibur flow cytometer was used to analyze the populations of cells in G0-G1, S and G2-M phases.
Transwell Invasion Assay
Transwell invasion assay was performed using 24-well transwell matrigel-coated chambers (0.8 μm; Corning New York, NY, USA). 4 x 104 MDA-MB-468 cells were suspended in 200 μL serum-free medium in the upper chamber. After that, 600 μL of DMEM medium containing 10% FBS was added into the lower chamber as the chemoattractant. After 24 h of incubation, cells adhering to the lower surface were fixed with 4% paraformaldehyde, and stained with 0.2% crystal violet at 24 h. The invaded cells were photographed under a laser confocal microscope (Olympus CX23 Tokyo, Japan).
Wound Healing Assay
MDA-MB-468 cells were seed at 5 × 105 cells per well in a 12-well culture plate and incubated overnight at 37°C. After that, a wound area was made using a 200 μL pipette tip by scratching cell monolayer. Later on, cells were infected with lenti-HOPX for 48 h. The width of the wound image was monitored by photographing at 0 h and 48 h using the fluorescence microscope (Olympus). Image J 1.47v software was used to determine the area of cell migration.
Western Blot Assay
Total proteins were quantified using BCA method (Beyotime Institute of Biotechnology), and proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Later on, the gels were transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After blocking in 5% skimmed milk in TBST for 1 h at room temperature, the membrane was incubated in primary antibodies against HOPX (1:1000), p21 (1:1000), cyclin D1 (1:1000), CDK4 (1:1000) and β-actin (1:1000) at 4°C overnight. Subsequently, the membranes were incubated with the secondary antibody (Abcam, 1: 5000) at room temperature for 1 h, and visualized using an electrochemiluminescence (Thermo Fisher Scientific). β-actin acted as the internal control. All antibodies were obtained from Abcam (Cambridge, MA, USA).
Animal Study
4–6-weeks old BALB/c nude mice were purchased from the Hubei Provincial center for Laboratory Animal. Animals were randomized into two groups: control, HOPX-OE group. MDA-MB-468 cells or MDA-MB-468 cells stably expressing lenti-HOPX cells (1 × 107 per mouse, in 100 μL of PBS) were injected subcutaneously into the left flank of nude mice respectively. Tumor volume was monitored every week with a digital caliper, and tumor volume were calculated using the formula V = (length x width2)/2 (Width < Length). Animals were sacrificed in 4 weeks, and the entire tumors were isolated and weighed. All animal experiments were approved by the Institutional Ethical Committee of Shanghai Pudong Hospital, and animals were housed with a 12-hr light/dark cycle following the guidelines of the Institutional Animal Care and Use Committee.
Immunohistochemistry (IHC) Assay
HOPX expression in tumor tissue was determined by IHC staining according to methods reported before.19 The slices were incubated with the primary antibodies overnight at 4°C, and then incubated with biotinylated goat anti-rabbit IgG for 30 min at room temperature. The IHC detection system (EnVision kit; Dako Japan) was used to visualize IHC reactions.
Statistical Analysis
All data were repeated in triplicate. Data are presented as the mean ± standard deviation (S.D.). All statistical analyses were performed using GraphPad Prism software (version 7.0, La Jolla, CA, USA). One-way analysis of variance (ANOVA) and Tukey’s tests were carried out for multiple group comparisons. For the comparison of two groups, Student’s t-test was applied. *P < 0.05 was considered to be statistically significant.
Results
Differentially Methylated CpG Sites Between Breast Cancer Tissues and Matched Adjacent Tissues
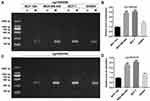
To investigate the DNA methylome in breast cancer, 13 breast cancer tissues and 13 matched adjacent tissues were profiled through Illumina Infinium Methylation EPIC BeadChip. The Infinium Methylation EPIC BeadChip included 840776 CpG sites. Champ package was used to screen differential methylated CpG sites. Finally, a total of 13 differentially methylated CpGs in the HOPX promoter region were found between breast cancer tissues and matched adjacent tissues, including 4 hypermethylated and 9 hypomethylated CpGs (Figure 1A and B). In addition, the CpG sites (cg218995965 and cg24862548) in the HOPX promoter region showed significantly higher methylation in breast cancer tissues than that in the adjacent tissues.
