Back to Journals » Neuropsychiatric Disease and Treatment » Volume 17
High Mobility Group Box 1/Toll-like Receptor 4 Signaling Increases GABRB3 Expression in Alcohol Exposure
Authors Sun G, Qi X, Wang W, Li X, Luo C, Bai S , Xu S, Zhong X, Huang C, Zhu X, Huang Z
Received 13 February 2021
Accepted for publication 14 May 2021
Published 1 June 2021 Volume 2021:17 Pages 1725—1732
DOI https://doi.org/10.2147/NDT.S306242
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yuping Ning
Guangtao Sun,1,* Xunzhong Qi,1,* Wei Wang,1,* Xintong Li,1,* Chunhua Luo,2,* Sunjie Bai,3 Shaohua Xu,2 Xiaogang Zhong,4 Chenglong Huang,5 Xiaofeng Zhu,6 Zuoyi Huang1
1Department of Neurology, The First Affiliated Hospital of Jiamusi University, Jiamusi, People’s Republic of China; 2Department of Laboratory Medicine, Yichang Central People’s Hospital, The First College of Clinical Medical Science, Three Gorges University, Hubei, People’s Republic of China; 3Department of Laboratory Medicine, The First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 4Key Laboratory of Psychoseomadsy, Stomatological Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 5Department of Laboratory Medicine, University-Town Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 6Mudanjiang Medical College, Mudanjiang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zuoyi Huang Email [email protected]
Introduction: Prefrontal cortex (PFC) and striatal neurotransmitter homeostasis is affected by alcohol dependence. In this study, the microarray dataset from the Gene Expression Omnibus (GEO) database were downloaded. The prefrontal and striatum data were cross-analyzed to reveal the co-effects of alcohol dependence on the two brain regions of mice.
Methods: The GSE123114 microarray profile was downloaded from the GEO database, and differentially expressed genes (DEGs) between the two groups were acquired by GEO2R. KEGG analyses were performed to identify the pivotal pathways of these DEGs. Key differential gene expressions and their mechanism associated with alcohol exposure were investigated by an intraperitoneal alcohol model.
Results: A total of 13 overlapping DEGs from the PFC and striatal datasets of the GSE123114 microarray profile were identified, and they were significantly enriched in the morphine addiction pathway. The transcript levels and protein expression of Gabrb3 were consistent with the microarray data both in the PFC and striatum. The transcript levels of HMGB1, TLR4, TNFα and IL-1β were upregulated in the PFC and striatum of mice in the alcohol group. The HMGB1 inhibitor decreased Gabrb3 transcript and protein levels as well as TNFα and IL-1β transcript levels both in the PFC and striatum in the intraperitoneal alcohol model mice.
Discussion: Through the reanalysis of GSE123114 microarray profile, we found that Gabrb3 is a key gene associated with alcohol exposure. In further experiments, our findings suggest that alcohol exposure modulates Gabrb3 expression through the HMGB1/TLR4 pathway. Moreover, inflammation-associated factors, such as IL-1β and TNFα, may be related to the HMGB1/TLR4-mediated regulation of GABRB3 expression in alcohol exposure.
Keywords: alcohol exposure, prefrontal cortex, striatum, Gabrb3, HMGB1/TLR4 pathway
Introduction
Alcohol dependence is a chronic recurrent brain disease, with negative health, economic, and social consequences for millions of adults worldwide.1–3 Emerging evidence suggests that alcohol dependence is related to dysregulation of limbic circuits, including pathways that connect with the prefrontal cortex (PFC) and striatum. It has been reported that repeated alcohol intake increases PFC dopamine release in mice, which plays a key role in facilitating learning associated with drug dependence.4 In particular, upregulation of GLT-1 and xCT expression in the PFC is related to attenuation of chronic and relapse-like alcohol drinking.1 In mammal, alcohol exposure is associated with reduced excitability in the prefrontal cortex.5
Several studies suggest that dysregulation of the striatal neurotransmitter system is closely associated with alcohol dependence. For example, extracellular glutamate levels are increased6 and GLT-1 expression is decreased in the striatum after alcohol withdrawal.7 Ceftriaxone-induced upregulation of GLT-1 in the striatum also leads to reduced alcohol consumption in mice. Furthermore, upregulation of GLT-1 in the striatum induced by ceftriaxone also results in reduced alcohol drinking in mice. Together, these studies suggest that PFC and striatal neurotransmitter homeostasis are closely associated with alcohol dependence.
