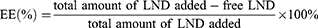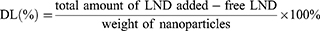Back to Journals » International Journal of Nanomedicine » Volume 18
Glutathione Programmed Mitochondria Targeted Delivery of Lonidamine for Effective Against Triple Negative Breast Cancer
Authors Wang Z, Qin Y, Wang X, Zhang T, Hu Y, Wang D, Zhang L , Zhu Y
Received 3 April 2023
Accepted for publication 11 July 2023
Published 24 July 2023 Volume 2023:18 Pages 4023—4042
DOI https://doi.org/10.2147/IJN.S413217
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Farooq A. Shiekh
Zhongjie Wang,1,* Yanru Qin,1,* Xueyuan Wang,2 Tianyu Zhang,1 Yixue Hu,2 Dongna Wang,1 Liefeng Zhang,1 Yongqiang Zhu1,2
1School of Food and Pharmaceutical Engineering, Nanjing Normal University, Nanjing, 210023, People’s Republic of China; 2College of Life Science, Nanjing Normal University, Nanjing, 210023, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liefeng Zhang; Yongqiang Zhu, Nanjing Normal University, Wenyuan Road 1#, Qixia District, Nanjing, 210023, People’s Republic of China, Tel/Fax +86-25-85898184 ; +86-25-85891591, Email [email protected]; [email protected]
Introduction: Mitochondria are a significant target of lonidamine (LND). However, its limited solubility and inability to specifically target mitochondria, LND can lead to hepatic toxicity and has shown only modest anticancer activity. The objective of this study is to establish a glutathione programmed mitochondria targeted delivery of LND for the effective treatment of triple negative breast cancer (TNBC).
Methods: In this study, LND was encapsulated in poly(lactic-co-glycolic acid) (PLGA) nanoparticles (NPs) wrapped with mitochondria-targeting short-chain triphenylphosphonium-tocopherol polyethylene glycol succinate (TPP-TPGS, TPS) and tumor-targeting long-chain 1, 2-distearoyl-sn-glycero-3-phosphoethanolamine-S-S-polyethylene glycol-R6RGD (DSPE-S-S-PEG2000-R6RGD, DSSR), which were designated as LND-PLGA/TPS/DSSR NPs. The release behavior was evaluated, and cellular uptake, in vitro and in vivo antitumor activity of nanoparticles were investigated. The mechanism, including apoptosis of tumor cells and mitochondrial damage and respiratory rate detection, was also further investigated.
Results: LND-PLGA/TPS/DSSR NPs were successfully prepared, and characterization revealed that they are globular particles with smooth surfaces and an average diameter of about 250 nm. Long-chain DSSR in LND-PLGA/TPS/DSSR NPs prevented positively charged LND-PLGA/TPS NPs from being cleared by the reticuloendothelial system. Furthermore, LND release rate from NPs at pH 8.0 was significantly higher than that at pH 7.4 and 5.5, which demonstrated specific LND release in mitochondria and prevented LND leakage in cytoplasm and lysosome. NPs could locate in mitochondria, and the released LND triggered apoptosis of tumor cells by damaging mitochondria and releasing apoptosis-related proteins. In addition, in TNBC mice model, programmed mitochondria targeted NPs improved efficacy and reduced LND toxicity.
Conclusion: LND-PLGA/TPS/DSSR NPs may be a useful system and provide an effective approach for the treatment of TNBC.
Keywords: alkaline-responsive drug release, GSH-responsive, programmed mitochondria targeting, triple negative breast cancer, lonidamine
Introduction
Triple negative breast cancer (TNBC), the most malignant subtype of breast cancer,1,2 lacks expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2),3 rendering current endocrine and anti-HER2 therapies ineffective.4,5 Chemotherapy remains the primary treatment method for TNBC. Lonidamine (LND), a small molecule chemotherapeutic drug, has shown promise in treating breast cancer by inducing apoptosis of tumor cells through depolarization of the mitochondrial membrane and release of cytochrome c.6 However, LND’s poor solubility and diffusion in the cytoplasm result in low bioavailability and severe side effects, such as hepatic toxicity.7–9 Therefore, it was necessary to design a novel drug delivery system to improve the efficacy of LND and reduce its toxicity.
Mitochondria play a vital role in regulating bioenergetics generation,10 cell proliferation,11 and mediating the intrinsic apoptotic pathway of tumor cells by releasing apoptotic proteins.12 The inner mitochondrial membrane (IMM) had a strong negative potential (ΔΨm) of −160~180 mV to support adenosine triphosphate synthesis.13 Delocalized lipophilic cations (DLCs) could effectively bind mitochondria through electrostatic and lipophilic interactions. Diquaternium (DQA) and triphenylphosphonium (TPP)14 were frequently used as DLC moieties and usually modified on the surface of nanoparticles to target mitochondria. While these mitochondrial targeting delivery systems may enhance therapeutic efficacy, several drawbacks need to be overcome. Firstly, positively charged nanoparticles could be recognized by the reticuloendothelial system in blood circulation and quickly cleared.15 Moreover, these strategies could not specifically target the mitochondria of cancer cells, and DLCs could also accumulate in normal cells.
To address these issues, dual-targeting nanoparticles were designed to actively target cancer cells and then target specific receptors of certain organs.16,17 Integrin ανβ3 was a crucial cell surface receptor specifically expressed in tumor-associated endothelial cells of various fast growing tumors.18,19 The most studied tripeptide of arginine-glycine-aspartate (RGD) could specifically bind integrin ανβ3.20 A smart core-shell nanostructure composed of RGD function shell and TPP core could effectively and specifically deliver drugs to tumor mitochondria and induce the mitochondria-associated apoptosis.21 However, the effectiveness of drugs must be released from the carrier, and the ability to release drugs at the target site is crucial for drug effectiveness.
To achieve tumor-specific drug release, researchers have utilized the unique characteristics of the tumor microenvironment, including the high concentration of glutathione (GSH), the acidic pH, hypoxia, and certain enzyme overexpression.22 Different concentrations of GSH were found in intracellular (2–10 mM) and extracellular (<20 μM) of tumor,23 with GSH expression in mitochondria high enough (10 mM) to reduce the disulfide bond.24,25 This was helpful in designing tumor-specific degradation nanoparticles by introducing the redox-responsive groups such as disulfide bond. Meanwhile, the pH value in tumor mitochondria was about 8.0,26 which is higher than that in normal tissue (pH 7.4) and lysosome (pH 5.5). The solubility of drugs with acidic groups is much higher in alkaline than in neutral and acidic pH environments,27 which can be used to design desirable nanoparticles with mitochondria-specific drug release to enhance antitumor efficacy.