Detection of Methylation Sites Through Pyrosequencing
Next, pyrosequencing was used to verify the DNA methylation status of HOPX promoter region in human normal breast cell line MCF-10A, and human breast cancer cell lines (MDA-MB-468, MCF-7, SKBR3, MDA-MB-231 and BT474). As shown in Figure 2A–D, and Supplementary Figure 1A and 1B, the methylation levels of cg218995965 and cg24862548 were higher in MDA-MB-468, MCF-7, MDA-MB-231 and BT474 cells than that in the normal breast cell line MCF-10A, indicating that promoter region of HOPX exhibited methylation in 4 breast cancer cell lines except SKBR3. In addition, previous report indicated that HOPX played an important role in the development of human cancers.20 Thus, HOPX gene was selected for the subsequently experiments. These data suggested that the methylation level in the HOPX promoter region was much higher in breast cancer cells than human normal breast cells.
Overexpression of HOPX Induced Apoptosis in Breast Cancer Cells
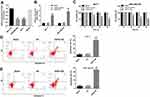
To determine the role of HOPX in breast cancer cells, we examined HOPX expression in human breast cancer cell lines (MCF-7, MDA-MB-468 and SKBR3 cells). Among these three breast cancer cell lines, HOPX expression was downregulated in MCF-7 and MDA-MB-468 cells compared with MCF-10A cells; in fact, there was no difference in HOPX level between SKBR3 cells and MCF-10A cells (Figure 3A).
Having known HOPX is downregulated in MCF-7 and MDA-MB-468 cells, we then investigate the potential anti-cancer role of HOPX in breast cancer. We established breast cancer cell lines (MCF-7 and MDA-MB-468) with HOPX stable overexpression. As shown in Figure 3B, the level of HOPX was markedly upregulated in MCF-7 and MDA-MB-468 cells following infection with lenti-HOPX. In addition, upregulation of HOPX notably inhibited proliferation of MCF-7 and MDA-MB-468 cells (Figure 3C). Moreover, the apoptosis rate was significantly increased in MCF-7 and MDA-MB-468 cells following infection with lenti-HOPX (Figure 3D and E). Meanwhile, upregulation of HOPX exhibited about 50.8 and 55.4% growth inhibition at 72 h in MCF-7 and MDA-MB-468 cells respectively (Figure 3C). MDA-MB-468 cells infected with lenti-HOPX exhibited lower cell viability at 72 h, compared with that of in MCF-7 cells. Therefore, MDA-MB-468 cells were utilized in the following experiments. These data indicated that upregulation of HOPX could inhibit proliferation and induce apoptosis of breast cancer cells.
Overexpression of HOPX Inhibited Migration and Invasion Abilities of Breast Cancer Cells
To investigate the effect of HOPX on migration and invasion of MDA-MB-468 cells, wound healing assay and transwell invasion assay were performed. As shown in Figure 4A, the migration ability of MDA-MB-468 cells was significantly reduced following infection with lenti-HOPX. In addition, overexpression of HOPX obviously suppressed invasion ability of MDA-MB-468 cells, compared with NC group (Figure 4B). These results suggested that upregulation of HOPX could inhibit migration and invasion of MDA-MB-468 cells.
