Back to Journals » OncoTargets and Therapy » Volume 14
High Expression of SLC41A3 Correlates with Poor Prognosis in Hepatocellular Carcinoma
Authors Li Q , Xiong DL, Wang H, Jin WL, Ma YY, Fan XM
Received 8 December 2020
Accepted for publication 16 March 2021
Published 5 May 2021 Volume 2021:14 Pages 2975—2988
DOI https://doi.org/10.2147/OTT.S296187
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tohru Yamada
Qian Li,1,2,* Dan-Lei Xiong,3,* Heng Wang,1 Wei-Li Jin,4 Ying-Yu Ma,5 Xiao-Ming Fan1,2
1Graduate Department, Bengbu Medical College, Bengbu, Anhui, 233000, People’s Republic of China; 2Department of Ultrasound, Zhejiang Provincial People’s Hospital, People’s Hospital of Hangzhou Medical College, Hangzhou, Zhejiang, 310014, People’s Republic of China; 3School of Medicine, Hangzhou Normal University, Hangzhou, Zhejiang, 310018, People’s Republic of China; 4Department of Gastroenterology, Nanxun People’s Hospital (Zhejiang Provincial People’s Hospital Nanxun Branch), Huzhou, 313009, Zhejiang Province, People’s Republic of China; 5Key Laboratory of Gastroenterology of Zhejiang Province, Zhejiang Provincial People’s Hospital, People’s Hospital of Hangzhou Medical College, Hangzhou, Zhejiang, 310014, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ying-Yu Ma
Key Laboratory of Gastroenterology of Zhejiang Province, Zhejiang Provincial People’s Hospital, People’s Hospital of Hangzhou Medical College, Hangzhou, Zhejiang, 310014, People’s Republic of China
Tel + 86-0571-85893781
Email [email protected]
Xiao-Ming Fan
Graduate Department, Bengbu Medical College, Bengbu, Anhui, 233000, People’s Republic of China
Tel + 86-0571-85893291
Email [email protected]
Purpose: SLC41A3 is a member of the solute carrier family 41 (SLC41) and is involved in many cellular processes as a magnesium ion transporter. Although it plays an important role in cancer formation and development, the correlation between the expression of SLC41A3 and the occurrence and prognosis of hepatocellular carcinoma (HCC) remains unclear. Therefore, this study was focused on the evaluation of the relationship between SLC41A3 and the development and prognosis of HCC.
Patients and Methods: Firstly, we collected the mRNA expression of SLC41A3 in HCC through the platform of Oncomine. Then, the subgroups of HCC were performed by the UALCAN website and the prognosis of HCC was analyzed by Kaplan–Meier Plotter database. Subsequently, immunohistochemistry (IHC) method was used to detect SLC41A3 expression in 323 clinically confirmed HCC samples and 184 non-cancerous liver tissues. Finally, function enrichment analysis was done using the LinkInterpreter module in LinkedOmics, and gene set enrichment analysis (GSEA) was performed using TCGA data set.
Results: The Oncomine database and immunohistochemical (IHC) showed higher SLC41A3 expression in HCC tissue compared to normal tissue. The expression of SLC41A3 was significantly correlated with tumor metastasis, Edmondson grade, microvascular invasion, and AFP level. Kaplan–Meier and Cox regression analyses verified that high SLC41A3 expression is a significant prognostic factor for reduced overall survival in HCC patients.
Conclusion: Our results demonstrated that high expression of SLC41A3 was the predictor of poor prognosis in HCC patients, suggesting that this protein may be a potential target for HCC therapy.
Keywords: HCC, solute carrier 41A3, immunohistochemistry, bioinformatics
Introduction
Liver cancer is the sixth most prevalent tumor in the world. With about 841,000 new cases and 782,000 deaths every year, it is one of the leading causes of cancer-related death globally.1 Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, accounting for 85~90% of all cases. Approximately 55% of HCC cases and HCC-related deaths occur in China.2,3 Risk factors for HCC include cirrhosis, hepatitis B virus (HBV) infection, hepatitis C virus (HCV) infection, alcoholism, smoking, and exposure to aflatoxin B1.4,5 The major methods for HCC treatment are surgical resection, liver transplant, trans-arterial chemoembolization (TACE), ablation, radiotherapy, and chemotherapy.6 However, most patients are diagnosed at advanced stages of liver cancer, and these therapies were relatively ineffective. Moreover, the high potential of HCC for metastasis and recurrence results in a five-year survival rate of less than 5%.7–9 Therefore, it is necessary to identify reliable biomarkers of HCC to ensure early diagnosis, accurate evaluation of prognosis, and effective treatment.