In this study, we developed programmed targeting nanoparticles to deliver LND into the mitochondria and release LND within the mitochondria for the treatment of TNBC (Scheme 1). The nanoparticles were composed of PLGA loaded with LND as the core structure, wrapped with functional short-chain TPP-TPGS (TPS) for mitochondrial targeting and long-chain DSPE-S-S-PEG2000-R6RGD (DSSR) for tumor targeting. After nanoparticles entered tumor cells, disulfide bonds of DSSR were cleaved by GSH, and LND-PLGA/TPS NPs were exposed, which targeted alkaline mitochondria to release the acid LND. The properties of LND-loaded NPs, including particle size, polydispersity index (PDI), zeta potential, entrapment, and loading efficiencies, were investigated. In addition, the specific uptake of NPs in ανβ3-overexpressing MDA-MB-231 cell mitochondria was studied, and LND specific releasing from NPs in mitochondria microenvironment was further researched. Furthermore, we assessed cell viability and its mitochondria-associated apoptosis mechanism after treatment with NPs. Finally, we verified that our programmed LND-PLGA/TPS/DSSR NPs effectively inhibited tumor growth in tumor-xenograft mice model of TNBC MDA-MB-231 cells.
Materials and Methods
R6RGD peptide was synthesized by GenScript Co., Ltd. (Nanjing, China). Poly(lactic-co-glycolic acid) (PLGA, lactide/glycolide = 50/50, Mw = 30 kDa) was purchased from Daigang Biomaterial Co., Ltd. (Jinan, China). Amine-PEG-maleimide (NH2-PEG2000-MAL) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE) were purchased from Shanghai Ponsure Biotech, Inc. (Shanghai, China). Triphenylphosphine (TPP) and tocopherol polyethylene glycol succinate (TPGS) were purchased from Innochem Science & Technology Co., Ltd. (Beijing, China). 3, 3’-dithiodipropionic acid, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) were all obtained from Aladdin Industries Inc. (Shanghai, China). Lonidamine was purchased from Bide Pharmatech Ltd. (Shanghai, China). The Annexin V-FITC/PI cell apoptosis kit was purchased from KeyGen Biotech Co., Ltd. (Nanjing, China). Fluorescein isothiocyanate (FITC) was acquired from Sigma-Aldrich. Cell Counting Kit-8 (CCK-8), MitoTracker Red, JC-1 and 4’,6-diamidino-2-phenylindole (DAPI) were acquired from Shanghai Biyuntian Biotechnology Co., Ltd. (Shanghai, China). Fetal bovine serum (FBS), RPMI 1640 cell culture medium (incomplete) and trypsin (0.25%) were procured from Nanjing Senbeijia Biological Technology Co., Ltd. All other chemicals were reagent grade and used as received.
Synthesis of TPP-TPGS (TPS, Triphenylphosphonium-Tocopherol Polyethylene Glycol Succinate)
TPP (1 equiv) was refluxed with 6-bromohexanoic acid (1 equiv) dissolved in dry acetonitrile in a pressure tube for 16 h, and 5-carboxypetyltriphenylphosphonium was crystallized after being cooled, the 1g of which product was then 80 °C refluxed with 6 mL sulfoxide chloride for 6 h. After solvent removal, the product (1 equiv) was dissolved in methylene chloride (DCM) and reacted with TPGS (2 equiv) for 12 h. DCM was removed and followed by dialysis (Mw: 1000 Da) for 3 days in distilled water to remove unconjugated reactants. Afterward, the product was freeze-dried, and the chemical structures were confirmed by 1H NMR.
Synthesis of DSPE-S-S-PEG2000-R6RGD (DSSR, 1, 2-Distearoyl-Sn-Glycero-3-Phosphoethanolamine-S-S-Polyethylene Glycol-R6RGD)
Dithiodipropionic anhydride (DTDPA) was prepared as described in reference,28 In a brief, 1g 3,3’-dithiodipropionic (DTDP) was refluxing in 10 mL of acetyl chloride at 70 ◦C for 5 h. Then, acetyl chloride was removed by distillation at reduced pressure. After being kept at 4°C, the solution was precipitated by excess ice ethyl ether, and the pale yellow solid of DTDPA was harvested by suction filtration and dried in a vacuum oven. DTDPA (2 equiv) reacted with the amino group of DSPE (1 equiv) to form DSPE containing disulfide bond (DSPE-S-S-COOH). NH2-PEG2000-R6RGD was synthesized from NH2-PEG2000-MAL (1 equiv) and R6RGD (1 equiv) by Michael addition in phosphate buffer solution (PBS, pH = 8.0). DSPE-S-S-PEG2000-R6RGD was prepared by amide reaction between the carboxylation of DSPE-S-S-COOH and the amino of NH2-PEG2000-R6RGD. DSPE-S-S-COOH (2 equiv) was dissolved in chloroform and activated with EDC (4 equiv) and NHS (1 equiv) for 4 h, and then NH2-PEG2000-R6RGD was added and reacted for 24 h. Chloroform was removed and followed by dialysis (Mw: 2000 Da) for 3 days in distilled water to remove unconjugated reactants. Afterward, the product was freeze-dried, and the chemical structures were confirmed by1 H NMR.
Preparation of the Nanoparticles
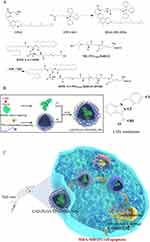
LND-loaded nanoparticles were prepared using oil-in-water (O/W) emulsion solvent evaporation.29 DSSR (4 mg) and TPS (4 mg) were added to 20 mL of water as the water phase, and TPGS (4 mg) was added to water as the emulsifier. PLGA (20 mg) and LND (8 mg) were dissolved in 1 mL acetone as the oil phase. Then, the oil phase was added dropwise to the water phase and the mixture was stirred at high speed for 4 h, the supernatant was discarded after centrifugation at 13,500 rpm for 30 min and washed twice with water to remove unused materials. LND-PLGA/TPS/DSSR NPs were obtained and suspended in water for direct use. LND-PLGA/TPS NPs and LND-PLGA/DSSR NPs were prepared with TPS or DSSR conjugate as the water phase, respectively. The FITC-loaded nanoparticles were prepared using the same process described above. The size and zeta potential of the nanoparticles were determined using dynamic light scattering. Transmission electron microscope (TEM) was used to observe the morphology of NPs. The entrapment efficiency (EE, %) and drug loading (DL, %) were calculated by the following equations, respectively.
