Back to Journals » Infection and Drug Resistance » Volume 16
Genotypic and Phenotypic Characterization of Some psms Hypervirulent Clinical Isolates of Staphylococcus aureus in a Tertiary Hospital in Hefei, Anhui
Authors Cao J , Zhang H, He Z , Piao Z, Zong X, Sun B
Received 30 November 2022
Accepted for publication 17 February 2023
Published 15 March 2023 Volume 2023:16 Pages 1471—1484
DOI https://doi.org/10.2147/IDR.S399688
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Jiaxin Cao,1,2 Huimin Zhang,1,2 Zhien He,2 Zhongwan Piao,1 Xianchun Zong,1 Baolin Sun1,2
1College of Life Science and Technology, Mudanjiang Normal University, Mudanjiang, People’s Republic of China; 2School of Life Science and Medicine, University of Science and Technology of China, Hefei, People’s Republic of China
Correspondence: Baolin Sun; Zhongwan Piao, Email [email protected]; [email protected]
Background: Staphylococcus aureus is a highly successful pathogen that can cause various infectious diseases, from relatively mild skin infections to life-threatening severe systemic diseases. The widespread pathogenicity of S. aureus is mainly due to its ability to produce many virulence factors that help destroy various host cells, causing disease. Our primary goal in this study was to explore the genes of highly virulent strains, to identify genes closely associated with high virulence, and to provide ideas for the treatment of infection by highly virulent clinical strains.
Results: This study collected 221 clinical strains from The First Affiliated Hospital Of The University of Science and Technology of China (USTC); their hemolytic abilities were tested. Eight isolates were selected based on their highly hemolytic ability and tested their hemolytic activity again; their phenotypes and gene sequences were also explored. Whole-genome sequencing (WGS) showed six plasmids (pN315, pNE131, pSJH901, pSJH101, SAP106B, and MSSA476), eight antibiotic resistance genes [blaR1, blaI, blaZ, mecA, erm(C), erm(T), tet(38), and fosB-Saur] and seventy-two virulence related genes. Three highly virulent strains, namely X21111206, 21092239, and 21112607, were found according the Galleria mellonella infection model. Therefore, we selected 10 representative virulence genes for qRT-PCR: psmα, psmβ, hlgA, hlgB, hlgC, hla, clfA, clfB, spa, and sak. Among them, the expression levels of psmα and psmβ, the three isolates, were significantly higher than the positive control NCTC8325.
Conclusion: Significant differences appear in the expression of virulence genes in the highly virulent strains, particularly the psmα and psmβ, It may be that the high expression of psm gene is the cause of the high virulence of Staphylococcus aureus. We can reduce the pathogenicity of Staphylococcus aureus by inhibiting the expression of psm gene, which may provide a strong basis for psm as a new target for clinical treatment of S. aureus infection.
Keywords: Staphylococcus aureus, virulence factors, qRT-PCR, next-generation sequencing, psm
Introduction
Staphylococcus aureus, a Gram-positive bacterium, is a highly successful pathogen.1 According to previous reports, permanent intranasal carriers make up approximately 20% to 30% of the human population, while 60% are intermittent carriers, which can cause different degrees of infection at different sites.2 The key to the success of S. aureus as a pathogen is its ability to colonize and persist in various parts of the host, not only through asymptomatic colonization in healthy hosts but also through the initiation of long-term chronic infections that resist conventional treatments.3
S. aureus infections are common in the community and hospital settings. There are three main types: superficial lesions (eg, wound infections and food poisoning), toxin poisoning (eg, toxic shock syndrome and scalded skin syndrome), and more serious life-threatening systemic diseases (eg, osteomyelitis, pneumonia, and bacteremia).4,5 Such a wide range of infections in mammalian hosts is the result of its ability to produce a variety of virulence factors. These play a role in the process of host attachment, invasion, colonization, avoidance of the host’s immune system, and toxin intoxication. Currently recognized toxins include α-, β-, γ-, and δ-hemolysins,6 all of which damage platelets, destroy lysosomes, and cause ischemic necrosis; leukocidins (Panton–Valentine leukocidin, PVL), which can destroy human leukocytes and giant cells;7 enterotoxins is a T-cell superantigen that can cause acute gastroenteritis and withstand temperatures of 100 °C for 30 minutes;8 phenol-soluble modulins (PSMs);9 exfoliative toxins (ET),10 which cause staphylococcal scalded skin syndrome (SSSS); and, finally, invasive enzymes, which include various extracellular proteases or coagulases, etc. coagulases is a plasminogen activator that deposit fibrin from blood or plasma on the surface of the bacteria, hindering the phagocytosis activity of phagocytes from the human body.11 A single factor does not regulate the expression of all these virulence factors; instead, it is controlled by a complex regulatory network. Therefore, while studying the pathogenic mechanism of S. aureus and the respective treatment plan, the evaluation of virulence factors and their regulatory pathways is an essential component that cannot be ignored.