Magnesium (Mg2+) is an essential ion in the organism and plays a key role in many cellular processes, such as the activation of enzymes, regulation of the synthesis of nucleic acids and proteins, and DNA repair.10 Additionally, it regulates the cell cycle, cell proliferation, and apoptosis. Experimental and clinical data suggest that magnesium deficiency may be associated with increased levels of inflammation and free radicals, which can lead to the formation of cancer.11 The solute carrier family 41 (SLC41) includes three members: A1, A2, and A3. Based on their distant homology with the Mg2+ channel MgtE of bacteria, they are all related to Mg2+ transporters.12 SLC41A3 was initially described as a part of the SLC41 family of Mg2+ transporters by Quamme and colleagues.13 Human SLC41A3 is located on chromosome 3q21.2-q2.3 and is predicted to produce seven alternative splicing variants. The full-length protein is composed of 507 amino acids and has a molecular weight of 54.7 kDa. While the molecular biology and exact function of SLC41A3 are poorly understood, it has been reported that its close homolog SLC41A1, which has a 52% identity at amino acid level, may be associated with tumor development.14
The present investigation evaluated the expression and biological functions of SLC41A3 in human HCC by bioinformatics analysis utilizing open databases. We then confirmed the role of SLC41A3 in 323 HCC cases by immunohistochemistry (IHC). The obtained results indicate that SLC41A3 may be a valuable candidate biomarker for the prognosis and treatment of HCC.
Materials and Methods
Oncomine
In the Oncomine database (https://www.oncomine.org), we acquired the data of the SLC41A3 gene in HCC by selecting the conditions of filtering and mining data. The criteria used were as follows: 1. “Cancer Type: Liver cancer”; 2. “Gene: SLC41A3”; 3. “Data Type: mRNA”; 4. “Analysis Type: Cancer vs Normal”; 5. Others: P-value<0. 0001, fold change>2, gene rank=top 10%.
Ualcan
UALCAN (http://ualcan.path.uab.edu) is a web site for analyzing the RNA level expression data in cancers, using datasets from the TCGA project. We used this site to explore the distribution of expression of SLC41A3 in between different types of tumors and normal tissues. Importantly, this site facilitates the analysis of relative levels of SLC41A3 expression between various subgroups, including tumor grade, tumor stage, tumor metastasis, and other clinicopathologic characteristics.
Survival Analysis
Kaplan–Meier Plotter database (http://kmplot.com/analysis/) is used to analyze gene expression and tumor prognosis. It can analyze the prognostic value of 54,675 genes in tumor samples of 10,461 patients. In this study, we used this site to evaluate the association between SLC41A3 expression and HCC prognosis. For this purpose, patients were divided into the high-expression group and the low-expression group according to the median expression, and overall survival was estimated by calculating log-rank P-value and hazard ratio (HR).
GO and KEGG Analysis
LinkedOmics (http://www.linkedomics.org/login.php) is based on the TCGA database, including 32 types of cancer. It can be used to analyze the expression of genes at the mRNA or protein level, the relationship with clinicopathological features, methylation, mutation sites, and other characteristics. LinkedOmics has three modules: LinkFinder, LinkInterpreter, and LinkCompare. In our study, we obtained information on SLC41A3 in HCC by setting conditions and used LinkFinder to identify genes co-expressed with SLC41A3.The filter criteria were set as: 1. “Gene: SLC41A3”; 2. “Analysis Type: Cancer vs Normal analysis”; 3. “Data Type: mRNA”; 4. “Cancer Type: Liver Cancer”; 5. “Gene Summary: P-value<0. 05, fold change=all, gene rank=top 10%”. Subsequently, we performed the GO/KEGG analysis of these genes using LinkInterpreter modules. The GO analysis of the genes includes molecular function, biological processes, and cellular component.
Gene Set Enrichment Analysis (GSEA)
Gene Set Enrichment Analysis (GSEA) is a computational method determining whether pre-defined sets of genes show statistically significant, concordant differences between two biological states. In addition, GSEA was utilized to identify the potential pathways associated with SLC41A3 expression and prognosis in HCC. To explore the mechanisms underlying the effects of SLC41A3, we downloaded data related to HCC from the TCGA database (https://portal.gdc.cancer.gov/), including 55,104 genes and 374 samples, and analyzed them using the GSEA software v4.1.0 (Broad Institute, MIT, Cambridge, MA, USA). Gene expression profiles of HCC patients were divided into the high expression group (n=221) and low expression group (n=153) based on the median value of SLC41A3 expression. The number of gene set permutations was set each time to 1000. A gene set was regarded as significantly enriched when the P-value and false discovery rate (FDR) were less than 0.05.
