Back to Journals » Journal of Inflammation Research » Volume 14
Gut and Cutaneous Microbiome Featuring Abundance of Lactobacillus reuteri Protected Against Psoriasis-Like Inflammation in Mice
Authors Chen HL, Zeng YB, Zhang ZY, Kong CY, Zhang SL, Li ZM, Huang JT, Xu YY, Mao Y, Cai PR, Han B, Wang WQ, Wang LS
Received 31 August 2021
Accepted for publication 6 November 2021
Published 24 November 2021 Volume 2021:14 Pages 6175—6190
DOI https://doi.org/10.2147/JIR.S337031
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Hui-Ling Chen,1,2,* Yi-Bin Zeng,3,* Zheng-Yan Zhang,1,2,* Chao-Yue Kong,1,2 Shi-Long Zhang,1,2 Zhan-Ming Li,1,2 Jia-Ting Huang,1,2 Ya-Yun Xu,1,2 Yu-Qin Mao,1,2 Pei-Ran Cai,1,2 Bing Han,1,2 Wu-Qing Wang,3 Li-Shun Wang1,2
1Center for Traditional Chinese Medicine and Gut Microbiota, Minhang Hospital, Fudan University, Shanghai, 201100, People’s Republic of China; 2Institute of Fudan-Minhang Academic Health System, Minhang Hospital, Fudan University, Shanghai, 201100, People’s Republic of China; 3Dermatological Department, Minhang Hospital, Fudan University, Shanghai, 201100, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Li-Shun Wang; Wu-Qing Wang
Center for Traditional Chinese Medicine and Gut Microbiota, Minhang Hospital, Fudan University, Shanghai, 201100, People’s Republic of China
Tel +86 21-64923400-6244
Email [email protected]; [email protected]
Background: Psoriasis is a chronic autoinflammatory skin disease, and its aetiology remains incompletely understood. Recently, gut microbial dysbiosis is found to be tightly associated with psoriasis.
Objective: We sought to reveal the causal role of gut microbiota dysbiosis in psoriasis pathogenesis and investigate the protective effect of healthy commensal bacteria against imiquimod -induced psoriasis-like skin response.
Methods: By using fecal microbial transplantation (FMT), 16S rRNA gene-based taxonomic profiling and Lactobacillus supplement, we have assessed the effect of FMT from healthy individuals on psoriasis-like skin inflammation and associated immune disorders in imiquimod-induced psoriasis mice.
Results: Here, by using psoriasis mice humanized with the stools from healthy donors and psoriasis patients, the imiquimod-induced psoriasis in mice with psoriasis patient stool was found to be significantly aggravated as compared to the mice with healthy donor stools. Further analysis showed fecal microbiota of healthy individuals protected against Treg/Th17 imbalance in psoriasis. Moreover, we found the gut and skin microbiome in mice receipted with gut microbiota of healthy individuals (HD) differed from those of mice receipted with gut microbiota of psoriasis patients (PSD). 16S rRNA sequencing revealed that Lactobacillus reuteri was greatly enriched in fecal and cutaneous microbiome of HD mice as compared to PSD mice. Intriguingly, supplement with Lactobacillus reuteri was sufficient to increase the expression of anti-inflammatory gene IL-10, reduce Th17 cells counts and confer resistance to imiquimod-induced inflammation on the mice with gut microbiota dysbiosis.
Conclusion: Our results suggested that the gut microbiota dysbiosis is the potential causal factor for psoriasis and the gut microbiota may serve as promising therapy target for psoriasis patients.
Keywords: gut microbiota, cutaneous microbiome, psoriasis, fecal microbiota transplantation, Lactobacillus reuteri, Th17
Introduction
Psoriasis is a T cell-mediated inflammatory skin disease, and affects 3% of the world’s population.1 The disorders of immune system are thought as the dominant cause for its pathogenesis.2 Genome-wide scans for psoriasis have identified its association with the mutations in predominant immune-related genes, which implicate the key role of genetics in psoriasis.3 However, the absence of complete concordance between monozygotic twins suggests a crucial role of environmental factors in this disease development.4 Besides, lifestyle also has the major implication in psoriasis.5–11 Overall, psoriasis is a multifactorial disease with an intimate interplay among the immune systems, genetic susceptibility, lifestyle and environment.2
Recently, several studies have been conducted on the correlation of gut microbiota dysbiosis with psoriasis.12–18 Cohort studies show a lower bacterial diversity and differences in β-diversity in the gut microbiome of psoriasis patients.12–14 The richness in gut microbiota taxa is significantly reduced in psoriasis patients, especially in psoriatic arthritis patients as compared to healthy controls.13 A lowering of the Bacteroidetes/Firmicutes ratio was observed in the gut microbiota of psoriasis patients as compared to the healthy ones.14 Besides, the reduced presence of several taxa is associated with the responses to anti-psoriasis treatment.19 In addition, the connection of the skin microbiome alterations and psoriasis20,21 focused the potential role of gut microbiota dysbiosis on the skin aberrant inflammatory response.
However, even that these recent studies suggested a disrupted gut microbiota composition in psoriasis, causative analysis is still absent. Here, we used a mouse model humanized with the stools from human donors to reveal the causal role of gut microbiota dysbiosis in psoriasis pathogenesis and investigate the protective effect of healthy commensal bacteria against imiquimod induced psoriasis-like skin response. 16S rRNA sequencing revealed that Lactobacillus reuteri was greatly enriched in fecal and cutaneous microbiome of HD mice as compared to PSD mice. Furthermore, we monocolonized mice with five well-known probiotic Lactobacillus species and their mixture to explore their protective effect of psoriasis.