Although several researchers have investigated the impact of alcohol dependence on PFC or striatal gene expression individually, the co-effects of alcohol dependence on these two brain regions have not been reported. In this study, the GSE123114 microarray profile was downloaded from the GEO (Gene Expression Omnibus) database. The PFC and striatal data were cross-analyzed to identify the co-effects of alcohol dependence on the two brain regions of mice to provide insight into the molecular pathogenesis of the disorder.
Materials and Methods
Microarray Data
The GSE123114 microarray profile datasets were obtained from GEO database which is based on the GPL1261 (Mouse430_2) platform of the Affymetrix Mouse Genome 430 2.0 Array that includes 12 mouse samples, including three inflexible drinkers PFC, three light drinkers PFC, three inflexible drinkers striatum and three light drinkers striatum. In this part of the experiment, the three-bottle free-choice method (5 and 10% of ethanol (v/v) and water for 16 weeks) was carried out on all experimental male mice. On the last two weeks of the experiment, the bitter and aversive substance, quinine was adulterated in both ethanol solutions. According to their individual ethanol intake and preference mice were classified in three groups: “light drinkers” (preference for water at all experimental stages); “heavy drinkers” (ethanol preference decreased after adulteration of the ethanol solution with quinine), and “inflexible drinkers” (preference for ethanol at all experimental stages).8,9
DEG Identification and Pathway Enrichment Analysis
Differentially expressed genes (DEGs) between the two groups were acquired by GEO2R. A P-value <0.05 and FC=1.5 were set as the cutoff criteria. KEGG pathway enrichment analyses were performed to identify the potential pathways of the DEGs, with P<0.01 as the threshold.
Animals
All experiments were approved by the Animal Ethics Committee of Jiamusi University and Chongqing Medical University and were performed in accordance with the National Institute of Health’s Guide for the Care and Use of Laboratory Animals (NIH Publication No. 80–23, revised in 1996). Six to eight week old C57BL/6J mice (males) were obtained from the Department of Laboratory Animal Science of Chongqing Medical University. All mice, with free access to food and water, were maintained in a temperature (21–22°C) and humidity-controlled room (55±5%) under a 12/12 h light/dark cycle.
Drug Treatment
All mice were randomly divided into control, alcohol and glycyrrhizin groups. Glycyrrhizic is a triterpenoid saponin which is present in large quantities in licorice root (Glycyrrhiza radix), and has been suggested to act as a direct HMGB1 antagonist. We used 0.9% saline to dissolve all drugs. Mice were treated with saline (control group), alcohol (20% v/v, 2 g/kg) or alcohol+glycyrrhizin (50 mg/kg)10,11 by intraperitoneal (ip) injection once per day for seven days.12–14 Glycyrrhizin was administered on days six and seven after alcohol injection. Glycyrrhizin and alcohol were administered as separate injections. On day eight, all mice were sacrificed and PFC and striatum were quickly isolated from mouse brain on ice and stored at −80°C.
Quantitative Real-time PCR (qRT-PCR) and Western Blot Analysis
TRIzol was used to isolate RNA from the PFC and striatal samples (Thermo Fisher Scientific, Rockford, IL, USA) and the PrimeScript RT master mix kit was used to reverse-transcribe the total RNA from the PFC and striatal samples (Takara, Otsu, Japan). The qRT-PCR primer sequences are listed in Supplementary materials Table S1. qRT-PCR which using SYBR Premix Ex Taq II (Takara) was performed on a LightCycler 96 System (Roche, Basel, Switzerland). Fold change was calculated using the ΔΔCt method. Transcript levels were calculated relative to β-actin.
As previously described,15 the PFC and striatum stored at −80°C were homogenized in ice-cold RIPA lysis buffer with the presence of phosphatase and protease inhibitors, then centrifuged at 4°C to extract total protein. Primary antibodies, GABRB3 (gamma-aminobutyric acid type A receptor subunit beta3; 1:1000) and GAPDH (1:10,000), were used for Western blotting analysis (Abcam, Cambridge, MA, USA). GAPDH was used as a loading control and enhanced chemiluminescence was used to detect signals.