Measurement of Glutathione and pH-Responsive Lonidamine Release of the Nanoparticles in vitro
The dialysis method was used to assess LND release from LND-PLGA/TPS/DSSR NPs. LND-PLGA/TPS/DSSR NP solution (1 mL) was placed in dialysis bag (MW = 3500 Da) and immersed in 20 mL of release medium (pH 5.5, pH 7.4, pH 8.0 PBS or pH 5.5 PBS+ 10 mM GSH, pH 8.0 PBS+ 5 mM GSH, pH 8.0 PBS+ 10 mM GSH). Then, 1 mL of release medium at different times was taken and replaced by 1 mL of the fresh release medium. UV/Vis spectrophotometer was used to measure the release content of LND in the medium, and then the concentration of LND was calculated according to the standard curve.
The Cell Culture Studies
MDA-MB-231 and MCF-7 cell lines were obtained from the National Center for Cell Science (NCCS). MDA-MB-231 cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS). MCF-7 cells were cultured in DMEM medium containing 10% FBS, 1% sodium pyruvate, 1% NEAA and 0.01 mg/mL insulin.
Cellular Uptake and Mitochondria Localization of Nanoparticles
Cellular uptake was visualized by confocal microscopy with FITC-labeled NPs. MCF-7 and MDA-MB-231 cells (3 × 105 cells/dish) were cultured for 24 h in confocal dishes with complete medium. After 24 h incubation, the cells were further treated with FITC-labeled NPs for 6 h under the same conditions. After that, the cells were washed with cold PBS to stop uptake, fixed with 1% paraformaldehyde for 30 min and stained with DAPI for 30 min and then washed with cold PBS.
Confocal laser scanning microscopy (CLSM) was also used to study the co-localization of nanoparticles with mitochondria. MDA-MB-231 cells were cultured in laser confocal dishes at a density of 3 × 105 cells/dish for 24 h. The cells were then treated with different FITC-loaded NPs and further cultured for 8 h. After incubation, cells were washed twice with PBS and stained with Mitotracker Red for 30 min, then cells were washed with PBS twice and fixed with 1% paraformaldehyde for 30 min, and nuclei were stained with DAPI and washed with cold PBS. Then, fluorescence images were obtained using CLSM.
Cell Viability
The cell viability of different NPs was evaluated using the CCK-8 kit following the manufacturer’s protocol. Cells were seeded at 4 × 103 cells/well in 96-well plate. After culture, cells were treated with LND, LND-PLGA/TPS NPs, LND-PLGA/DSSR NPs and LND-PLGA/TPS/DSSR NPs or blank NPs at different concentrations, respectively. The culture medium was used as a blank control. After 24 h, 10 μL of CCK-8 was added to each well and incubated for 1 h, and absorbance was obtained by using microplate reader (Bio Tek microplate reader) at 450 nm. Cell viability was calculated as follows:
Measurement of Mitochondrial Membrane Potential and Mitochondrial Damage in vitro
The mitochondrial membrane potential (MMP, ΔΨm) was evaluated by mitochondrial membrane potential assay kit JC-1. Briefly, MDA-MB-231 cells (3 × 105 cells/dish) were cultured for 24 h in confocal dishes, followed by treatment with LND, and three LND-loaded NPs (40 μg/mL LND) for 12 h, respectively. The culture medium was used as the control group. Afterwards, cells were stained with JC-1 working solution for 20 min and imaged by CLSM.
Cultured cells were treated with LND and LND-PLGA/TPS/DSSR NPs for 12 h and centrifuged to collect cells. The harvested cells were fixed by electron microscopic solution and further characterized by TEM for damage to mitochondrial nanostructure.
In vitro Cell Apoptosis Study
The Annexin V-FITC/PI kit was used to quantify nanoparticle induced apoptosis. Briefly, cells were put in 6-well plates and exposed to LND and LND-loaded NPs (40 μg/mL LND) for 24 h. Thereafter, cells were collected by digestion of trypsin. Then, 500 μL of binding buffer was added to suspend the cells, and the cells were stained with 5 μL of PI and 5 μL of Annexin V-FITC for 30 min in dark, respectively. Finally, the cells were subjected to flow cytometry to quantify stained cells.
The activity of caspase-3 and caspase-9 in MDA-MB-231 cells was determined using Western blot analysis. MDA-MB-231 cells (1 × 106 cells/dish) were cultured in cell dishes for 24 h, followed by treatment of LND and LND-loaded NPs (40 μg/mL LND) for 24 h. Afterwards, total proteins were extracted from cultured cells, mixed with 5 × SDS loading buffer, denatured in boiling water for 5 min and stored in −80 °C refrigerator. Western blotting was performed according to the manufacturer’s instructions. The protein bands were detected using an enhanced chemiluminescence reagent. β-Actin was used as the internal control. The grey intensity of protein bands was quantified by the Image J software and normalized using β-actin protein expressions.
Mitochondrial Oxygen Consumption Rates
Cell respiration rate was measured using an XF96 Cell Mito Stress Test Kit (Agilent). MDA-MB-231 cells were seeded on 96-well plates. To measure oxygen consumption rate (OCR), the cells were treated with various concentrations of nanoparticles for different times, and then the cells were washed and changed to seahorse assay medium in a non-CO2 incubator for 1 h. After that, mitochondrial inhibitors (1 μM of oligomcin, 0.5 μM of FCCP, the mixture of 0.5 μM of antimycin A and 0.5 μM of rotenone) were sequentially added to investigate the changes of OCR by the Seahorse XF96 Extracellular Flux Analyzer.