Materials and Methods
Participants
A total of 16 psoriasis patients and 22 sex- and age-matched healthy controls were recruited from the medical staff at the Minhang Hospital. The information of all participants was summarized in the Supplementary Table S1. The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Minhang Hospital, Fudan University and informed consent was obtained from all participants.
Imiquimod-Induced Psoriasis-Like Skin Inflammation Model
Three to 4 weeks Balb/c female mice (Beijing Vital River Laboratory Animal Technology Co., Ltd.) were bred and maintained in plastic flexible film gnotobiotic isolators under a strict 12 h light cycle and fed autoclaved water and diet at the Centralab of Minhang Hospital, Fudan University. To establish a psoriasis-like skin inflammation model, the mice were treated with a daily topical dose of 62.5 mg of imiquimod cream (5%) (Sichuan Medshine Pharmaceutical) on the shaved back and on the left ear (5 mg) for 6 consecutive days. The severity of erythema and scaling was monitored daily, using a scale based on the Psoriasis Area and Severity Index (PASI).22 Ear swelling and skin thickening were measured at the end of the experiment, as previously described.16 On day 8, All mice were anaesthetized with ketamine (200 mg/kg) and xylazine (10 mg/kg), and blood was collected by cardiac puncture. Histopathological examinations were performed in 4 μm sections stained with HE. The scoring system describing the degree of imiquimod-induced skin inflammation on a scale of 0–2.17
FMT Experiments
Fresh stools from each participant were immediately suspended in an equal volume (w/v) of PBS containing 20% glycerol in PBS, snap-frozen in liquid nitrogen, and stored in −80°C refrigerator until use. FMT was performed according to the modified method described previously.18,23,24 Briefly, mice were received with a broad-spectrum cocktail of antibiotics containing vancomycin (100 mg/kg), neomycin sulfate (200 mg/kg), metronidazole (200 mg/kg), and ampicillin (200 mg/kg) once a day for 7 days by oral gavage to remove indigenous gut microbiota. Then, stool samples for human were thawed and suspended in an equal volume of PBS, vortexed and the supernatant was centrifuged. 200 µL of the fecal suspension was administered to mice by oral gavage three times during the following two weeks. All mice from experimental groups received FMT from either healthy or psoriasis donors, with each donor sample delivered to 10 mice. One day after the last FMT, mice were challenged with imiquimod. All animal experiments were approved by the Ethical Committee of Minhang Hospital, Fudan University.
Lactobacillus Administration
The commercially available bacterial strains, including LA: Lactobacillus acidophilus, LP: Lactobacillus plantarum, LR: Lactobacillus reuteri, LRH: Lactobacillus rhamnosus, LC: Lactobacillus casei, MIX: the union of the five different species Lactobacillus were gained from Yaodan Biotechnology Co., Ltd (Shanghai, China). Each mouse was treated with twice a week 1×109 CFU for 6 weeks by oral gavage before imiquimod challenge.
Gut Microbiota Analysis
Total DNA in fecal was isolated using the DNA extraction kit (TIANGEN, China) according to the manufacturer’s instructions. For analysis of the taxonomic composition of the microbiota, the hypervariable regions V3-V4 region of 16S rRNA genes was amplified and sequenced at the Illumina MiSeq 2500 platform (Illumina MiSeq, USA). Reads were trimmed and classified using QIIME (V 1.8.0). After quality filtering and chimera removal, clean sequences with 97% similarity were phylogenetically sorted to the same operational taxonomic units (OTUs). The phylogenetic affiliation of each 16S rRNA gene sequence was assigned using RDP classifier. Gut microbiota α- and β-diversity, were analyzed according to OTU information using QIIME25 and displayed with R software R V.3.5.0 (under RStudio V.1.1.453). Linear discriminant analysis effect size (LEfSe) was performed to determine the features most likely to explain the differences between groups.26 The co-occurrence networks were generated in Gephi software (version 0.9.2).
Flow Cytometry
Spleen were mechanically dissociated to yield a single-cell suspension and were treated with ammonium chloride–potassium buffer for red blood cell lysis. After isolation, cell suspensions were passed through 40μm cell strainers (BD Pharmingen) and cell populations were characterized by flow cytometry. The cells were then blocked with Fc-block (BD Biosciences) and stained with anti-mouse antibodies according to the manufacturer’s instructions. Stained samples were analyzed on Cyan ADP 9 colors cytometer (Beckman Coulter) and analyses were performed with FlowJo software version 10. For intracellular staining of produced cytokines, cells were first stimulated with 1 μL PMA/Ionomycin Mixture (250×) and 1 μL BFA/Monensin Mixture (250×). With mononuclear cells only as control. Mix wells were incubated in CO2 incubator at 37°C for 4–6 hours and vortexed every 1–2 hours during incubation, according to the manufacturer’s instructions. Data were acquired using LSRII (BD Bioscience) and analyzed by FlowJo software v9.6.2. (Tree Star, Inc., Ashland, OR).
RT-qPCR
Total RNA was extracted from skin samples using TRIzol reagent (Invitrogen) and a Microplate Readers (Cytation 5, Biotek) was used for RNA quality control. mRNA was reverse-transcribed with the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa Biotech Co.) and each test consumed 1 μg of total RNA according to the manufacturer’s instructions. qPCR was carried out with the SYBR Premix Ex Taq II (TliRNaseH Plus) (TaKaRa Biotech Co.) using a QuantStudio™ 7 Flex Real-Time PCR System (ThermoFisher). Primers were purchased from Sangon Biotech. The used primers were listed in Supplementary Table S2. Actin was used as the internal control and the relative gene expression was determined compared with the expression of internal control.