Overexpression of HOPX Induced Cell Cycle Arrest in Breast Cancer Cells
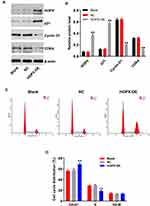
Next, the protein expression and cell cycle distribution in HOPX-overexpression MDA-MB-468 cells were investigated by Western blot and flow cytometry assays. As indicated in Figure 5A and B, the expressions of HOPX and p21 were upregulated, while the levels of cyclin D1 and CDK4 were downregulated in MDA-MB-468 cells following infection with lenti-HOPX. In addition, flow cytometry assay indicated that MDA-MB-468 cells infected with lenti-HOPX displayed a significant increase in the cell percentages of G0/G1 phase but decreased proportions in S phase compared with MDA-MB-468 cells (Figure 5C and D). These data indicated that overexpression of HOPX could induce G0/G1 cell cycle arrest in breast cancer cells.
Upregulation of HOPX Inhibited Tumor Growth in MDA-MB-468 Xenografts in vivo
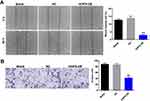
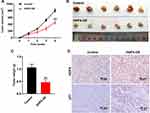
Next, to investigate the role of HOPX in the regulation of breast cancer tumor growth in vivo, a mouse MDA-MB-468 subcutaneous xenograft model was established. As indicated in Figure 6A–C, overexpression of HOPX markedly suppressed the tumor volume and tumor weight, compared with control group. Moreover, IHC assay indicated that lenti-HOPX obviously increased the expressions of HOPX and p21 in tumor tissues (Figure 6D). These results suggested that upregulation of HOPX could inhibit the tumor growth of MDA-MB-468 xenografts in vivo.
Discussion
Evidences have been shown that the molecular and cellular pathogenesis of tumor is commonly associated with tumor suppressor gene inactivation and oncogene activation.21,22 In addition, inactivation of tumor suppressor gene is associated with epigenetic modification, including DNA methylation.23 DNA methylation biomarkers can be used as prognostic markers for the treatment of human cancers.24 Therefore, exploring potential DNA methylation biomarkers is very important and urgently needed.
In this study, differentially methylated CpG sites between breast cancer tissues and adjacent tissues were selected. The data indicated that a total of 13 differentially methylated CpGs in the HOPX promoter region were found between breast cancer tissues and adjacent tissues, including 4 hypermethylated and 9 hypomethylated CpGs. In addition, the CpG sites (cg218995965 and cg24862548) in the HOPX promoter region showed significantly higher methylation in breast cancer tissues. Kikuchi et al indicated that HOPX was downregulated in human breast cancer cell lines, and promoter methylation of HOPX is frequent in breast cancer cells.25 In addition, Ooizumi et al indicated that HOPX is silenced by promoter DNA hypermethylation in papillary thyroid cancer.26 Our data verified that promoter region of HOPX in CpG islands exhibited higher methylation in four breast cancer cell lines except SKBR3, which was consistent with previous study.25 In addition, the expression of HOPX was downregulated in MCF-7 and MDA-MB-468 cells, indicating that the epigenetic silencing is involved in suppression of HOPX expression. These data indicated that the methylation state of the HOPX promoter was much higher in breast cancer cells than that in human normal breast cells, and breast cancer cells with low expression of HOPX was due to the hypermethylation of the HOPX promoter.
However, the mechanism by which HOPX regulates the proliferation, apoptosis and invasion of breast cancer remains unknown. Our data found that overexpression of HOPX significantly inhibited proliferation, and induced apoptosis of estrogen receptor (ER) positive MCF-7 and ER-negative MDA-MB-468 cells. These data indicated that anti-tumor effect of HOPX on breast cancer is associated with the methylation status of HOPX promoter, but is not related to intracellular ER expression. Moreover, overexpression of HOPX markedly suppressed migration and invasion abilities of breast cancer cells. Yap et al reported that overexpression of HOPX could inhibit the proliferation and migration in head and neck cancer cells.27 Ooki et al studied that HOPX promoter hypermethylation was found in gastric cancer as well, and upregulation of HOPX obviously inhibited proliferation and induced apoptosis of gastric cancer cells.28 These data indicated that HOPX functioned as a tumor suppressor in breast cancer.