Statistical Analysis
For qRT-PCR and Western blot analysis, one-way ANOVA with the LSD post hoc test was used to assess the differences among more than two groups; Student’s t-test was used to assess the differences between two groups. P<0.05 was considered statistically significant.
Results
Alcohol Exposure Increases Expression of Morphine Addiction-related Genes in the PFC and Striatum
Using the cutoff criteria described above, we extracted 229 DEGs between inflexible drinker mice and light drinker mice from the PFC datasets of the GSE123114 microarray profile (Figure 1C). The most enriched KEGG pathways related to these 229 DEGs were “morphine addiction” (mmu05032, P=0.0000), “amphetamine addiction” (mmu05031, P=0.0000) and “dopaminergic synapse” (mmu04728, P=0.0000) (Figure 1A). Using the same cutoff criteria, we extracted 32 DEGs between inflexible drinkers and light drinkers from the striatum datasets of the GSE123114 microarray profile (Figure 1C). The most enriched KEGG pathways related to the 32 DEGs were “arrhythmogenic right ventricular cardiomyopathy (ARVC)” (mmu05412, P=0.0032), “GABAergic synapse” (mmu04727, P=0.0043) and “morphine addiction” (mmu05032, P=0.0045) (Figure 1B). The 13 overlapping DEGs from the PFC and striatum were subjected to KEGG enrichment analysis (Figure 1C). Again, the most enriched KEGG pathway was “morphine addiction” (mmu05032, P=0.0006) (Figure 1D). More data about KEGG analysis results were shown in supplementary materials Table S2-S4.
Alcohol Exposure Increases Gabrb3 Expression in the PFC and Striatum
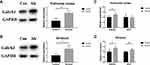
According to KEGG enrichment analysis (Figure 1D), two genes, Gabrb3 and Drd1, among the 13 overlapping DEGs were enriched in the morphine addiction pathway. The effect of alcohol exposure on the expression of Gabrb3 and Drd1 was investigated by intraperitoneal alcohol model. The mRNA expression of Gabrb3 was consistent with the microarray data, both in the PFC and striatum (Gabrb3: F1, 10=0.011, P=0.024; Figure 2C) (Gabrb3: F1, 10=1.429, P=0.015; Figure 2D). The transcript levels of Drd1 in the striatum were increased in the alcohol group mice, but no difference was detected in the PFC (Drd1: F1, 10=0.230, P=0.013; Figure 2D) (Drd1: F1, 10=0.168, P=0.745; Figure 2C). The protein expression of Gabrb3 were both increased in the PFC and striatum of the Alcohol group mice (F1, 4=1.198, P=0.015, Figure 2A) (F1, 4=3.467, P=0.037, Figure 2B). These results indicate that alcohol exposure regulates Gabrb3 signaling in the PFC and striatum.
Alcohol Exposure Upregulates HMGB1 and TLR4 in the PFC and Striatum
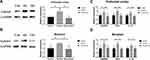
It has been reported that the HMGB1/TLR4 pathway is related to the alcohol-mediated regulation of GABRB3 expression in the mouse brain,16 and HMGB1 induces TNFα and IL-1β expression through TLR4 activation in alcohol dependence.17 To analyze the mechanisms by which alcohol exposure regulates Gabrb3 expression in the PFC and striatum, we examined HMGB1/TLR4 signaling, which is closely associated with alcohol exposure. The transcript levels of HMGB1 and TLR4 were both upregulated in the PFC and striatum of the alcohol group mice (HMGB1: F1, 10=11.673, P=0.017; TLR4: F1, 10=0.779, P=0.030; Figure 3A) (HMGB1: F1, 10=6.003, P=0.009; TLR4: F1, 10=1.857, P=0.004; Figure 3B). Also, the expression levels of inflammatory response-related factors, IL-1β and TNFα, were significantly increased in the PFC and striatum of mice in the alcohol group [TNFα: (F2, 15=7.465, P=0.006); control vs alcohol, P=0.005; IL-1β: (F2, 15=7.399, P=0.006); control vs alcohol, P=0.011; Figure 4C] [TNFα: (F2, 15=7.519, P=0.005); control vs alcohol, P=0.005; IL-1β: (F2, 15=4.492, P=0.030); control vs alcohol, P=0.017; Figure 4D]. These results indicate that alcohol exposure regulates HMGB1/TLR4 signaling in the PFC and striatum.