In vivo Efficacy of Lonidamine-Loaded Nanoparticles
Xenograft TNBC models were established in female BALB/c nude mice (6–8 weeks age) by hypodermically inoculating 5 × 106 MDA-MB-231 cells into the armpits. When the tumor volume reached ~50 mm3, the mice were randomly divided into five groups (5 mice for each group). LND and LND-loaded NPs were intravenously administered with 4 mg/kg LND every other day for 18 days, and physiological saline was used as control. Weight and tumor volume were monitored three times a week. The volume of tumor was calculated: Volume = 0.5 × L × W2, where L and W stand for the length and width of tumor tissues, respectively. After 18 days of treatment, the mice were euthanized. Tumors were removed, weighted and performed for tunnel analysis, and tissues were harvested for histopathologic analysis. The tumor inhibition rate was calculated as the relative tumor weight for each group. Blood samples (n = 3) were collected from the retro orbital vein in heparin-containing tubes, and blood routine examinations were performed. All animal procedures were conducted in accordance with the Guidelines for Care and Use of Laboratory Animals of Nanjing Normal University and approved by the Animal Management and Ethics Committee of Nanjing Normal University (Nanjing, China).
Statistical Analysis
Results were given as mean ± standard deviation (SD). One-way analysis of variance (ANOVA) was used for the statistical analysis of the data. p-value <0.05 was considered statistically significant.
Results and Discussion
Synthesis and Characterization of the Functional Molecules
Synthetic routes of DSSR and TPS were illustrated in Scheme 1A, and the synthetic routes of starting materials DSPE-S-S-COOH, NH2-PEG2000-R6RGD and TPP-COCl were shown in Figure S1. Chemical structures were determined by 1H NMR. As shown in Figures 1 and S2, characteristic chemical shifts of 1.26, 1.77, 2.49 and 3.52 ppm of DSSR compound attributed to -CH2- of DSPE, R6RGD, -CH2-S- and -CH2-O- of PEG, respectively, indicated that DSSR was successfully synthesized. 1H NMR also indicated that the bonding ratio of R6RGD to NH2-PEG2000-MAL was 1:1, which was compatible with the R6RGD content in NH2-PEG2000-R6RGD (50%); and the bonding ratio of NH2-PEG2000-R6RGD to DSPE-S-S-COOH is 0.7:1, which is compatible with the NH2-PEG2000-R6RGD in DSPE-S-S-PEG2000-R6RGD (41%). Figure S3 illustrated the 1H NMR spectra of TPP-TPGS. The characteristic chemical shifts of aromatic protons in TPP-COOH were 7.70–7.80 ppm while those of polyethylene glycol in TPGS were 3.52 ppm. The 1H-NMR spectra showed that these functional molecules (DSSR and TPS) were successfully prepared.
 |
Figure 1 The 1H NMR spectra of DSPE-S-S-COOH, NH2-PEG2000-R6RGD and DSPE-S-S-PEG2000-R6RGD (DSSR). |
Preparation and Characterization of Nanoparticles
The preparation of LND-PLGA/TPS/DSSR NPs and their internalization, mitochondrial localization and apoptosis in MDA-MB-231 cells were depicted in Schemes 1B and C. Nanoparticles were successfully prepared using o/w emulsion solvent evaporation. Its uptake, delivery, biodistribution and clearance were affected by physicochemical properties, such as particle size and surface properties.30 The TEM image showed that the LND-loaded nanoparticles were spherical in shape with a smooth surface morphology, which had uniform particle size of about 200–350 nm (Figure 2A). As shown in Figure S4A, the average diameters of LND-PLGA/TPS/DSSR NPs were 319.93 ± 16.66 nm and were larger than those of LND-PLGA/TPS NPs and LND-PLGA/DSSR NPs, which was due to the co-wrapped effects of DSSR and TPS. The polydispersity index (PDI) was about 0.3 (Figure S4B), indicating good dispersibility of nanoparticles. Figure S4C showed that the zeta potentials of LND-PLGA/TPS NPs were positive, while both LND-PLGA/DSSR NPs and LND-PLGA/TPS/DSSR NPs were negative. Positively charged NPs would bind the negatively charged surface of blood vessels and could be cleared rapidly from blood circulation.18 Our results revealed that DSSR conjugates could effectively prevent positively charged TPS from being rapidly cleared by the reticuloendothelial system (RES) and circulation lasted much longer. Meanwhile, NPs loaded with LND showed good stability in size and PDI in one week (Figure S5).
To determine the drug loading capacity of NPs, encapsulation efficiency (EE) and drug loading (DL) were determined by UV–Vis spectrometry. The EE and DL values of LND-PLGA/TPS/DSSR NPs were 87.51% and 18.82%, respectively, and the corresponding values for LND-PLGA/TPS NPs and LND-PLGA/DSSR NPs were 78.48%, 16.98% and 85.24%, 18.42%, respectively (Table S1). The high EE of LND-PLGA/TPS/DSSR NPs was due to emulsification of TPGS31 and PEGylated modification,32 which could increase the solubility of LND.
Measurement of Glutathione and pH-Triggered Drug Release in vitro
The LND release rate of LND-PLGA/TPS/DSSR NPs was investigated under different conditions to simulate normal tissue and tumor microenvironments. At 37 °C, NPs were exposed to different simulated solutions with pH of 5.5, 7.4 and 8.0 for 24 h. As shown in Figure 2B, LND release was pH dependent. 46% and 55% of LND were released at pH 5.5 and pH 7.4, respectively. However, nearly 70% of LND was released at pH 8.0 imitating the pH value in tumor mitochondria. It was suggested that the release of LND containing carboxylic acid was much higher in weakly alkaline environments than in neutral or acidic environments, which avoided drug leakage into cytoplasm and lysosome and was beneficial to the specific release of LND from NPs into mitochondria. In addition, the concentration of GSH in cancer cells was about 10 mM,33 which was at least four times that of normal ones.34,35 LND release was also investigated at pH 5.5 with 10 mM of GSH, pH 8.0 with 5 and 10 mM of GSH, respectively. The LND release was increased by the presence of glutathione to reduce disulfide bonds. After 24 h, approximately 100% and 76% of LND were released from NPs at pH 8.0 with 10 and 5 mM of GSH, respectively, which was significantly higher than that at pH 5.5 in the presence of 10 mM of GSH (64%) (Figure 2C). Indeed, the GSH could further contribute to the release of LND in NPs. It was more beneficial to LND release at pH 8.0 than in acid or neutral conditions with the presence of 5 or 10 mM of GSH. It was demonstrated that LND-PLGA/TPS/DSSR NPs could specifically respond to both weakly alkaline and high concentrations of GSH in the tumor cell mitochondria to reduce LND and decrease drug leakage in cytoplasm and lysosome.