It is well documented that cell cycle dysregulation is one of the most important characteristics of tumor cells.29 Waraya et al found that overexpression of HOPX not only induce cell cycle arrest in G1 stage in pancreatic cancer cells but also inhibit invasive ability of cells.30 The arrest of cancer cells at G0/G1 phase could prevent cell proliferation.31 Our data found that overexpression of HOPX markedly induced G0/G1 phase arrest in MDA-MB-468 cells via upregulation of p21 and downregulation of cyclin D1 and CDK4, which was consistent with previous study. These data indicated that HOPX functioned as a tumor suppressor in breast cancer via inducing cell cycle arrest.
Conclusion
Collectively, our study indicated that HOPX, a tumor suppressor that is epigenetically silenced in breast cancer, can suppress cell proliferation, and induce apoptosis and cell cycle arrest in breast cancer cells. Therefore, HOPX might be a potential biomarker for the treatment of breast cancer.
Disclosure
The authors declare no competing financial interests.
References
1. DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69(6):438–451. doi:10.3322/caac.21583
2. Thordarson G, Lee AV, McCarty M, et al. Growth and characterization of N-methyl-N-nitrosourea-induced mammary tumors in intact and ovariectomized rats. Carcinogenesis. 2001;22(12):2039–2047. doi:10.1093/carcin/22.12.2039
3. Liu CY, Lau KY, Hsu CC, et al. Combination of palbociclib with enzalutamide shows in vitro activity in RB proficient and androgen receptor positive triple negative breast cancer cells. PLoS One. 2017;12(12):e0189007. doi:10.1371/journal.pone.0189007
4. Tian J, Wang Y, Zhang X, et al. Calycosin inhibits the in vitro and in vivo growth of breast cancer cells through WDR7-7-GPR30 Signaling. J Exp Clin Cancer Res. 2017;36(1):153. doi:10.1186/s13046-017-0625-y
5. Sharma GN, Dave R, Sanadya J, Sharma P, Sharma KK. Various types and management of breast cancer: an overview. J Adv Pharm Technol Res. 2010;1(2):109–126.
6. Buranrat B, Bootha S. Antiproliferative and antimigratory activities of bisphosphonates in human breast cancer cell line MCF-7. Oncol Lett. 2019;18(2):1246–1258. doi:10.3892/ol.2019.10438
7. Wang Y, Zhang Y, Pan C, Ma F, Zhang S. Prediction of poor prognosis in breast cancer patients based on microRNA-21 expression: a meta-analysis. PLoS One. 2015;10(2):e0118647. doi:10.1371/journal.pone.0118647
8. Mazzone R, Zwergel C, Artico M, et al. The emerging role of epigenetics in human autoimmune disorders. Clin Epigenetics. 2019;11(1):34. doi:10.1186/s13148-019-0632-2
9. Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. doi:10.1016/j.cell.2007.02.006
10. de Ruijter TC, van der Heide F, Smits KM, et al. Prognostic DNA methylation markers for hormone receptor breast cancer: a systematic review. Breast Cancer Res. 2020;22(1):13. doi:10.1186/s13058-020-1250-9
11. Liu Z, Han S, Shen X, et al. The landscape of DNA methylation associated with the transcriptomic network in layers and broilers generates insight into embryonic muscle development in chicken. Int J Biol Sci. 2019;15(7):1404–1418. doi:10.7150/ijbs.35073
12. Papathanasiou I, Trachana V, Mourmoura E, Tsezou A. DNA methylation regulates miR-140-5p and miR-146a expression in osteoarthritis. Life Sci. 2019;228:274–284. doi:10.1016/j.lfs.2019.05.018
13. Jeschke J, Collignon E, Fuks F. DNA methylome profiling beyond promoters - taking an epigenetic snapshot of the breast tumor microenvironment. FEBS J. 2015;282(9):1801–1814. doi:10.1111/febs.13125
14. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi:10.1038/npp.2012.112