HMGB1 Inhibitor Decreases the Inflammatory Response and Gabrb3 Expression in the PFC and Striatum of Alcohol Exposure Mice
In order to further analyze the regulation mechanism of GABRB3 expression mediated by alcohol exposure, the effect of HMGB1 inhibitor on GABRB3 expression was detected by qRT-PCR and Western blotting. The HMGB1 inhibitor decreased Gabrb3 protein expression and transcript levels both in the PFC and striatum of alcohol exposure mice [Gabrb3: (F2, 6=7.747, P=0.022); alcohol vs glycyrrhizin, P=0.011; Figure 4A] [Gabrb3: (F2, 15=5.982, P=0.012); alcohol vs glycyrrhizin, P=0.005; Figure 4C] [Gabrb3: (F2, 6=5.226, P=0.048); alcohol vs glycyrrhizin, P=0.026; Figure 4B] [Gabrb3: (F2, 15=3.731, P=0.048); alcohol vs glycyrrhizin, P=0.045; Figure 4D]. Also, the expression of inflammatory response-related factors, IL-1β and TNFα, was significantly inhibited by HMGB1 inhibitor in the PFC and striatum of the alcohol group mice [TNFα: (F2, 15=7.465, P=0.006); alcohol vs glycyrrhizin, P=0.004; IL-1β: (F2, 15=7.399, P=0.006); alcohol vs glycyrrhizin, P=0.002; Figure 4C] [TNFα: (F2, 15=7.518, P=0.005); alcohol vs glycyrrhizin, P=0.004; IL-1β: (F2, 15=7.337, P=0.006); alcohol vs glycyrrhizin P=0.024; Figure 4D]. These results indicate that HMGB1/TLR4 signaling may be related to the regulation of Gabrb3 expression by alcohol exposure in the PFC and striatum.
Discussion
By analyzing GSE123114 microarray profile, we found that the morphine addiction pathway was significantly different in both the PFC and striatum. The chronic use of morphine is characterized by adaptive changes in neurons and neuronal communication. Morphine can cause indirect excitation of dopamine neurons by reducing inhibitory synaptic transmission mediated by GABAergic neurons. Differences in the morphine addiction pathway suggested that there are similar pathological processes and corresponding gene expression changes in alcohol-dependent mice. Furthermore, we identified Gabrb3 of morphine addiction pathway as a key gene. GABRB3 is a gamma-aminobutyric acid receptor, a major inhibitory neurotransmitter in human central nervous system.18 As a GABA-gated chloride ion channel,19 it plays an important role in the formation of functional inhibitory GABAergic synapses and participates in synaptic inhibition.19 Also, it participates in mediating the cellular response to histamine,20 somatosensation, and the production of antinociception.21
Alleles of GABRB3 in Caucasian (non-Hispanic) subjects have been associated with alcohol dependence.22 Alcohol dependence patients with the GABRB3 G1+ allele reported significantly lower drinking refusal self-efficacy in situations of stronger mood-related alcohol expectancy (AE) and social pressure relating to negative affective change.23 In Korea, the GABRB3 G1− alleles are more commonly in children of alcoholics (COAs) than in non-COAs.24 In African Americans, CpGs in GABRB3 are hypermethylated in alcohol dependence patients compared with controls.25 Together, these studies indicate that GABRB3 is closely related to alcohol dependence. In in vitro experiments, alcohol increases the expression of the GABRB3 gene in human embryonic stem cells.26 In in vivo experiments, adult male offspring born to Sprague Dawley dams that received ethanol had elevated expression of the Gabrb3 gene in the hippocampus.27 These studies provide further evidence that GABRB3 is related to the regulation of brain function by alcohol. In this research, functional analysis of DEGs from GSE123114 microarray profile revealed that Gabrb3 is the key gene of alcohol dependence in both the PFC and striatum. In the PFC and striatum of alcohol exposure mice, GABRB3 expression was significantly increased at both the transcript and protein levels, consistent with the microarray profile. Together, all these findings suggest that Gabrb3 is a critical novel gene in alcohol exposure.
It has been reported that the expression of Gabrb3 is elevated in the male offspring frontal cortex of ethanol-fed wild-type dams, but is unaffected in the offspring of ethanol-fed TLR4(-/-) dams.16 These results indicate that the TLR4 pathway is related to the alcohol-mediated regulation of GABRB3 expression in the mouse brain.