Statistical Analysis
According to the different data, statistical analysis between groups were analysed using the Wilcoxon’s rank-sum test, Student’s t-test or one –way analysis of variance. Correlation analyses were performed based on spearman’s rho statistic. Rate comparisons were performed with Pearson’s x2 test or Fisher’s exact test. The data are presented as mean ± standard deviation (SD). The tests for statistical comparisons were clarified in related legends. P < 0.05 was considered to be statistically significant. Statistical analyses and data visualization were performed using R V.3.5.0 (under RStudio V.1.1.453) and GraphPad prism 8.
Result
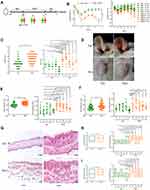
Dysbiosis of Gut Microbiota in Psoriasis Patients
A total of 22 healthy control (HD) and 16 psoriasis patients (PsO) were included in the present study (Supplementary Table S1). Alpha-diversity indices, including Shannon index, ACE, Chao1 and Observed species, were markedly decreased in the PsO group (Figure 1A). According to PCoA, the gut microbiome of the PsO group differed significantly from that of the healthy group using the unweighted UniFrac distance (Figure 1B). Next, we examined the relative abundance of the taxa that constitute the multivariate distribution of the gut microbiota. Skyline plots demonstrated generally similar compositions of gut microbiota in terms of phylum level relative abundance (Figure 1C) and heatmap showed the compositions of gut microbiota of family level relative abundance (Figure 1D). We observed that two phyla (Firmicutes and Bacteroidota) and four families (Bacteroidaceae, Lachnospiraceae, Prevotellaceae and Ruminococcaceae) dominate the gut microbiota communities in both HD and PsO (Figure 1C and D). Many taxa were found to be associated with psoriasis status, despite high variance in each group (Figure 1E). These results indicated the serious dysbiosis of gut microbiota in psoriasis patients.
Fecal Microbiota Transplantation from Healthy Individuals Protected Against Psoriasis-Like Inflammation in Mice
To further test whether gut microbiota from healthy donors could protect against the progression of psoriasis, human feces from five healthy donors (HD) and five psoriasis donors (PSD) were colonized in psoriasis mice, respectively (Figure 2A). The information of the ten donors was listed in Supplementary Table S3. Following application of imiquimod, the body weights in PSO mice were significantly lower than HD mice from the second day to the 7th day (Figure 2B). The psoriatic skin inflammation according to the psoriasis area and severity index (PASI) scores was significantly increased in PSD group (P < 0.001) (Figure 2C). Specifically, as compared to HD mice, PSD mice displayed more severe erythema and scaling (Figure 2D), more severe ear thickening (Figure 2E) and skin thickening (Figure 2F) and more severe hyperkeratosis, acanthosis and leukocyte infiltration into the dermis (Figure 2G–I). Of note, in the PSD group, all five individual humanized mice resulted in the exacerbation of skin lesions (Figure 2B and C, E and F, H and I), while in the HD group, two individual humanized mice (HD2 and HD5, especially HD 5), almost completely abrogated skin lesion progression (Figure 2B and C, E and F, H and I). Besides, there were intra-group variations among the HD mice and PSD mice, indicating the outcome of anti-psoriatic skin inflammation in the context of gut microbiota might be donor-dependent (Figure 2B and C, E and F, H and I). Taken together, these data demonstrated that the gut microbiota of psoriasis patients were functionally involved in skin lesions of psoriasis while these microbiota from healthy individuals harbored dramatically anti-psoriatic skin inflammation effect.
Fecal Microbiota Transplantation from Healthy Individuals Beneficially Modulated the Treg/Th17 Balance in Psoriasis Mice
The imbalance of Treg/Th17 is known to play an important role in psoriasis pathogenesis.27 To determine whether the gut microbiota contributes to the imbalance of Treg/Th17, spleen cells of these mice were analyzed for the percentages and counts of Th17 (Figure 3A and B) and Treg (Figure 3C and D) cells by flow cytometry. We found that PSD mice had significantly higher frequencies and counts of Th17 cells (Figure 3A and B). In contrast, significantly lower frequencies and counts of Th17 cells were detected in HD mice, which was in accordance with the attenuated skin inflammation (Figure 3A and B). The frequencies and counts of Treg cells in the spleen were not significantly changed by the FMT (Figure 3C and D). However, the Treg/Th17 cell ratios were lower in PSD mice than in HD mice (Figure 3E). PSD mice exhibited significantly higher mRNA level of cytokine and chemokine, including IL17A, IL1b, IL23, IL6, Cxcl2, S100A8, S100A9 and CCL2 in skin lesion (Figure 3F and G) than HD mice, which indicated an augmented inflammation in the skin of PSD mice. Importantly, we found that the transcript levels of IL10 in the skin lesion were significantly increased in the HD mice (Figure 3H), which was reported as a well-known anti-inflammatory factor.28 Collectively, these data indicated that the fecal microbiota from psoriasis patients disturbed the skin and systemic immunological response, while healthy microbiota demonstrated a beneficial modulation on it.