Measurement of Uptake and Mitochondrial Localization of Nanoparticles in vitro
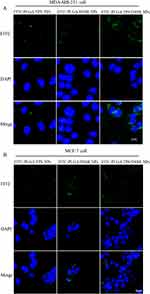
The αVβ3 integrin is overexpressed in MDA-MB-231 cells and lowly expressed in MCF-7 cells.36,37 The cellular uptakes of different nanoparticles were investigated on the MDA-MB-231 and MCF-7 cells, respectively. To visualize the cell internalization of NPs, LND was replaced by the fluorescent tracker FITC. After incubation with FITC-loaded NPs, the cells showed remarkable green FITC fluorescence, indicating efficient internalization of these nanoparticles into cells. As a blue fluorescent tracker, DAPI could accurately locate the nuclei. As shown in Figure 3A, after incubation with nanoparticles for 6 h, MDA-MB-231 cells treated with FITC-PLGA/TPS/DSSR NPs showed obvious green fluorescence. However, MCF-7 cells showed negligible FITC fluorescence compared with MDA-MB-231 cells (Figure 3B). These results indicated that FITC-PLGA/TPS/DSSR NPs modified by conjugate DSSR could be specifically accumulated and recruited by MDA-MB-231 cell with integrin αVβ3-overexpressing, which was mainly attributed to the targeting and penetration ability of R6RGD modification. In addition, the mechanism of absorption of different FITC-loaded NPs was also carried out. As shown in Figure S6, it was obvious that the uptake of FITC-PLGA/TPS/DSSR NPs was energy dependent.
To determine the subcellular location of NPs in cells, NPs were incubated with MDA-MB-231 cells for 8 h, followed by mitochondrial staining with MitoTracker red. The merged orange signal was imaged by CLSM and shown in Figure 4, which were overlapping results of green fluorescent FITC-loaded NPs and red fluorescent-tagged mitochondria. The orange color of FITC-PLGA/TPS/DSSR NPs was much deeper than that of the other two NP groups, indicating that FITC-PLGA/TPS/DSSR NPs were efficiently absorbed by MDA-MB-231 cells and located in mitochondria. These results demonstrated that co-wrapped NPs by TPS and DSSR could specifically target the mitochondria of TNBC MDA-MB-231 cells, and it could also avoid being cleared by RES with DSSR protection.
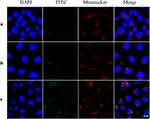 |
Figure 4 Representative CLSM images of MDA-MB-231 cells incubated for 8 h with (a) FITC-PLGA/TPS NPs, (b) FITC-PLGA/DSSR NPs and (c) FITC-PLGA/TPS/DSSR NPs locating in mitochondria. Scale bar: 10 μm. |
Measurement of Cytotoxicity of Lonidamine-Loaded Nanoparticles in vitro
CCK-8 cell assay was used to evaluate the inhibitory effects of LND-loaded NPs or LND-unloaded NPs on MDA-MB-231 cells. The results are shown in Figure 5. It showed that cytotoxicity of LND and LND-loaded NPs was concentration-dependent. In addition, LND-PLGA/TPS/DSSR NPs co-wrapped by DSSR and TPS exhibited the highest cytotoxicity among NPs, possibly due to its good absorption by tumor cells. Meanwhile, cell survival rate was higher than 80% with blank carriers, indicating that the nanoparticles were safe and non-toxic to MDA-MB-231 cells (Figure S7). The IC50 values (Table S2) of NPs formed by TPS, DSSR and TPS/DSSR were 130.71, 115.90 and 106.07 μM, respectively, which showed an enhanced drug efficacy compared with the LND that showed an IC50 = 408.42 μM.
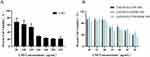 |
Figure 5 CCK-8 assay for MDA-MB-231 cells after treatment with (A) LND or (B) LND-loaded NPs at different concentrations (n=3). |
Measurement of Mitochondrial Membrane Potential (MMP) and Mitochondrial Damage in vitro
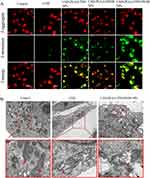
The decrease in MMP was analyzed by JC-1 assay to indicate mitochondrial dysfunction, which reflected early apoptosis in cancer cells. JC-1 formed J-aggregates with red fluorescence in normal cells, which showed high MMP. However, in apoptotic or unhealthy cells, JC-1 was in the monomer form with green fluorescence, indicating the low MMP.38 After treatment with LND-PLGA/TPS/DSSR NPs, cell MMP decreased dramatically (Figure 6A), indicating obvious cell apoptosis. However, there was less damage to cell mitochondria treated with LND-PLGA/TPS NPs or LND-PLGA/DSSR NPs. Meanwhile, LND-PLGA/TPS NPs could target mitochondria and cause relatively higher damage than LND-PLGA/DSSR NPs. Compared with the NPs groups, LND showed little green fluorescence, which indicated a high MMP and little cell apoptosis.
Mitochondrial structural damage in MDA-MB-231 cells was also directly visualized by TEM images after 12 h of exposure to LND-PLGA/TPS/DSSR NPs or LND. There was no mitochondrial morphology damage in the control group, but abnormal mitochondria resulting from LND or LND-PLGA/TPS/DSSR NPs were observed (Figure 6B). In addition, LND-PLGA/TPS/DSSR NPs-treated cells showed more significant mitochondrial damage with incomplete mitochondrial ridge structure than LND. These results indicated that LND-PLGA/TPS/DSSR NPs could destroy the integrity of the IMM structure. Since the damage was irreversible, it would lead to excessive fragmentation, mass decay and loss of mitochondria.39 Changes in mitochondrial morphology could regulate many mitochondrial functions, such as respiratory activity of the electron transport chain and apoptosis,40 and the proliferation of MDA-MB-231 cells would be affected.