Toll-like receptors are the most well characterized pattern recognition receptors that recognize PAMPs and DAMPs, including HMGB1 and heat shock proteins.28 TLR4 has been identified as a critical receptor related to ethanol-induced neuropathology.28,29 HMGB1, a nuclear chromatin binding protein, is a endogenous TLR4 agonist which has been identified as a key immune mediator in alcohol dependence.28 Extracellular HMGB1 released as an innate immune mediator activates innate immune responses through direct binding to TLR4 or RAGE.30 It has been reported that the expression of TLR4 and HMGB1 is increased both in chronic ethanol-administered mice and the human postmortem alcoholic brain.31 In this experiment, the expression levels of HMGB1 and TLR4 were significantly increased in the PFC and striatum of mice in the alcohol group. These results are consistent with previous research indicating increased HMGB1/TLR4 signaling in the alcoholic brain. To examine whether changes in HMGB1/TLR4 signaling are associated with increased Gabrb3 gene expression in alcoholic brain, we assessed the effects of an HMGB1 inhibitor on Gabrb3 expression in alcohol exposure mice. The HMGB1 inhibitor decreased Gabrb3 expression in both the PFC and striatum of these mice. Together, these results reveal that alcohol exposure modulates Gabrb3 expression through the HMGB1/TLR4 pathway.
Previous studies have reported that HMGB1 induces TNFα and IL-1β expression through TLR4 activation in alcohol dependence,17 and that inflammation inhibits GABA transmission in CNS diseases.32 In this study, the HMGB1 inhibitor decreased TNFα and IL-1β expression both in the PFC and striatum of alcohol exposure mice, consistent with previous studies. These results indicate that HMGB1 may regulate Gabrb3 expression through the inflammatory response pathway. It has been reported that activation of Akt is involved in GABA(A) receptor anchoring in neurons,33 and that Akt1 deficiency modulates GABAergic functions and reduces GABAAR subunit expression in the hippocampus.34 In glioma cells, TNFα-induced oxidative stress-dependent Akt signaling affects actin cytoskeletal organization,35 and in the CNS, IL-1β enhances the expression of GABAA receptor in cell surface by the Akt pathway.36 Collectively, these findings suggest that inflammation-related factors in the PFC and striatum of alcohol-dependent mice, such as TNFα and IL-1β, may regulate Gabrb3 expression through the Akt pathway. However, further study is needed to test this hypothesis.
Conclusion
Our findings suggest that Gabrb3 is the key gene associated with alcohol exposure in mice, and reveal that alcohol exposure modulates Gabrb3 expression through the HMGB1/TLR4 pathway. Moreover, inflammation-related factors, such as IL-1β and TNFα, may be related to the regulation of GABRB3 expression by the HMGB1/TLR4 pathway in alcohol exposure.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was supported by Doctoral Initiation Project of Jiamusi University (grant numbers: JMSUBZ2019-06); talent training project for basic scientific research of Heilongjiang Province Educational Commission of China (grant numbers: 2019-KYYWF-1357); The National Key R&D Program of China (grant numbers: 2018YFC1314404); National Natural Science Foundation of China (grant numbers: 81901398). Natural Science Foundation of Chongqing, China (grant numbers: cstc2019jcyj-msxmX0025).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Alasmari F, Goodwani S, McCullumsmith RE, Sari Y. Role of glutamatergic system and mesocorticolimbic circuits in alcohol dependence. Prog Neurobiol. 2018;171:32–49.