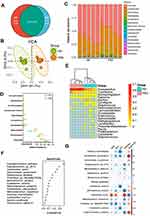
Analysis of Fecal Samples Revealed Taxonomic Signatures After FMT
Next, we examined the OTUs abundance in the human donor samples, murine fecal samples pre-FMT (basal condition after antibiotic treatment) and post-FMT at the end point. Interrogation of bacterial origin showed that 41% of murine gut microbiome in recipient mice post-FMT were found as human donor origin bacteria (Figure 4A). In addition, alpha-diversity indices were markedly decreased in the mice that received psoriasis patient feces as compared to the mice gavaged with healthy individual feces (Figure 4B). According to the no-contaminant filtered data, PCoA revealed the significant separation of two clusters of PSD and HD groups (Figure 4C). Additionally, fecal samples of the two groups showed a different taxonomic signature (Figure 4D and E). HD group showed a predominance of Alistipes and unidentified Clostridiales (P < 0.05) at genus level. In contrast, Klebsiella, Phascolarctobacterium, Lachnoclostridium and Bilophila (P < 0.05) were predominant in the intestine of PSD group (Figure 4E). Furthermore, we then assessed at a lower taxonomic level, and the top 10 abundance genus were shown in each individual group (Figure 4F). In addition, a significant increase of Bacteroides/Lactobacillus ratio was observed in PSD group (Figure 4G). These findings suggested that gut microbiota of healthy donors and psoriasis patients possessed different taxonomic signatures.
Fecal Microbiota Transplantation from Healthy Individuals Created a Gut Microbial Community with High Abundance of Lactobacillus reuteri in Mice
Then, we constructed a co-abundance network to illustrate the potential interactions among the 50 OTUs that were significantly different between the HD and PSD mice. The genus Lactobacillus and Akkermansia were significantly enriched (Figure 5A). To identify functionally relevant bacterial taxa responsible for anti-psoriatic skin inflammation, the correlation between the most important genera in fecal samples and clinical manifestations was analyzed by Spearman’s rank correlation analysis (Figure 5B). Notably, negative correlations were observed between Lactobacillus and most clinical parameters (Figure 5B). The correlation coefficients of Akkermansia were just opposite (Figure 5B). Subsequently, the mice in the PSD were dominated by Bacteroidaceae at the family level, Proteobacteria at phylum level. In contrast, HD group showed a predominance of Alistipes at the genus level, and Lactobacillus reuteri at the species level (Figure 5C). To determine the microbiota variables which most influenced the protective effect of HD, multivariate tests including RandomFroest (Figure 5D) and Simper analysis (Figure 5E) were carried out. Then, we found that Lactobacillus reuteri was the most contributed taxa. Collectively, we hypothesized that member of Lactobacillus might represent a major component underlying the improved anti-inflammation effects of gut microbiota.
The Skin Microbiota in HD and PSD are Highly Distinct
Studies of the skin microbiota of psoriasis patients showed that skin dysbiosis contributed to the development of inflammatory skin diseases, as their gut dysbiosis.29–31 We next asked whether the skin microbiome contributed to the protective effect of HD. Skin tissue was collected from the psoriasis mice of both HD and PSD. An overall analysis of skin microbiota indicated clear differences between HD and PSD (Figure 6A and B). The skin OTUs were markedly decreased in PSD mice as compared to HD mice (Figure 6A). PCA revealed the significant separation of two clusters of PSD and HD groups (Figure 6B). We observed that the Firmicutes dominate the skin microbiota communities in HD and Proteobacteria dominate the skin microbiota communities in PSD (Figure 6C and D). In addition, heatmap showing that the skin microbiota in HD and PSD mice were quite different at the genus level relative abundance (Figure 6E). RandomFroest analysis indicated Lactobacillus reuteri was dominated in HD group and contributed to the protective effect (Figure 6F). Notably, negative correlations were observed between Lactobacillus reuteri and most clinical parameters (Figure 6G).
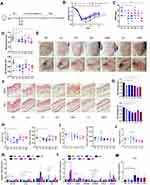
Lactobacillus reuteri Alleviated the Skin Inflammation by Reducing Th17 Cells
As mentioned above, the abundance of Lactobacillus, especially Lactobacillus reuteri was correlated with the alleviated skin inflammation in HD mice. Next, we evaluated if this effect would be verified by Lactobacillus supplement. We used the imiquimod-induced psoriasis model to evaluate the anti-inflammatory properties of these HD-enriched Lactobacillus, including Lactobacillus acidophilus (LA), Lactobacillus plantarum (LP), Lactobacillus reuteri (LR), Lactobacillus rhamnosus (LRH), Lactobacillus casei (LC) and their mixture (MIX). Following antibiotics treatment, the recipient mice were randomly divided into 7 groups and orally inoculated with prepared Lactobacillus twice a week for 6 consecutive weeks and the mice were sacrificed at day 7 (Figure 7A). As shown in Figure 7B, compared with the mice who were gavaged with saline only (PS), Lactobacillus supplement groups showed less weight loss when challenged with imiquimod (Figure 7B). In LR receiving mice group, the PASI score (Figure 7C), skin and ear thickness (Figure 7D), the psoriasis mice skin appearance (Figure 7E) and skin inflammation (Figure 7G) were considerably alleviated. Instead, other Lactobacillus, including LA, LP, LRH, LC and MIX could alleviate these parameters partially (Figure 7C–G). Then, Th17 cells was detected for each mouse treated with Lactobacillus. We observed a significant reduction of Th17 cells (Figure 7H), but no change of Treg cells (Figure 7I) in LR group compared with those in the PS group. Surprisingly, we did not find a reduction of the Treg/Th17 cell ratios (Figure 7C) were observed in these groups. As shown in Figures 7K and L, qPCR analysis revealed significantly decreased gene expression of IL1b, IL17, IL6, CXCL2 and S100A9, but not IL23, CXCL1, S100A8, CCl2 and CCl20 in LR mice compared to PS mice. Besides, the MIX treatment resulted in significantly decreased gene expression of IL-17 and IL-1b in skin (Figure 7K). Notably, the transcript level of IL10 in the skin lesion was increased in the Lactobacillus reuteri recipient mice (Figure 7M). Collectively, Lactobacillus, especially L.reuteri, might partially contribute to the alleviation of imiquimod induced psoriasis-like skin inflammation via the reduction of Th17 cell and inflammatory cytokines.