Protein–Protein Interaction Networks (PPI)
The regulation of the expression of genes related to SLC41A3 was analyzed using the search tool of the Retrieval of Interacting Genes database (http://string-db.org/) and the GeneMANIA (http://genemania.org/) web portal. The necessary data were obtained from the website by entering the protein name, species, and other required information.
Patients and Tissue Samples
All 323 samples of cancerous liver tissue were collected from patients undergoing surgical resection at the Zhejiang Provincial People’s Hospital, Hangzhou, China, between January 2010 and September 2016. The presence of the tumor was confirmed by a pathologist. The patient population included 262 males and 61 females aged 25 to 90 years (median age, 56.95 years). None of the patients received radiotherapy, chemotherapy, or other treatment before the surgery. In addition, 184 specimens of non-cancerous liver tissue were collected from patients with no hepatocellular carcinoma during the same period. The samples were used to prepare tissue microarrays (TMAs), which were constructed by Shanghai Biochip Co., Ltd. (Shanghai, China). The clinical information of patients, including age, gender, tumor size, tumor number, TNM stage, tumor metastasis, and survival time, was retrieved from patient records. The survival time was calculated as the time from the surgery to the patient’s death or the end of follow-up. The study was approved by the Ethics Committee of the Zhejiang Provincial People’s Hospital (2021QT077), and had been proceeded in accordance with the Helsinki Declaration. And written informed consent was provided by all patients.
Immunohistochemistry (IHC)
The IHC staining of TMAs was performed using a standard method. Briefly, TMA was heated at 70°C for 2 hours, and the samples were deparaffinized with xylene and rehydrated in a graded alcohol series (95, 85, 75%). Antigen retrieval was performed by boiling the TMA in the TE buffer in a pressure cooker at 120°C for 3 minutes. Subsequently, samples were incubated with 3% H2O2 for 15 minutes to block the activity of endogenous peroxidase and with normal goat serum for 15 minutes to block the non-specific binding; these steps were done at room temperature. The serum was removed and, without washing in-between, specimens were incubated with a rabbit anti-SLC41A3 polyclonal antibody (PA5-50763, Invitrogen, Carlsbad, CA, USA) diluted 1:1000 at 4°C overnight. Next, sections were incubated with biotin-labeled goat anti-mouse IgG for 15 minutes at room temperature, and then with streptavidin-biotinylated horseradish peroxidase-conjugated antibody for another 15 minutes at room temperature. Finally, the color was developed using freshly prepared 3,3-diaminobenzidine (DAB), and the reaction was stopped before the color became too dark. TMAs were counterstained with hematoxylin for 5 minutes, dehydrated for 45 minutes, mounted in resin under a cover glass, and allowed to dry.
Stained samples were scored by two pathologists working independently, who were blinded to the clinical or clinicopathological status of the specimens. The expression level of SLC41A3 was evaluated using a scoring system based on the intensity and the proportion of positively stained cells. The staining intensity was scored as: 0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining. The fraction of stained cells was scored as: 0, no cell stained; 1, 1–25% of cells stained; 2, 2–50% of cells stained; 3, 51–75% of cells stained; 4, >75% of cells stained. The total score was obtained by multiplying intensity scores and the percentage scores, and ranged from 0 to 12. Total scores ranging from 0 to 6 were considered as low SLC41A3 expression, and higher scores were considered as high SLC41A3 expression.
Statistical Analysis
Statistical analysis was performed using SPSS 17.0 (IBM SPSS, Chicago, IL, USA) and GraphPad Prism version 8.0 (GraphPad Software, San Diego, CA, USA). The χ2 test was used to evaluate the correlation between the expression of SLC41A3 protein and various clinicopathological factors. The overall survival curves were plotted using the Kaplan–Meier method, and survival data were analyzed by a Log rank test and univariate and multivariate Cox regression analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
The Expression Level of SLC41A3 in HCC Was Significantly Higher Than in Normal Tissues
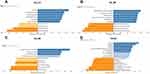
The difference in the level of SLC41A3 mRNA expression between various tumor samples and normal tissues was evaluated using UALCAN on the basis of TCGA data (Figure 1A). We found that SLC41A3 is expressed in many cancers, and there is a significant difference between tumor tissues and normal tissues in liver hepatocellular cancer (LIHC) and cholangiocarcinoma (CHOL), especially. In addition, we further used Oncomine to extract the information necessary to evaluate the expression of SLC41A3 in normal and HCC tissues. According to the screening criteria, the 4 databases connected with our study included a total of 408 samples. Among them, 3 databases (Roessler Liver data set, Roessler Liver 2 data set, and Wurmbach Liver data set) showed that SLC41A3 expression was significantly higher in HCC than in normal tissues (P<0.05) (Figure 1B–D). However, no statistically significant difference was found in the Mas Liver data set, indicating that additional studies involving a larger number of samples are still needed to demonstrate the significance of SLC41A3 in HCC (Figure 1E). A meta-analysis of these 4 results showed that SLC41A3 was highly expressed in HCC (P<0.05) (Figure 1F). Furthermore, we used UALCAN to analyze the expression of SLC41A3 in 50 normal liver tissue and 371 HCC included in the TCGA database. The results showed that the level of SLC41A3 expression was higher in HCC patients with high Edmondson grade and advanced TNM stage than in those with low Edmondson grade and early TNM stage, respectively (Figure 2).