2. Vornholt E, Drake J, Mamdani M, et al. Network preservation reveals shared and unique biological processes associated with chronic alcohol abuse in NAc and PFC. PLoS One. 2020;15(12):e0243857. doi:10.1371/journal.pone.0243857
3. Grant BF, Goldstein RB, Saha TD, et al. Epidemiology of DSM-5 alcohol use disorder: results from the national epidemiologic survey on alcohol and related conditions III. JAMA Psychiatry. 2015;72(8):757–766. doi:10.1001/jamapsychiatry.2015.0584
4. Trantham-Davidson H, Chandler LJ. Alcohol-induced alterations in dopamine modulation of prefrontal activity. Alcohol. 2015;49(8):773–779. doi:10.1016/j.alcohol.2015.09.001
5. Kahkonen S, Wilenius J, Nikulin VV, Ollikainen M, Ilmoniemi RJ. Alcohol reduces prefrontal cortical excitability in humans: a combined TMS and EEG study. Neuropsychopharmacology. 2003;28(4):747–754. doi:10.1038/sj.npp.1300099
6. Rossetti ZL, Carboni S. Ethanol withdrawal is associated with increased extracellular glutamate in the rat striatum. Eur J Pharmacol. 1995;283(1–3):177–183. doi:10.1016/0014-2999(95)00344-K
7. Abulseoud OA, Camsari UM, Ruby CL, Kasasbeh A, Choi S, Choi DS. Attenuation of ethanol withdrawal by ceftriaxone-induced upregulation of glutamate transporter EAAT2. Neuropsychopharmacology. 2014;39(7):1674–1684. doi:10.1038/npp.2014.14
8. da Silva ESDA, Frozino Ribeiro A, Damasceno S, et al. Inflexible ethanol intake: a putative link with the Lrrk2 pathway. Behav Brain Res. 2016;313:30–37. doi:10.1016/j.bbr.2016.07.001
9. de Paiva Lima C, da Silva ESDA, Damasceno S, et al. Loss of control over the ethanol consumption: differential transcriptional regulation in prefrontal cortex. J Neurogenet. 2017;31(3):170–177. doi:10.1080/01677063.2017.1349121
10. Zhu JR, Lu HD, Guo C, et al. Berberine attenuates ischemia-reperfusion injury through inhibiting HMGB1 release and NF-κB nuclear translocation. Acta Pharmacol Sin. 2018;39(11):1706–1715. doi:10.1038/s41401-018-0160-1
11. Le K, Wu S, Chibaatar E, Ali AI, Guo Y. Alarmin HMGB1 plays a detrimental role in hippocampal dysfunction caused by hypoxia-ischemia insult in neonatal mice: evidence from the application of the HMGB1 inhibitor glycyrrhizin. ACS Chem Neurosci. 2020;11(6):979–993. doi:10.1021/acschemneuro.0c00084
12. Li J, Li J, Liu X, et al. MicroRNA expression profile and functional analysis reveal that miR-382 is a critical novel gene of alcohol addiction. EMBO Mol Med. 2013;5(9):1402–1414. doi:10.1002/emmm.201201900
13. Okhuarobo A, Igbe I, Yahaya A, Sule Z. Effect of caffeine on alcohol consumption and alcohol-induced conditioned place preference in rodents. J Basic Clin Physiol Pharmacol. 2018;30(1):19–28. doi:10.1515/jbcpp-2018-0068
14. Adermark L, Söderpalm B, Burkhardt JM. Brain region specific modulation of ethanol-induced depression of GABAergic neurons in the brain reward system by the nicotine receptor antagonist mecamylamine. Alcohol. 2014;48(5):455–461. doi:10.1016/j.alcohol.2014.06.004
15. Zeng L, Zeng B, Wang H, et al. Microbiota modulates behavior and protein kinase C mediated cAMP response element-binding protein signaling. Sci Rep. 2016;6(1):29998. doi:10.1038/srep29998
16. Shukla PK, Meena AS, Rao R, Rao R. Deletion of TLR-4 attenuates fetal alcohol exposure-induced gene expression and social interaction deficits. Alcohol. 2018;73:73–78. doi:10.1016/j.alcohol.2018.04.004
17. Zou JY, Crews FT. Release of neuronal HMGB1 by ethanol through decreased HDAC activity activates brain neuroimmune signaling. PLoS One. 2014;9(2):e87915. doi:10.1371/journal.pone.0087915
18. Tanaka M, Olsen RW, Medina MT, et al. Hyperglycosylation and reduced GABA currents of mutated GABRB3 polypeptide in remitting childhood absence epilepsy. Am J Hum Genet. 2008;82(6):1249–1261.