Discussion
Psoriasis is a systemic inflammatory disorder that involves the innate and adaptive immune system.2 The prediction, prevention and treatment of the psoriasis are still insufficient due to unclear aetiology of this disease.32 Several studies have revealed the tight correlation of gut microbiota dysbiosis with psoriasis.12–18 However, it is still unclear whether the dysbiosis should be considered as a driving step in the pathogenesis. In this study, the IMO-induced psoriasis like-inflammation was found to be aggravated by the administration of stool samples from psoriasis patients in comparison to samples from healthy donors. This indicates a potential role of the dysbiotic gut microbiota in worsening psoriasis symptoms, whereas protective effects of the healthy gut microbiome could be expected. To date, the humanization mouse by gavage the gut microbiota from the donor was commonly used to explore the role of gut microbiota on disease,33–35 and we followed this method. Interrogation of bacterial origin showed that the colonized gut microbiome of the mice is 41% of donor origin, after FMT, which was consistent with the previous report.36
Excessive skin inflammation is commonly involved in psoriasis pathogenesis by an imbalance of Treg/Th17.37–39 Gastrointestinal bacteria were reported to regulate the immune response by driving the differentiation of naive T cells into either Tregs or Th17 cells.40 In addition, the increased inflammatory cytokines of IL17, IL1b, CXCL2, IL23 are well known to be involved in the development of psoriasis.41,42 In the current study, we found that the PSD mice receiving feces from patients with psoriasis exhibited an imbalance of Treg and Th17 cells, increased inflammatory cytokines, which were tightly correlated with a severe hyperkeratosis, acanthosis and leukocyte infiltration into the dermis.
So far, great interest was posed on the gut microbiota dysbiosis of psoriasis. Exogenous factors, including drugs, antibiotics and diet, which influence the gut microbiota, were reported to be associated with the psoriasis skin inflammation.5–9,16–18,43,44 It has been reported that the germ-free, broad-spectrum antibiotic-treated or metronidazole-treated alone mice were more resistant to psoriasis than the conventional mice in adults,17,18 whereas mice treated neonatally with these antibiotics develop exacerbated psoriasis.16 Dietary habits can impact the gut microbiota and strongly influence the psoriasis skin inflammation.5–9 These findings indicate that a correction of the gut microbiome dysbiosis in patients with psoriasis would mitigate the disease symptoms. Therefore, FMT from healthy individuals served as an efficient and safe way to reconstitution of the gut microbiota, with great potential of clinical implications in psoriasis. FMT has shown efficacy in treating recurrent Clostridium difficile infection45–48 and is increasingly being applied to other gastrointestinal disorders.48–54 To date, one case report for psoriasis FMT has been documented.55
It was reported that the skin microbiota from psoriatic lesions were similar to those of unaffected or healthy skin, which were compositions of Corynebacterium, Propionibacterium, Staphylococcus and Streptococcus.20 Although some studies have suggested a potential contribution of the skin microbiome of psoriasis,29–31 the interaction of the microbiota between gut and skin contributes to the development of inflammation in psoriasis has not yet been shown. Here, we found the skin microbiota in HD and PSD are highly distinct. We observed that the Firmicutes dominate the skin microbiota communities in HD and Proteobacteria dominate the skin microbiota communities in PSD. Notably, the skin microbiome featuring abundance of Lactobacillus reuteri in HD mice associated with the protective effect. Overall, our study showed that the gut microbiota for healthy donor associated with reshaping the skin microbiome and contributed to protect the psoriasis-like skin inflammation in HD mice. Our results indicated that the “gut-skin” axis based on bacteria could be a potential clue in understanding the aetiology of psoriasis.
In addition, supplement containing a probiotic mixture including Lactobacillus was shown to be beneficial in the alleviation of psoriasis symptoms.56,57 The ability of Lactobacillus to suppress the IL23/Th17 axis may mediate this immune-modulatory effect,58 which is tightly associated with the pathogenesis of psoriasis. Intriguingly, we found that Lactobacillus supplement, especially L. reuteri supplement could partially alleviate the psoriasis skin inflammation outcome by reducing the numbers of Th17 cells. L. reuteri was reported exclusively able to modulate reduced Th1/Th17 cells and their associated cytokines IFNγ/IL-17 in autoimmune encephalomyelitis mice model.59 Recently, imiquimod-induced psoriasis was reported to cause severe DSS (dextran sulfate sodium) colitis, the decreased L. reuteri under psoriasis do not contribute to the DSS colitis.60 However, the potential effect of L. reuteri supplement on psoriasis was not mentioned. Our data showed that the relative abundance of Lactobacillus was decreased in mice colonized by feces from patients with psoriasis. We observed moreover that the administration of certain Lactobacillus members could protect against the psoriasis-like induced inflammation and reduce of Th17 cells count.