IHC was used to further evaluate the expression of SLC41A3 protein in HCC and normal tissues. This method showed both high and low expression of SLC41A3 in HCC tissue (Figure 3A–C). High expression of SLC41A3 was detected in 232 (71.83%) of 323 HCC samples and 72 (39.13%) of 184 normal tissue samples, while low expression of SLC41A3 was found in 91 (21.87%) of 323 HCC samples and 112 (60.87%) of 184 normal tissue samples. These differences were statistically significant (χ2=51.20, P<0.001). The correlation between SLC41A3 protein level and various clinicopathological features is listed in Table 1. We found that high expression of SLC41A3 is positively related to the Edmondson grade (P=0.02), metastasis (P=0.04), microvascular invasion (P=0.01), and the level of AFP (P=0.04), but is not significantly associated with age, gender, tumor size, cirrhosis, and other clinical variables.
 |
Table 1 Expression of SLC41A3 in Hepatocellular Carcinoma Tissues (n=323) |
Clinical Significance of SLC41A3 Expression in the Prognosis of HCC
Next, we used the Kaplan–Meier Plotter database to determine the clinical significance of SLC41A3 expression in the prognosis of HCC. The result showed that overall survival was shorter in the high-expression group than in the low-expression group (Figure 4A). In our clinical samples, the survival analysis indicated that the mean length of survival of HCC patients with high expression of SLC41A3 was 40.40±2.90 months, which was significantly shorter than in patients with low expression of SLC41A3, 59.40±3.46 months (P<0.001; Figure 4B). In addition, univariate and multivariate analyses were performed to determine whether high expression of SLC41A3 was an independent factor affecting the prognosis of HCC patients. Univariate analysis demonstrated that the overall survival correlated with tumor size (HR=1.95; 95% CI 1.22–3. 13; P<0.01), Edmondson grade (HR=2.78; 95% CI 1.74–4.46; P<0.01), metastasis (HR=4.82; 95% CI 2.55–9.11; P<0.01), microvascular invasion (HR=2.11; 95% CI 1.25–3.56; P<0.01), AFP level (HR=2.51; 95% CI 1.40–4.52; P<0.01), and SLC41A3 expression (HR=2.56; 95% CI 1.43–4.60; P<0.01). Multivariate analysis was used to exclude confounding factors and documented that the Edmondson grade (HR=2.55; 95% CI 1.21–5.36; P=0.01) and metastasis (HR=3.02; 95% CI 1.11–8.22; P=0.03; Table 2) were independent prognostic factors of overall survival in HCC patients. Although SLC41A3 was not identified as an independent factor, it negatively affected the development of HCC, which may be one of the factors affecting the poor prognosis.
 |
Table 2 Univariate and Multivariate Cox Regression Survival Analysis of Clinicopathological Parameters and SLC41A3 Expression in Hepatocellular Carcinoma Patients |
GO and KEGG Pathway Analyses of Genes Co-Expressed with SLC41A3 in HCC
The genes co-expressed SLC41A3 in HCC were analyzed in 371 TCGA database samples using the LinkedOmics online tool. The volcano plot (Figure 5A) shows 3590 genes positively correlated with SLC41A3 (red spots; FDR<0.05) and 2712 genes negatively correlated with SLC41A3 (green spots; FDR<0.05). The top 50 genes positively and negatively correlated with SLC41A3 are shown in the heat map (Figure 5B and C). The top three positively correlated genes are HDAC11, ELOVL1, and NEU1 (Figure 5D–F), and the top three negatively correlated genes are CYP4V2, USP38, and SOCS2 (Figure 5G–I).