Measurement of Cell Apoptosis in vitro
To investigate the effects of LND-loaded NPs on the necrosis and apoptosis of cancer cells, double staining was performed using Annexin V-FITC and PI. As shown in Figure 7A, LND-PLGA/TPS/DSSR NPs resulted in a higher percentage of early and late apoptotic cells (43.6% and 11.6%, respectively) than LND (34.3% and 9.15%, respectively), whereas MDA-MB-231 cells treated by LND-PLGA/TPS NPs and LND-PLGA/DSSR NPs only caused 36.5% and 9.53%, 40.1% and 10.7% of early and late apoptosis, respectively. The cell apoptosis results were consistent with cytotoxicity and mitochondrial membrane damage, which indicated that the high cytotoxicity of LND-PLGA/TPS/DSSR NPs was due to their damage to mitochondria, which could further lead to cell apoptosis.
The opening of mitochondrial permeability transition pore (mPTP) resulted in decreasing ΔΨm. Subsequently, caspase-3 and caspase-9 were activated, and mitochondria-mediated apoptotic processes occurred.41 To explain the molecular mechanism of tumor cell apoptosis caused by LND-loaded NPs, Western blot was used to analyze caspase-9 and caspase-3 expression level. Results in Figure 7B shows that LND-PLGA/TPS/DSSR NPs successfully upregulated cleaved caspase-9 and caspase-3 expression level in MDA-MB-231 cells compared with LND, LND-PLGA/DSSR NPs, and LND-PLGA/TPS NPs, which validated that LND-PLGA/TPS/DSSR NPs induced intrinsic apoptosis by activating caspase-3 and caspase-9.
Measurement of Mitochondrial Oxygen Consumption Rates
To evaluate the effects of NPs on mitochondrial function, Seahorse XF Cell Mito Stress Test Kit was used to measure oxygen consumption rate (OCR) of MDA-MB-231 cells treated with NPs for 3 and 6 h, respectively. After measuring basal respiration, specific mitochondrial inhibitors of oligomycin (ATP synthase inhibitor), FCCP (uncoupling agent) and antimycin A/rotenone (electron transport chain inhibitor) were added at different time. This approach was frequently used to measure basal oxygen consumption, ATP production, maximal respiration, and spare respiratory capacity, all of which were essential parameters for investigating mitochondrial respiration levels. As shown in Figure 8, after 3 h of NPs or LND treatment, there were no significant decreases in OCR in basal and maximal respiration, ATP production, and spare respiration capacity. However, treatment of cancer cells with LND-PLGA/TPS/DSSR NPs for 6 h, followed by the addition of specific mitochondrial inhibitors, significantly decreased ATP production and reduced spare respiratory capacity, indicating a decline in mitochondrial capacity to meet high energy demands under pressure. Compared with LND, LND-PLGA/TPS/DSSR NPs did significantly impair mitochondrial respiration function, which further affected cancer cell proliferation.
 |
Figure 8 Oxygen consumption rate (OCR) profile for MDA-MB-231 cells treated with LND-loaded NPs (80 μg/mL of LND). *p<0.05, **p<0.01, ***p<0.001. |
The concentration of NPs significantly affected mitochondrial respiratory functions. As shown in Figure 9, cells were treated by NPs (40 and 80 μg/mL of LND) or LND (100 or 200 μg/mL) for 6 h followed by Seahorse XF96 analysis. Compared with the control, 40 μg/mL of LND in NPs did not affect maximal respiration and spare respiration capacities, while it significantly decreased basal respiration and ATP production. LND-PLGA/TPS/DSSR NPs (40 μg/mL of LND) were significantly lower than those in 100 μg/mL LND group. Remarkably, when the LND concentration of LND-PLGA/TPS/DSSR NPs was increased from 40 to 80 μg/mL, the damage of mitochondrial respiratory chain was increased, and this injury was much more serious than other NP groups and 200 μg/mL of LND group. It was suggested that LND-PLGA/TPS/DSSR NPs had better inhibition of mitochondrial respiration in MDA-MB-231 cells than LND.
Experimental results showed that LND could alter the bioenergetics of cancer cells by inhibiting mitochondrial respiration. This was consistent with previous results that LND could inhibit fumarate and malate formation and suppress succinate-induced respiration of isolated mitochondria.42 Importantly, LND-PLGA/TPS/DSSR NPs indeed enhanced the influence on mitochondria respiration compared with LND or other NPs. This further demonstrated the results of cytotoxicity, mitochondrial membrane damage and cell apoptosis. Accordingly, mitochondrial dysfunction could release apoptosis-associated proteins and induce apoptosis in cancer cells through mitochondria-mediated apoptotic pathways.
In vivo Anti-Tumor Study
MDA-MB-231-bearing BALB/c nude mice were employed to investigate the in vivo anti-tumor activities of NPs. Drug administration was performed as shown in Figure 10A. When the tumor volume of nude mice reached 50 mm3, test drugs (saline, LND, LND-PLGA/TPS NPs, LND-PLGA/DSSR NPs and LND-PLGA/TPS/DSSR NPs) were intravenously injected into tumor-bearing mice every other day for 18 days. The results showed that tumor growth in the LND-PLGA/TPS/DSSR NP group was the slowest of all groups (Figures 10B–D, F and G). In addition, the tumor growth inhibition rate was calculated based on relative tumor weight. The tumor inhibition rates of LND-PLGA/TPS/DSSR NPs were also significantly higher than those in other groups. The nanoparticles modified by TPS/DSSR enabled LND to be specifically delivered to the mitochondria of tumor cells and induced apoptosis of tumor cells. Cell apoptosis levels were assessed by tunnel staining, and results revealed that apoptotic cells treated by the LND-PLGA/TPS/DSSR NP group significantly increased (Figure S8). Thus, LND-PLGA/TPS/DSSR NPs exhibited a significant ability to efficiently inhibit TNBC development by inducing cell apoptosis.
The body weight change curve showed that the NPs groups were as safe as the control group during the treatment period (Figure 10E), while diarrhea occurred during the administration of LND and body weight lost and did not recover at the end of the experiment compared with the saline group. Major mice organs including heart, liver, spleen, lung and kidney were also collected for H&E staining (Figure 11). Results showed that the liver was primarily impaired by hepatocyte watery degeneration and kidney tissue with slight degeneration of renal tubular epithelial cells in the LND group. However, no obvious toxicities were found in all organs of NP groups. Therefore, the NPs significantly reduced the toxicities of LND. Blood routine tests, including WBC (White blood cell count), Lymph (Lymphocyte), MPV (Mean platelet volume), Gran (Granulocyte), RBC (Red blood cell), HGB (Hemoglobin), HCT (Hematocrit), MCV (Mean corpuscular volume), MCH (Mean corpuscular hemoglobin), MCHC (Mean corpuscular hemoglobin concentration), RDW (Red blood cell volume distribution width) and PLT (Platelet count) were normal in all groups except for relatively lower RBC and PLT levels in LND-PLGA/DSSR NP group (Figure S9). Collectively, LND-PLGA/TPS/DSSR NPs improved the efficacy of TNBC in vivo and reduced the side effects of LND by targeting tumor cell mitochondria in programmed and specifically releasing LND in mitochondria, causing effective intrinsic apoptosis of TNBC cells.