15. Yari K, Rahimi Z. Promoter methylation status of the retinoic acid receptor-beta 2 gene in breast cancer patients: a case control study and systematic review. Breast Care. 2019;14(2):117–123. doi:10.1159/000489874
16. Boyne DJ, O’Sullivan DE, Olij BF, et al. Physical activity, global DNA methylation, and breast cancer risk: a systematic literature review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2018;27(11):1320–1331. doi:10.1158/1055-9965.EPI-18-0175
17. Michels KB, Binder AM, Dedeurwaerder S, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods. 2013;10(10):949–955. doi:10.1038/nmeth.2632
18. Heiss JA, Brennan KJ, Baccarelli AA, et al. Battle of epigenetic proportions: comparing Illumina’s EPIC methylation microarrays and TruSeq targeted bisulfite sequencing. Epigenetics. 2020;15:174–182. doi:10.1080/15592294.2019.1656159
19. Henri O, Pouehe C, Houssari M, et al. Selective stimulation of cardiac lymphangiogenesis reduces myocardial edema and fibrosis leading to improved cardiac function following myocardial infarction. Circulation. 2016;133(15):
20. Liu Y, Zhang W. The role of HOPX in normal tissues and tumor progression. Biosci Rep. 2020;40(1). doi:10.1042/BSR20191953
21. Chen P, Duan X, Li X, et al. HIPK2 suppresses tumor growth and progression of hepatocellular carcinoma through promoting the degradation of HIF-1α. Oncogene. 2020;39(14):2863–2876. doi:10.1038/s41388-020-1190-y
22. Wang Z, Zhang Q, Sun Y, Shao F. Long non-coding RNA PVT1 regulates BAMBI to promote tumor progression in non-small cell lung cancer by sponging miR-17-5p. Onco Targets Ther. 2020;13:131–142. doi:10.2147/OTT.S217335
23. Li L, Yuan S, Zhao X, Luo T. ADAMTS8 is frequently down-regulated in colorectal cancer and functions as a tumor suppressor. Biochem Biophys Res Commun. 2020;524:663–671.
24. Yi L, Luo P, Zhang J. Identification of aberrantly methylated differentially expressed genes in breast cancer by integrated bioinformatics analysis. J Cell Biochem. 2019;120(9):16229–16243. doi:10.1002/jcb.28904
25. Kikuchi M, Katoh H, Waraya M, et al. Epigenetic silencing of HOPX contributes to cancer aggressiveness in breast cancer. Cancer Lett. 2017;384:70–78. doi:10.1016/j.canlet.2016.10.017
26. Ooizumi Y, Katoh H, Yokota M, Watanabe M, Yamashita K. Epigenetic silencing of HOPX is critically involved in aggressive phenotypes and patient prognosis in papillary thyroid cancer. Oncotarget. 2019;10(57):5906–5918. doi:10.18632/oncotarget.27187
27. Yap LF, Lai SL, Patmanathan SN, et al. HOPX functions as a tumour suppressor in head and neck cancer. Sci Rep. 2016;6:38758. doi:10.1038/srep38758
28. Ooki A, Yamashita K, Kikuchi S, et al. Potential utility of HOP homeobox gene promoter methylation as a marker of tumor aggressiveness in gastric cancer. Oncogene. 2010;29(22):3263–3275. doi:10.1038/onc.2010.76
29. Li T, Li L, Wu X, Tian K, Wang Y. The oncogenic role of GNL3 in the progression and metastasis of osteosarcoma. Cancer Manag Res. 2019;11:2179–2188. doi:10.2147/CMAR.S195360
30. Waraya M, Yamashita K, Katoh H, et al. Cancer specific promoter CpG Islands hypermethylation of HOP homeobox (HOPX) gene and its potential tumor suppressive role in pancreatic carcinogenesis. BMC Cancer. 2012;12:397. doi:10.1186/1471-2407-12-397
31. Zhang C, Zhou SS, Feng LY, et al. In vitro anti-cancer activity of chamaejasmenin B and neochamaejasmin C isolated from the root of Stellera chamaejasme L. Acta Pharmacol Sin. 2013;34(2):262–270. doi:10.1038/aps.2012.158
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.