The results of Gene Ontology (GO) enrichment analysis of LinkedOmics showed that genes co-expressed with SLC41A3 were located mostly in ribosomes, spliceosomal complex, chromosomal region, cell-substrate junction, microtubules, and vacuolar membrane. These genes primarily participated in ribonucleoprotein complex biogenesis, chromosome segregation, ncRNA processing, protein-DNA complex subunit organization, protein targeting, and RNA splicing. They acted as structural constituents in nucleosome binding, unfolded protein binding, and ribosomes (Figure 6A–C). KEGG pathway analysis indicated that the function of genes co-expressed with SLC41A3 were mainly enriched in the ribosome, spliceosome, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, RNA transport, and proteasome (Figure 6D). These results suggest that SLC41A3 has extensive effects on the transcriptome.
GSEA
To investigate the potential functional mechanisms of SLC41A3 in HCC, we performed Gene Set Enrichment Analysis (GSEA) using TCGA data to identify KEGG pathways enriched in the samples with high SLC41A3 expression. Significantly enriched pathways were selected based on their normalized enrichment score (NSE). The outcomes of GSEA showed that cancer-related signaling pathways, including “PATHWAYS_IN_CANCER”, “WNT_SIGNALING-PATHWAY”, “MTOR_SIGNALING-PATHWAY”, “TGF_BETA_SIGNALING-PATHWAY”, “APOPTOSIS” and “CELL_CYCLE”, were mostly enriched in samples with high expression of SLC41A3 (Figure 7A–F and Table 3). These findings suggest that SLC41A3 may contribute to the progression of HCC by engaging in several cancer-associated signaling pathways.
 |
Table 3 Gene Sets Enriched in High SLC41A3 Expression Phenotype in HCC |
Protein–Protein Interaction Network (PPI)
To predict potential interactions between SLC41A3 and other cancer-related proteins, the STRING database was used to construct a protein–protein interaction network. Results showed that SLC41A3 interacts with SLC35A4, NIPAL3, PIMS1, CHST13, NIPAL1, MAGT1, TRPM6, TRPM7, CNNM2, and MRS2 (Figure 8A). Furthermore, we used GeneMANIA to analyze the interaction among these proteins. The analysis of the protein–protein interaction network revealed an intricate interaction between SLC41A3 and SLC41A1, SLC41A2, PDCD6, HSDL1, PDZD3, MAP2K5, COPS8, TOX4, FAM206A, PPP3CB, LRP11, SERAC1, ZACN, RNF25, REPS1, AMZ2, PNMAL1, POLR2G, GIT1, and SPRYD7 (Figure 8B).
 |
Figure 8 Protein–protein interaction network of SLC41A3. Interactions between SLC41A3 and other genes were obtained from the web portals STRING (A) and GeneMANIA (B). |
Discussion
Liver cancer is one of the most common malignancies of the digestive system in the world. In 2015, 466,100 people were diagnosed with liver cancer, and 422,100 died of liver cancer in China alone.15 Currently, surgical resection is the main method for the treatment of HCC. However, when HCC metastasizes, the efficacy of surgical treatment decreases dramatically.16 At present, an increased serum concentration of AFP is used as a marker for early screening for HCC, but only about 70% of HCC patients have an elevated level of AFP.17 Small-molecule tyrosine kinase inhibitors (TKIs) belonging to a class of molecule-targeted drugs have become an important focus of anti-liver cancer research. TKIs produce anti-tumor effects primarily by inhibiting cancer cell proliferation and angiogenesis.18 However, TKIs generate several adverse side effects, such as hand–foot skin reaction (HFSR), diarrhea, hypertension, nausea and vomiting, decreased appetite, and weight loss, and their safety and effectiveness still require careful consideration.19 Therefore, it is particularly important to find new biomarkers for HCC.
SLC41A3, together with SLC41A1 and SLC41A2, is a member of the SLC41 family. Currently, studies on SLC41A1 and SLC41A2 are relatively extensive and mainly focused on Mg2+ transport.20,21 However, it has been shown that SLC41A3 is also involved in the transport of Mg2+.22 Magnesium ions play an important role in regulating biological processes such as cell cycle and DNA damage and repair. Castiglioni and coworkers demonstrated that magnesium deficiency might potentiate inflammation and increase the level of free radicals.11 Animals deficient in magnesium are more sensitive to oxidative stress, and their cells are more sensitive to peroxidation in vitro. Inflammatory mediators and free radicals produce DNA damage that can lead to cancer. SLC41A1 was reported to inhibit pancreatic ductal adenocarcinoma (PDAC) via the Akt/mTOR pathway.14 However, Liu and collaborators found that SLC41A3, one of the genes co-expressed with AATF, is associated with HCC prognosis.23
In our present study, the analysis of several online databases showed that the expression of SLC41A3 mRNA was significantly higher in HCC than in normal liver tissues and positively correlated with tumor stage and grade. The use of IHC to determine the expression of SLC41A3 protein showed that the level of this protein was significantly higher in HCC than in normal liver tissue. Thus, the IHC data on the protein level were consistent with the mRNA data obtained by bioinformatics analysis. In addition, the UALCAN database showed that the expression of SLC41A3 was higher in patients with high grade and high stage of the tumor. Together, these data revealed that the upregulated expression of SLC41A3 might promote tumor progression.