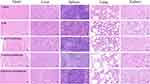 |
Figure 11 Histopathological results of the H&E staining images (40×). |
Conclusion
TNBC is a type of breast cancer that accounts for approximately 10% to 20% of all breast cancer cases and has limited treatment options and a poor prognosis. Herein, we developed a drug delivery system LND-PLGA/TPS/DSSR NPs modified with R6RGD and TPP to target the mitochondria in a programmed manner and release lonidamine (LND) specifically in the mitochondria to induce cell apoptosis. Such programmed mitochondria targeted NPs exhibited excellent tumor targeting, mitochondrial localization, disulfide bond breaking by GSH, and ability to release LND in the weakly alkaline mitochondrial microenvironment. Taken together, the results suggest that LND-PLGA/TPS/DSSR NPs are an effective LND delivery system due to their mitochondrial targeting and release capabilities. These advantages indicate that the drug delivery system may be useful for other drugs that target mitochondria.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (21877061).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no competing interests in this work.
References
1. Patel N, Weekes D, Drosopoulos K, et al. Integrated genomics and functional validation identifies malignant cell specific dependencies in triple negative breast cancer. Nat Commun. 2018;9(1):1–16. doi:10.1038/s41467-018-03283-z
2. Xu X, Zhang L, He X, et al. TGF-β plays a vital role in triple-negative breast cancer (TNBC) drug-resistance through regulating stemness, EMT and apoptosis. Biochem Biophys Res Commun. 2018;502(1):160–165. doi:10.1016/j.bbrc.2018.05.139
3. Bhattarai S, Klimov S, Mittal K, et al. Prognostic role of androgen receptor in triple negative breast cancer: a multi-institutional study. Cancers. 2019;11(7):1–9. doi:10.3390/cancers11070995
4. Goel S, Wang Q, Watt AC, et al. Overcoming therapeutic resistance in HER2-positive breast cancers with CDK4/6 inhibitors. Cancer Cell. 2016;29(3):255–269. doi:10.1016/j.ccell.2016.02.006
5. Zhao H, Li D, Zhang B, et al. PP2A as the main node of therapeutic strategies and resistance reversal in triple-negative breast cancer. Molecules. 2017;22(12):1–17. doi:10.3390/molecules22122277
6. Cohen-Erez I, Issacson C, Lavi Y, et al. Antitumor effect of lonidamine-polypeptide-peptide nanoparticles in breast cancer models. ACS Appl Mater Interfaces. 2019;11(36):32670–32678. doi:10.1021/acsami.9b09886
7. Zhang BF, Xing L, Cui PF, et al. Mitochondria apoptosis pathway synergistically activated by hierarchical targeted nanoparticles co-delivering siRNA and lonidamine. Biomaterials. 2015;61:178–189. doi:10.1016/j.biomaterials.2015.05.027
8. Wu C, Liu J, Tang X, Zhai Z, Xu K, Zhong W. An enzyme-assisted self-delivery system of lonidamine-peptide conjugates for selectively killing cancer cells. Chem Commun. 2019;55(98):14852–14855. doi:10.1039/c9cc06204a
9. Lahiani-Skiba M, Bounoure F, Fessi H, Skiba M. Effect of cyclodextrins on lonidamine release and in-vitro cytotoxicity. J Incl Phen Macro. 2010;69:481–485. doi:10.1007/s10847-010-9872-7
10. Cannino G, Ciscato F, Masgras I, Sánchez-Martín C, Rasola A. Metabolic plasticity of tumor cell mitochondria. Front Oncol. 2018;8:1–21. doi:10.3389/fonc.2018.00333
11. Fernández-Tussy P, Fernández-Ramos D, Lopitz-Otsoa F, et al. miR-873-5p targets mitochondrial GNMT-complex II interface contributing to non-alcoholic fatty liver disease. Mol Metab. 2019;29:40–54. doi:10.1016/j.molmet.2019.08.008
12. Wang YH, Scadden DT. Harnessing the apoptotic programs in cancer stem-like cells. EMBO Rep. 2015;16:1084–1098. doi:10.15252/embr.201439675
13. Ramadass R, Bereiter-Hahn J. How DASPMI reveals mitochondrial membrane potential: fluorescence decay kinetics and steady-state anisotropy in living cells. Biophys J. 2008;95(8):4068–4076. doi:10.1529/biophysj.108.135079
14. Zielonka J, Joseph J, Sikora A, et al. Mitochondria-targeted triphenylphosphonium-based compounds: syntheses, mechanisms of action, and therapeutic and diagnostic applications. Chem Rev. 2017;117(15):10043–10120. doi:10.1021/acs.chemrev.7b00042
15. He B, Sui X, Yu B, Wang S, Shen Y, Cong H. Recent advances in drug delivery systems for enhancing drug penetration into tumors. Drug Deliv. 2020;27(1):1474–1490. doi:10.1080/10717544.2020.1831106
16. López V, Villegas MR, Rodríguez V, et al. Janus mesoporous silica nanoparticles for dual targeting of tumor cells and mitochondria. ACS Appl Mater Interfaces. 2017;9(32):26697–26706. doi:10.1021/acsami.7b06906
17. He Y, Lei L, Cao J, et al. A combinational chemo-immune therapy using an enzyme-sensitive nanoplatform for dual-drug delivery to specific sites by cascade targeting. Sci Adv. 2021;7(6):1–13. doi:10.1126/sciadv.aba0776
18. Dai Y, Xu C, Sun X, Chen X. Nanoparticle design strategies for enhanced anticancer therapy by exploiting the tumour microenvironment. Chem Soc Rev. 2017;46(12):3830–3852. doi:10.1039/c6cs00592f
19. Arriortua OK, Insausti M, Lezama L, et al. RGD-functionalized Fe3O4 nanoparticles for magnetic hyperthermia. Colloids Surf B. 2018;165:315–324. doi:10.1016/j.colsurfb.2018.02.031