L. reuteri is a well-known probiotic and has been reported to inhibit Th17-mediated inflammatory responses and enhance the immunoregulatory cells-mediated immunosuppression through the increase of anti-inflammatory cytokines IL10.61–63 IL10 has a potent immunomodulatory capacity and is known to play an important role in psoriasis.64–67 Lower cutaneous IL-10 mRNA expression in psoriasis compared with other inflammatory dermatoses was observed and IL10 administration is a successful antipsoriatic therapy.64,67 In fact, imiquimod-applied IL10 deficient mice showed more persistent psoriasis-like inflammation and higher severity index.68 Consist to the previous study, we found the m RNA expression level of IL10 in the skin was higher in the L. reuteri receipted mice, indicated that the Lactobacillus reuteri administration increase the expression level of IL10 in the skin may partly be the mechanism of the gut microbiome on alleviating psoriasis-like mouse model and decreasing the number of Th17 cells.
Conclusion
Our results suggested the gut microbiota dysbiosis is the potential causal factor for psoriasis and the gut microbiota may serve as promising therapy target for psoriasis patients.
Data Sharing Statement
All the data supporting the findings of this study are available within the article and its Supplementary Materials.
Ethics Approval and Consent to Participate
Animal care and experiments were carried out in strict accordance with the Animal Care and Use Guide and approved by the ethics committee of Fudan University.
Funding
This work was supported by grants from National Natural Science Foundation of China (81872245 and 81803601), Research Project of Shanghai Municipal Health Commission (20214Y0326), Open Research Fund of State Key Laboratory of Genetic Enigineering, Fudan Univeristy (SKLGE-2112), Natural Science Research Project of Shanghai Minhang District (No.2020MHZ090 and No.2021MHZ086) and High-level Backbone Physicians Training Programme of Minhang District (2020MZYS18).
Disclosure
The authors declare no conflicts of interest.
References
1. Parisi R, Symmons DPM, Griffiths CEM, et al. Global epidemiology of psoriasis: a systematic review of incidence and prevalence. J Invest Dermatol. 2013;133(2):377–385. doi:10.1038/jid.2012.339
2. Boehncke WH, Schon MP. Psoriasis. Lancet. 2015;386(9997):983–994. doi:10.1016/S0140-6736(14)61909-7
3. Elder JT, Bruce AT, Gudjonsson JE, et al. Molecular dissection of psoriasis: integrating genetics and biology. J Invest Dermatol. 2010;130(5):1213–1226. doi:10.1038/jid.2009.319
4. Perera GK, Di Meglio P, Nestle FO. Psoriasis. Annu Rev Pathol. 2012;7(1):385–422. doi:10.1146/annurev-pathol-011811-132448
5. Jensen P, Christensen R, Zachariae C, et al. Long-term effects of weight reduction on the severity of psoriasis in a cohort derived from a randomized trial: a prospective observational follow-up study. Am J Clin Nutr. 2016;104(2):259–265. doi:10.3945/ajcn.115.125849
6. Nakamizo S, Honda T, Adachi A, et al. High fat diet exacerbates murine psoriatic dermatitis by increasing the number of IL-17-producing gammadelta T cells. Sci Rep. 2017;7(1):14076. doi:10.1038/s41598-017-14292-1
7. Herbert D, Franz S, Popkova Y, et al. High-fat diet exacerbates early psoriatic skin inflammation independent of obesity: saturated fatty acids as key players. J Invest Dermatol. 2018;138(9):1999–2009. doi:10.1016/j.jid.2018.03.1522
8. Castaldo GG, Aufiero FR, Cereda E. Very low-calorie ketogenic diet may allow restoring response to systemic therapy in relapsing plaque psoriasis. Obes Res Clin Pract. 2016;5:348–352.
9. Damiani G, Watad A, Bridgewood C, et al. The impact of Ramadan fasting on the reduction of PASI score, in moderate-to-severe psoriatic patients: a real-life multicenter study. Nutrients. 2019;11(2):277. doi:10.3390/nu11020277
10. Armstrong AW, Harskamp CT, Dhillon JS, Armstrong EJ. Psoriasis and smoking: a systematic review and meta-analysis. Br J Dermatol. 2014;170(2):304–314. doi:10.1111/bjd.12670
11. Brenaut E, Horreau C, Pouplard C, et al. Alcohol consumption and psoriasis: a systematic literature review. J Eur Acad Dermatol Venereol. 2013;27(Suppl 3):30–35. doi:10.1111/jdv.12164
12. Chen YJ, Ho HJ, Tseng CH, Lai ZL, Shieh JJ, Wu CY. Intestinal microbiota profiling and predicted metabolic dysregulation in psoriasis patients. Exp Dermatol. 2018;27(12):1336–1343. doi:10.1111/exd.13786
13. Scher JU, Ubeda C, Artacho A, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67(1):128–139. doi:10.1002/art.38892
14. Hidalgo-Cantabrana C, Gómez J, Delgado S, et al. Gut microbiota dysbiosis in a cohort of patients with psoriasis. Br J Dermatol. 2019;181(6):1287–1295. doi:10.1111/bjd.17931
15. Sikora M, Stec A, Chrabaszcz M, et al. Gut microbiome in psoriasis: an updated review. Pathogens. 2020;9(6):463. doi:10.3390/pathogens9060463
16. Zanvit P, Konkel JE, Jiao X, et al. Antibiotics in neonatal life increase murine susceptibility to experimental psoriasis. Nat Commun. 2015;6(1):8424. doi:10.1038/ncomms9424
17. Zakostelska Z, Málková J, Klimešová K, et al. Intestinal microbiota promotes psoriasis-like skin inflammation by enhancing Th17 response. PLoS One. 2016;11(7):e0159539. doi:10.1371/journal.pone.0159539