To evaluate the prognostic value of SLC41A3 in HCC, tumor size, Edmondson grade, metastasis, microvascular invasion, SLC41A3 expression and AFP level may influence prognosis of HCC by univariate survival analysis, but only Edmondson grade and metastasis were independent prognostic factors suggested by multivariate survival analysis. This due to that the mechanism of tumor occurrence and development is complex, and multiple factors may affect HCC prognosis. Non-independent prognostic factors may interact or synergize with other factors in HCC development, such as tumor size and microvascular invasion. In addition, we performed survival analysis and found that HCC patients with high expression of SLC41A3 have a poorer prognosis than patients with low expression. This result was consistent with the survival analysis based on the Kaplan–Meier Plotter database. Therefore, we concluded that SLC41A3 might promote the development of HCC, and its elevated expression might be used for the prediction of HCC tumor metastasis and progression.
To further understand the mechanism of SLC41A3 in HCC, GO and KEGG analyses of the genes co-expressed with SLC41A3 were conducted. The results documented that SLC41A3 plays an important role primarily in the structure and function of ribosome, spliceosome, DNA, RNA, and protein. This analysis also showed that the top three genes positively correlated with SLC41A3 expression are HDAC11, ELOVL1, and NEU1. The expression of HDAC11 mRNA in liver cancer cells is significantly higher than that in pancreatic cells, and HDAC11 protein forms a complex with Egr1, which induced deacetylation of Egr1 and reduced the p53 expression in HCC cells. Moreover, p53 is an important molecule in induction of cancer cell apoptosis, suggestion that HDAC11 contributes to the tumorigenesis in HCC.24,25 ELOVL1 is highly expressed in breast cancer, and siRNA-mediated ELOVL1 silencing significantly affects cell survival.26 Guojun and coworkers demonstrated that NEU1 is highly expressed in liver cancer and may promote tumor cell proliferation and metastasis through the spliceosome pathway.27 Therefore, we hypothesize that SLC41A3 may promote the onset and development of HCC.
GSEA indicated that SLC41A3 is mainly involved in the WNT, mTOR, and TGF-β signaling pathways as well as in the regulation of apoptosis and cell cycle. Vilchez and collaborators showed that the WNT/β-catenin pathway promotes cell proliferation, migration, and invasion and is associated with the aggressiveness of HCC.28 The mTOR cascade has a variety of regulatory functions during the development of HCC, affecting proliferation, survival, invasion, metabolism, and other characteristics of cancer cells.29 Furthermore, Lu and colleagues found that SLC7A5/SLC3A2 is crucial to the activation of mTORC1.30 The TGF-β signaling pathway tends to inhibit the proliferation of precancerous hepatocytes in the early stage of HCC, but promotes the epithelial-mesenchymal transformation (EMT) and tumor invasion at later stages, suggesting that this pathway plays an essential role in HCC progression and determines poor prognosis.31 In addition, in terms of immune regulation, WNT and TGF-β signaling pathways are related to tumor genesis and immune response, and Mg2+ homeostasis may also affect immune function, such as immune cell adhesion and macrophage response. Therefore, SLC41A3, as the Mg2+ transporter, may affect the overall prognosis by changing the cancer microenvironment and immune response through these signaling pathways.32–34
Finally, the analysis of the protein interaction network showed that genes associated with SLC41A3, among them TRPM6, TRPM7, and NIPAL1, also have a crucial function in tumors. The upregulation of TRPM6 promotes neuroblastoma cell proliferation.35 TRPM7 is highly expressed in liver cancer, pancreatic cancer, and other cancer types.36,37 Moreover, increased TRPM7 level is significantly associated with the recurrence, metastasis, and prognosis of bladder cancer.38 NIPAL1 may be a novel factor promoting oral squamous cell carcinoma (OSCC) and a useful molecular marker of this tumor.39
In conclusion, our study demonstrates the relevance of SLC41A3 in the pathogenesis of HCC and documents that the evaluation of SLC41A3 expression may be employed as an auxiliary test in HCC diagnosis. In addition, since the upregulation of SLC41A3 in HCC is associated with shorter survival time and reduced survival rate, it may open up new approaches for the prevention and treatment of HCC. Finally, we predicted the signaling pathways and interacting proteins involved in the effects of SLC41A3, facilitating future research on the elucidation of the specific role of SLC41A3 in the pathogenesis of HCC.