20. Guo X, Wei X, Chen Z, Zhang X, Yang G, Zhou S. Multifunctional nanoplatforms for subcellular delivery of drugs in cancer therapy. Prog Mater Sci. 2020;107(92):1–24. doi:10.1016/j.pmatsci.2019.100599
21. Hu H, Deng X, Song Q, et al. Mitochondria-targeted accumulation of oxygen-irrelevant free radicals for enhanced synergistic low-temperature photothermal and thermodynamic therapy. J Nanobiotechnol. 2021;19(1):1–20. doi:10.1186/s12951-021-01142-6
22. Franco MS, Gomes ER, Roque MC, Oliveira MC. Triggered drug release from liposomes: exploiting the outer and inner tumor environment. Front Oncol. 2021;11:1–23. doi:10.3389/fonc.2021.623760
23. Ko NR, Van SY, Hong SH, et al. Dual pH- and GSH-responsive degradable PEGylated graphene quantum dot-based nanoparticles for enhanced HER2-positive breast cancer therapy. Nanomaterials. 2020;10(1):1–15. doi:10.3390/nano10010091
24. Cheng Y, Ji Y. Mitochondria-targeting nanomedicine self-assembled from GSH-responsive paclitaxel-ss-berberine conjugate for synergetic cancer treatment with enhanced cytotoxicity. J Control Release. 2020;318:38–49. doi:10.1016/j.jconrel.2019.12.011
25. Wang X, Yang L, Fang Q, et al. GLUT1-targeting and GSH-responsive DOX/L61 nanodrug particles for enhancing MDR breast cancer therapy. Part Part Syst Char. 2020;37(9):1–13. doi:10.1002/ppsc.202000165
26. Chen Y, Zhu C, Cen J. Ratiometric detection of pH fluctuation in mitochondria with a new fluorescein/cyanine hybrid sensor. Chem Sci. 2015;6(5):3187–3194. doi:10.1039/c4sc04021j
27. Tan Y, Zhu Y, Zhao Y, et al. Mitochondrial alkaline pH-responsive drug release mediated by Celastrol loaded glycolipid-like micelles for cancer therapy. Biomaterials. 2018;154:169–181. doi:10.1016/j.biomaterials.2017.07.036
28. Zhao Y, Liu B, Lou R, et al. Construction of hyperbranched polysiloxane-based multifunctional fluorescent prodrug for preferential cellular uptake and dual-responsive drug release. Biomaterials Advances. 2022;137:1–12. doi:10.1016/j.bioadv.2022.212848
29. Ospina-Villa JD, Gómez-Hoyos C, Zuluaga-Gallego R, Triana-Chávez O. Encapsulation of proteins from Leishmania panamensis into PLGA particles by a single emulsion-solvent evaporation method. J Microbiol Methods. 2019;162:1–7. doi:10.1016/j.mimet.2019.05.004
30. Duan X, Li Y. Physicochemical characteristics of nanoparticles affect circulation, biodistribution, cellular internalization, and trafficking. Small. 2013;9(9–10):1521–1532. doi:10.1002/smll.201201390
31. Yang C, Wu T, Qi Y, Zhang Z. Recent advances in the application of vitamin E TPGS for drug delivery. Theranostics. 2018;8(2):464–485. doi:10.7150/thno.22711
32. Zhao T, Liu Y, Gao Z, et al. Self-assembly and cytotoxicity study of PEG-modified ursolic acid liposomes. Mater Sci Eng C. 2015;53:196–203. doi:10.1016/j.msec.2015.04.022
33. Ma Y, Mou Q, Wang D, Zhu X, Yan D. Dendritic polymers for theranostics. Theranostics. 2016;6(7):930–947. doi:10.7150/thno.14855
34. Liu J, Li F, Zheng J, Li B, Zhang D, Jia L. Redox/NIR dual-responsive MoS2 for synergetic chemo-photothermal therapy of cancer. J Nanobiotechnol. 2019;17(1):1–16. doi:10.1186/s12951-019-0510-2
35. Bhavsar DB, Patel V, Sawant KK. Design and characterization of dual responsive mesoporous silica nanoparticles for breast cancer targeted therapy. Eur J Pharm Sci. 2020;152:1–15. doi:10.1016/j.ejps.2020.105428
36. Zhong P, Gu X, Cheng R, Deng C, Meng F, Zhong Z. αVβ3 integrin-targeted micellar mertansine prodrug effectively inhibits triple-negative breast cancer in vivo. Int J Nanomed. 2017;12:7913–7921. doi:10.2147/IJN.S146505
37. Ou Z, Wu B, Xing D, Zhou F, Wang H, Tang Y. Functional single-walled carbon nanotubes based on an integrin αVβ3 monoclonal antibody for highly efficient cancer cell targeting. Nanotechnology. 2009;20(10):1–7. doi:10.1088/0957-4484/20/10/105102
38. Chen H, He C, Chen T, Xue X. New strategy for precise cancer therapy: tumor-specific delivery of mitochondria-targeting photodynamic therapy agents and in situ O2-generation in hypoxic tumors. Biomater Sci. 2020;8(14):3994–4002. doi:10.1039/d0bm00500b
39. Zhang S, Che L, He C, et al. Drp1 and RB interaction to mediate mitochondria-dependent necroptosis induced by cadmium in hepatocytes. Cell Death Dis. 2019;10(7):1–17. doi:10.1038/s41419-019-1730-y
40. Alam MM, Sohoni S, Kalainayakan SP, Garrossian M, Zhang L. Cyclopamine tartrate, an inhibitor of Hedgehog signaling, strongly interferes with mitochondrial function and suppresses aerobic respiration in lung cancer cells. BMC Cancer. 2016;16:1–10. doi:10.1186/s12885-016-2200-x
41. Nagahara Y, Ikekita M, Shinomiya T. T cell selective apoptosis by a novel immunosuppressant, FTY720, is closely regulated with Bcl-2. Br J Pharmacol. 2002;137(7):953–962. doi:10.1038/sj.bjp.0704970
42. Guo L, Shestov AA, Worth AJ, et al. Inhibition of mitochondrial complex II by the anticancer agent lonidamine. J Biol Chem. 2016;291(1):42–57. doi:10.1074/jbc.M115.697516
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.