18. Stehlikova Z, Kostovcikova K, Kverka M, et al. Crucial role of microbiota in experimental psoriasis revealed by a gnotobiotic mouse model. Front Microbiol. 2019;10:236. doi:10.3389/fmicb.2019.00236
19. Yeh NL, Hsu CY, Tsai TF, Chiu HY. Gut microbiome in psoriasis is perturbed differently during secukinumab and ustekinumab therapy and associated with response to treatment. Clin Drug Investig. 2019;39(12):1195–1203. doi:10.1007/s40261-019-00849-7
20. Alekseyenko AV, Perez-Perez GI, De Souza A, et al. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;1(1):31. doi:10.1186/2049-2618-1-31
21. Chang HW, Yan D, Singh R, et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome. 2018;6(1):154. doi:10.1186/s40168-018-0533-1
22. Wu R, Zeng J, Yuan J, et al. MicroRNA-210 overexpression promotes psoriasis-like inflammation by inducing Th1 and Th17 cell differentiation. J Clin Invest. 2018;128(6):2551–2568. doi:10.1172/JCI97426
23. Benakis C, Brea D, Caballero S, et al. Commensal microbiota affects ischemic stroke outcome by regulating intestinal gammadelta T cells. Nat Med. 2016;22(5):516–523. doi:10.1038/nm.4068
24. Tang TWH, Chen H-C, Chen C-Y, et al. Loss of gut microbiota alters immune system composition and cripples postinfarction cardiac repair. Circulation. 2019;139(5):647–659. doi:10.1161/CIRCULATIONAHA.118.035235
25. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi:10.1038/nmeth.f.303
26. Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi:10.1186/gb-2011-12-6-r60
27. Diani M, Altomare G, Reali E. T helper cell subsets in clinical manifestations of psoriasis. J Immunol Res. 2016;2016:7692024. doi:10.1155/2016/7692024
28. Ouyang W, O’Garra A. IL-10 family cytokines IL-10 and IL-22: from basic science to clinical translation. Immunity. 2019;50:871–891.
29. Belkaid Y, Tamoutounour S. The influence of skin microorganisms on cutaneous immunity. Nat Rev Immunol. 2016;16(6):353–366. doi:10.1038/nri.2016.48
30. Fyhrquist N, Muirhead G, Prast-Nielsen S, et al. Microbe-host interplay in atopic dermatitis and psoriasis. Nat Commun. 2019;10(1):4703. doi:10.1038/s41467-019-12253-y
31. Langan EA, Griffiths CEM, Solbach W, Knobloch JK, Zillikens D, Thaci D. The role of the microbiome in psoriasis: moving from disease description to treatment selection? Br J Dermatol. 2018;178(5):1020–1027. doi:10.1111/bjd.16081
32. Armstrong AW, Read C. Pathophysiology, clinical presentation, and treatment of psoriasis: a review. JAMA. 2020;323(19):1945–1960. doi:10.1001/jama.2020.4006
33. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi:10.1126/science.aan3706
34. Feehley T, Plunkett CH, Bao R, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med. 2019;25(3):448–453. doi:10.1038/s41591-018-0324-z
35. Chen X, Li P, Liu M, et al. Gut dysbiosis induces the development of pre-eclampsia through bacterial translocation. Gut. 2020;69(3):513–522. doi:10.1136/gutjnl-2019-319101
36. Riquelme E, Zhang Y, Zhang L, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795–806 e712. doi:10.1016/j.cell.2019.07.008
37. Lochner M, Wang Z, Sparwasser T. The special relationship in the development and function of T helper 17 and regulatory T cells. Prog Mol Biol Transl Sci. 2015;136:99–129.
38. Sulaimani J, et al. Dimethyl fumarate modulates the Treg-Th17 cell axis in patients with psoriasis. Br J Dermatol. 2020;184(3):495–503.
39. Alzahrani KS, Nadeem A, Ahmad SF, et al. Inhibition of spleen tyrosine kinase attenuates psoriasis-like inflammation in mice through blockade of dendritic cell-Th17 inflammation axis. Biomed Pharmacother. 2019;111:347–358. doi:10.1016/j.biopha.2018.12.060
40. Omenetti S, Pizarro TT. The Treg/Th17 axis: a dynamic balance regulated by the gut microbiome. Front Immunol. 2015;6:639. doi:10.3389/fimmu.2015.00639
41. Muromoto R, Hirao T, Tawa K, et al. IL-17A plays a central role in the expression of psoriasis signature genes through the induction of IkappaB-zeta in keratinocytes. Int Immunol. 2016;28(9):443–452. doi:10.1093/intimm/dxw011
42. Nograles KE, Zaba LC, Guttman‐Yassky E, et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol. 2008;159:1092–1102.
43. Ramirez-Bosca A, Navarro-López V, Martínez-Andrés A, et al. Identification of bacterial DNA in the peripheral blood of patients with active psoriasis. JAMA Dermatol. 2015;151(6):670–671. doi:10.1001/jamadermatol.2014.5585
44. Drago F, Ciccarese G, Indemini E, Savarino V, Parodi A. Psoriasis and small intestine bacterial overgrowth. Int J Dermatol. 2018;57(1):112–113. doi:10.1111/ijd.13797
45. Debast SB, Bauer MP, Kuijper EJ; European Society of Clinical M, Infectious D. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2014;20(Suppl 2):1–26. doi:10.1111/1469-0691.12418
46. McDonald LC, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018;66(7):987–994. doi:10.1093/cid/ciy149
47. Quraishi MN, Widlak M, Bhala N, et al. Systematic review with meta-analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory Clostridium difficile infection. Aliment Pharmacol Ther. 2017;46(5):479–493. doi:10.1111/apt.14201
48. Moayyedi P, Surette MG, Kim PT, et al. Fecal microbiota transplantation induces remission in patients with active Ulcerative Colitis in a randomized controlled trial. Gastroenterology. 2015;149(1):102–109 e106. doi:10.1053/j.gastro.2015.04.001
49. Reinshagen M, Stallmach A. [Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial]. Z Gastroenterol. 2017;55:779–780.