Acknowledgments
This work was supported by the Natural Science Foundation of Zhejiang Province (grant no. GF19H160085).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. doi:10.1053/j.gastro.2007.04.061
3. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi:10.3322/canjclin.55.2.74
4. Chedid MF, Kruel CRP, Pinto MA, et al. Hepatocellular carcinoma: diagnosis and operative management. Arq Bras Cir Dig. 2017;30(4):272–278. doi:10.1590/0102-6720201700040011
5. Keating GM. Sorafenib: a review in hepatocellular carcinoma. Target Oncol. 2017;12(2):243–253. doi:10.1007/s11523-017-0484-7
6. Yegin EG, Oymaci E, Karatay E, Coker A. Progress in surgical and nonsurgical approaches for hepatocellular carcinoma treatment. Hepatobiliary Pancreat Dis Int. 2016;15(3):234–256. doi:10.1016/S1499-3872(16)60097-8
7. Tokumitsu Y, Sakamoto K, Tokuhisa Y, et al. A new prognostic model for hepatocellular carcinoma recurrence after curative hepatectomy. Oncol Lett. 2018;15(4):4411–4422. doi:10.3892/ol.2018.7821
8. Zhu Q, Li N, Zeng X, et al. Hepatocellular carcinoma in a large medical center of China over a 10-year period: evolving therapeutic option and improving survival. Oncotarget. 2015;6(6):4440–4450. doi:10.18632/oncotarget.2913
9. Louafi S, Boige V, Ducreux M, et al. Gemcitabine plus oxaliplatin (GEMOX) in patients with advanced hepatocellular carcinoma (HCC): results of a Phase II study. Cancer. 2007;109(7):1384–1390. doi:10.1002/cncr.22532
10. Chakraborti S, Chakraborti T, Mandal M, Mandal A, Das S, Ghosh S. Protective role of magnesium in cardiovascular diseases: a review. Mol Cell Biochem. 2002;238(1–2):163–179. doi:10.1023/A:1019998702946
11. Castiglioni S, Maier JA. Magnesium and cancer: a dangerous liason. Magnes Res. 2011;24(3):S92–S100. doi:10.1684/mrh.2011.0285
12. Schweigel-Röntgen M, Kolisek M. SLC41 transporters–molecular identification and functional role. Curr Top Membr. 2014;73:383–410.
13. Quamme GA. Molecular identification of ancient and modern mammalian magnesium transporters. Am J Physiol Cell Physiol. 2010;298(3):C407–429. doi:10.1152/ajpcell.00124.2009
14. Xie J, Cheng CS, Zhu XY, et al. Magnesium transporter protein solute carrier family 41 member 1 suppresses human pancreatic ductal adenocarcinoma through magnesium-dependent Akt/mTOR inhibition and bax-associated mitochondrial apoptosis. Aging (Albany NY). 2019;11(9):2681–2698. doi:10.18632/aging.101940
15. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
16. Toyoda H, Kumada T, Tada T. Changes in patient backgrounds may increase the incidence of HCC after SVR in the era of IFN-free therapy for HCV. Hepatology. 2016;64(5):1818–1819. doi:10.1002/hep.28632
17. Guan L, Luo Q, Liang N, Liu H. A prognostic prediction system for hepatocellular carcinoma based on gene co-expression network. Exp Ther Med. 2019;17(6):4506–4516. doi:10.3892/etm.2019.7494
18. Berretta M, Rinaldi L, Di Benedetto F, et al. Angiogenesis inhibitors for the treatment of hepatocellular carcinoma. Front Pharmacol. 2016;7:428. doi:10.3389/fphar.2016.00428
19. Rimassa L, Danesi R, Pressiani T, Merle P. Management of adverse events associated with tyrosine kinase inhibitors: improving outcomes for patients with hepatocellular carcinoma. Cancer Treat Rev. 2019;77:20–28. doi:10.1016/j.ctrv.2019.05.004
20. Mandt T, Song Y, Scharenberg AM, Sahni J. SLC41A1 Mg(2+) transport is regulated via Mg(2+)-dependent endosomal recycling through its N-terminal cytoplasmic domain. Biochem J. 2011;439(1):129–139. doi:10.1042/BJ20110807
21. Sahni J, Nelson B, Scharenberg AM. SLC41A2 encodes a plasma-membrane Mg2+ transporter. Biochem J. 2007;401(2):505–513. doi:10.1042/BJ20060673
22. Mastrototaro L, Smorodchenko A, Aschenbach JR, Kolisek M, Sponder G. Solute carrier 41A3 encodes for a mitochondrial Mg(2+) efflux system. Sci Rep. 2016;6:27999. doi:10.1038/srep27999
23. Liu J, Lu J, Ma Z, Li W. A nomogram based on a three-gene signature derived from AATF coexpressed genes predicts overall survival of hepatocellular carcinoma patients. Biomed Res Int. 2020;2020:7310768.