19. Brown LE, Fuchs C, Nicholson MW, Stephenson FA, Thomson AM, Jovanovic JN. Inhibitory synapse formation in a co-culture model incorporating GABAergic medium spiny neurons and HEK293 cells stably expressing GABAA receptors. J Vis Exp. 2014;93:e52115. doi:10.3791/52115
20. Saras A, Gisselmann G, Vogt-Eisele AK, et al. Histamine action on vertebrate GABAA receptors: direct channel gating and potentiation of GABA responses. J Biol Chem. 2008;283(16):10470–10475. doi:10.1074/jbc.M709993200
21. Ugarte SD, Homanics GE, Firestone LL, Hammond DL. Sensory thresholds and the antinociceptive effects of GABA receptor agonists in mice lacking the beta3 subunit of the GABA(A) receptor. Neuroscience. 2000;95(3):795–806. doi:10.1016/S0306-4522(99)00481-9
22. Noble EP, Zhang X, Ritchie T, et al. D2 dopamine receptor and GABA(A) receptor beta3 subunit genes and alcoholism. Psychiatry Res. 1998;81(2):133–147. doi:10.1016/S0165-1781(98)00084-5
23. Young RM, Lawford BR, Feeney GF, Ritchie T, Noble EP. Alcohol-related expectancies are associated with the D2 dopamine receptor and GABAA receptor beta3 subunit genes. Psychiatry Res. 2004;127(3):171–183. doi:10.1016/j.psychres.2003.11.004
24. Namkoong K, Cheon KA, Kim JW, Jun JY, Lee JY. Association study of dopamine D2, D4 receptor gene, GABAA receptor beta subunit gene, serotonin transporter gene polymorphism with children of alcoholics in Korea: a preliminary study. Alcohol. 2008;42(2):77–81. doi:10.1016/j.alcohol.2008.01.004
25. Zhang H, Herman AI, Kranzler HR, et al. Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcohol Clin Exp Res. 2013;37(Suppl 1):E108–115.
26. Krishnamoorthy M, Gerwe BA, Scharer CD, et al. GABRB3 gene expression increases upon ethanol exposure in human embryonic stem cells. J Recept Signal Transduct Res. 2011;31(3):206–213.
27. Tunc-Ozcan E, Ullmann TM, Shukla PK, Redei EE. Low-dose thyroxine attenuates autism-associated adverse effects of fetal alcohol in male offspring’s social behavior and hippocampal gene expression. Alcohol Clin Exp Res. 2013;37(11):1986–1995. doi:10.1111/acer.12183
28. Crews FT, Lawrimore CJ, Walter TJ, Coleman LG
29. Alfonso-Loeches S, Pascual-Lucas M, Blanco AM, Sanchez-Vera I, Guerri C. Pivotal role of TLR4 receptors in alcohol-induced neuroinflammation and brain damage. J Neurosci. 2010;30(24):8285–8295. doi:10.1523/JNEUROSCI.0976-10.2010
30. Janko C, Filipovic M, Munoz LE, et al. Redox modulation of HMGB1-related signaling. Antioxid Redox Signal. 2014;20(7):1075–1085. doi:10.1089/ars.2013.5179
31. Crews FT, Qin L, Sheedy D, Vetreno RP, Zou J. High mobility group box 1/Toll-like receptor danger signaling increases brain neuroimmune activation in alcohol dependence. Biol Psychiatry. 2013;73(7):602–612. doi:10.1016/j.biopsych.2012.09.030
32. Rossi S, Studer V, Motta C, et al. Inflammation inhibits GABA transmission in multiple sclerosis. Mult Scler. 2012;18(11):1633–1635. doi:10.1177/1352458512440207
33. Margolis EB, Mitchell JM, Hjelmstad GO, Fields HL. A novel opioid receptor-mediated enhancement of GABAA receptor function induced by stress in ventral tegmental area neurons. J Physiol. 2011;589(17):4229–4242.
34. Chang CY, Chen YW, Wang TW, Lai WS. Akting up in the GABA hypothesis of schizophrenia: akt1 deficiency modulates GABAergic functions and hippocampus-dependent functions. Sci Rep. 2016;6(1):33095. doi:10.1038/srep33095
35. Ghosh S, Tewari R, Dixit D, Sen E. TNFalpha induced oxidative stress dependent Akt signaling affects actin cytoskeletal organization in glioma cells. Neurochem Int. 2010;56(1):194–201. doi:10.1016/j.neuint.2009.10.003
36. Serantes R, Arnalich F, Figueroa M, et al. Interleukin-1beta enhances GABAA receptor cell-surface expression by a phosphatidylinositol 3-kinase/Akt pathway: relevance to sepsis-associated encephalopathy. J Biol Chem. 2006;281(21):14632–14643. doi:10.1074/jbc.M512489200
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.