50. Johnsen PH, Hilpüsch F, Cavanagh JP, et al. Faecal microbiota transplantation versus placebo for moderate-to-severe irritable bowel syndrome: a double-blind, randomised, placebo-controlled, parallel-group, single-centre trial. Lancet Gastroenterol Hepatol. 2018;3(1):17–24. doi:10.1016/S2468-1253(17)30338-2
51. Bajaj JS, Kassam Z, Fagan A, et al. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: a randomized clinical trial. Hepatology. 2017;66(6):1727–1738. doi:10.1002/hep.29306
52. Engen PA, Zaferiou A, Rasmussen H, et al. Single-arm, non-randomized, time series, single-subject study of fecal microbiota transplantation in multiple sclerosis. Front Neurol. 2020;11:978. doi:10.3389/fneur.2020.00978
53. Kang DW, Adams JB, Gregory AC, et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. 2017;5(1):10. doi:10.1186/s40168-016-0225-7
54. Kootte RS, Levin E, Salojärvi J, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017;26(4):611–619 e616. doi:10.1016/j.cmet.2017.09.008
55. Yin G, Li JF, Sun YF, et al. [Fecal microbiota transplantation as a novel therapy for severe psoriasis]. Zhonghua Nei Ke Za Zhi. 2019;58:782–785. Chinese.
56. De Pessemier B, Grine L, Debaere M, Maes A, Paetzold B, Callewaert C. Gut-skin axis: current knowledge of the interrelationship between microbial dysbiosis and skin conditions. Microorganisms. 2021;9(2):353. doi:10.3390/microorganisms9020353
57. Navarro-Lopez V, Martínez-Andrés A, Ramírez-Boscá A, et al. Efficacy and safety of oral administration of a Mixture of probiotic strains in patients with psoriasis: a randomized controlled clinical trial. Acta Derm Venereol. 2019;99:1078–1084.
58. Xu M, Lu H, Lee Y-H, et al. An interleukin-25-mediated autoregulatory circuit in keratinocytes plays a pivotal role in psoriatic skin inflammation. Immunity. 2018;48(4):787–798 e784. doi:10.1016/j.immuni.2018.03.019
59. He B, Hoang TK, Tian X, et al. Lactobacillus reuteri reduces the severity of experimental autoimmune encephalomyelitis in mice by modulating gut microbiota. Front Immunol. 2019;10:385. doi:10.3389/fimmu.2019.00385
60. Kiyohara H, Sujino T, Teratani T, et al. Toll-like receptor 7 agonist-induced dermatitis causes severe Dextran Sulfate Sodium Colitis by altering the gut microbiome and immune cells. Cell Mol Gastroenterol Hepatol. 2019;7(1):135–156. doi:10.1016/j.jcmgh.2018.09.010
61. Alghetaa H, Mohammed A, Zhou J, Singh N, Nagarkatti M, Nagarkatti P. Resveratrol-mediated attenuation of superantigen-driven acute respiratory distress syndrome is mediated by microbiota in the lungs and gut. Pharmacol Res. 2021;167:105548. doi:10.1016/j.phrs.2021.105548
62. Mu Q, Zhang H, Liao X, et al. Control of lupus nephritis by changes of gut microbiota. Microbiome. 2017;5(1):73. doi:10.1186/s40168-017-0300-8
63. Hu R, Lin H, Wang M, et al. Lactobacillus reuteri-derived extracellular vesicles maintain intestinal immune homeostasis against lipopolysaccharide-induced inflammatory responses in broilers. J Anim Sci Biotechnol. 2021;12(1):25. doi:10.1186/s40104-020-00532-4
64. Al-Robaee AA, Al-Zolibani AA, Al-Shobili HA, Kazamel A, Settin A. IL-10 implications in psoriasis. Int J Health Sci. 2008;2:53–58.
65. Asadullah K, Sabat R, Friedrich M, Volk HD, Sterry W. Interleukin-10: an important immunoregulatory cytokine with major impact on psoriasis. Curr Drug Targets Inflamm Allergy. 2004;3(2):185–192. doi:10.2174/1568010043343886
66. Saxena A, Khosraviani S, Noel S, Mohan D, Donner T, Hamad AR. Interleukin-10 paradox: a potent immunoregulatory cytokine that has been difficult to harness for immunotherapy. Cytokine. 2015;74(1):27–34. doi:10.1016/j.cyto.2014.10.031
67. Asadullah K, Sterry W, Stephanek K, et al. IL-10 is a key cytokine in psoriasis. Proof of principle by IL-10 therapy: a new therapeutic approach. J Clin Invest. 1998;101(4):783–794. doi:10.1172/JCI1476
68. Jin SP, Koh SJ, Yu DA, et al. Imiquimod-applied Interleukin-10 deficient mice better reflects severe and persistent psoriasis with systemic inflammatory state. Exp Dermatol. 2018;27(1):43–49. doi:10.1111/exd.13403
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.