24. Freese K, Seitz T, Dietrich P, et al. Histone deacetylase expressions in hepatocellular carcinoma and functional effects of histone deacetylase inhibitors on liver cancer cells in vitro. Cancers (Basel). 2019;11(10):1587. doi:10.3390/cancers11101587
25. Gong D, Zeng Z, Yi F, Wu J. Inhibition of histone deacetylase 11 promotes human liver cancer cell apoptosis. Am J Transl Res. 2019;11(2):983–990.
26. Hilvo M, Denkert C, Lehtinen L, et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011;71(9):3236–3245. doi:10.1158/0008-5472.CAN-10-3894
27. Hou G, Liu G, Yang Y, et al. Neuraminidase 1 (NEU1) promotes proliferation and migration as a diagnostic and prognostic biomarker of hepatocellular carcinoma. Oncotarget. 2016;7(40):64957–64966. doi:10.18632/oncotarget.11778
28. Vilchez V, Turcios L, Marti F, Gedaly R. Targeting Wnt/β-catenin pathway in hepatocellular carcinoma treatment. World J Gastroenterol. 2016;22(2):823–832. doi:10.3748/wjg.v22.i2.823
29. Lu X, Paliogiannis P, Calvisi DF, Chen X. Role of the mTOR pathway in liver cancer: from molecular genetics to targeted therapies. Hepatology. 2020. doi:10.1002/hep.31310
30. Liu P, Ge M, Hu J, et al. A functional mammalian target of rapamycin complex 1 signaling is indispensable for c-Myc-driven hepatocarcinogenesis. Hepatology. 2017;66(1):167–181. doi:10.1002/hep.29183
31. Dituri F, Mancarella S, Cigliano A, Chieti A, Giannelli G. TGF-β as multifaceted orchestrator in HCC progression: signaling, EMT, immune microenvironment, and novel therapeutic perspectives. Semin Liver Dis. 2019;39(1):53–69. doi:10.1055/s-0038-1676121
32. Liu J, Zhang S, Dai W, Xie C, Li JC. A comprehensive prognostic and immune analysis of SLC41A3 in pan-cancer. Front Oncol. 2021;10:586414. doi:10.3389/fonc.2020.586414
33. Perugorria MJ, Olaizola P, Labiano I, et al. Wnt-β-catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol. 2019;16(2):121–136. doi:10.1038/s41575-018-0075-9
34. Caja L, Dituri F, Mancarella S, et al. TGF-β and the tissue microenvironment: relevance in fibrosis and cancer. Int J Mol Sci. 2018;19(5):1294. doi:10.3390/ijms19051294
35. Zhang Z, Faouzi M, Huang J, et al. N-Myc-induced up-regulation of TRPM6/TRPM7 channels promotes neuroblastoma cell proliferation. Oncotarget. 2014;5(17):7625–7634. doi:10.18632/oncotarget.2283
36. Mishra R, Rao V, Ta R, Shobeiri N, Hill CE. Mg2+- and MgATP-inhibited and Ca2+/calmodulin-sensitive TRPM7-like current in hepatoma and hepatocytes. Am J Physiol Gastrointest Liver Physiol. 2009;297(4):G687–694. doi:10.1152/ajpgi.90683.2008
37. Yee NS, Zhou W, Liang IC. Transient receptor potential ion channel Trpm7 regulates exocrine pancreatic epithelial proliferation by Mg2+-sensitive Socs3a signaling in development and cancer. Dis Model Mech. 2011;4(2):240–254. doi:10.1242/dmm.004564
38. Gao SL, Kong CZ, Zhang Z, Li ZL, Bi JB, Liu XK. TRPM7 is overexpressed in bladder cancer and promotes proliferation, migration, invasion and tumor growth. Oncol Rep. 2017;38(4):1967–1976. doi:10.3892/or.2017.5883
39. Sasahira T, Nishiguchi Y, Kurihara-Shimomura M, Nakashima C, Kuniyasu H, Kirita T. NIPA-like domain containing 1 is a novel tumor-promoting factor in oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2018;144(5):875–882. doi:10.1007/s00432-018-2612-x
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.