In our experiment, we selected 10 representative genes, all of which play important roles in Staphylococcus aureus infection. The clfA and clfB genes encode important adhesin fibrinogen-binding proteins involved in biofilm formation.12 The clfA is the major virulence factor responsible for S. aureus aggregation in plasma, with the overwhelming majority of clinical strains of S. aureus carrying the clfA gene.13 The clfb gene promotes the interaction of S. aureus with fibrinogen in the early exponential phase, being proteolytically degraded in stationary phase cultures; on the other hand, clfA is present throughout the growth cycle.14 The hlgA, hlgB, hlgC, and hla genes encode hemolysin. The hla encodes α-hemolysin, which can lead to the lysis of various cells, such as erythrocytes, platelets, endothelial or epithelial cells, and certain leukocytes, by forming heptameric β-barrel pores in the target cell membrane.15–19 The hlgA and hlgC genes encode two S subunits of leukotoxins, and hlgB encodes F subunits, constituting two leukotoxins: γ-hemolysin HlgAB and HlgCB.20 Similar to α-hemolysin, γ-hemolysin is a pore-forming toxin that forms octameric pores on the surface of a variety of cells, leading to cell lysis.21,22 Both psmα and psmβ encode phenol-soluble modulin (PSM), responsible for cell lysis, biofilm formation, and immune regulation during S. aureus infection. After S. aureus is exposed to phagocytes, the PSM is produced to lyse them, mediating the intracellular escape of S. aureus. Sub-lytic concentrations of PSM have immunomodulatory effects on host cells. PSM can also form biofilms by forming channels required for nutrient transport and dissemination.9,23–26 The spa gene encodes the protein A (SpA), the only known B-cell antigen produced by S. aureus that regulates phagocytosis by binding to immunoglobulin (IG) and blocking specific anti-S. aureus antibodies.27 Finally, the sak gene encodes staphylokinase (Sak), a cofactor produced by S. aureus, which by itself has no enzymatic activity, but can form a 1:1 complex with plasmin in serum (Sak–plasmin). This complex can convert plasminogen to plasmin.28,29 Sak reduces the formation of biofilms by activating plasminogen to infection severity and promoting the detachment of mature biofilms to spread bacteria.30
A large number of antibiotics used in clinical treatment result in the emergence of bacterial antibiotics resistance, and different antibiotics resistance genes act on different antibiotics. The blaR1 gene encodes a transmembrane protein whose ectodomain senses the stressful effects of penicillin in the environment. The blaI encodes a repressor protein that inhibits the expression of the bla operon. The blaZ gene encodes penicillinase, which can hydrolyze the amide bond in the penicillin beta-lactam ring, hindering the bactericidal effect of penicillin. As such, the blaR1-blaI-blaZ operon regulates the mechanism of penicillin resistance in S. aureus.31 Additionally, the mecA gene encodes a penicillin-binding protein, PBP2a, which has a low affinity for β-lactam antibiotics, allowing bacteria to become resistant and cells to synthesize normally.32 The erm gene can make S. aureus resistant to macrolide antibiotics. It encodes a synthetic methylase that methylates the adenine of the 23S rRNA located in the 50S subunit of the ribosome, impeding antibiotics from binding to the target site.33 The tet(48) gene mainly ensures bacterial resistance to tetracycline antibiotics, by encoding efflux pump proteins that expel tetracycline and metal ion complexes by proton exchange.34 Finally, the fosB can encode a Mn2+-dependent fosfomycin-inactivating enzyme in Gram-positive bacteria such as S. aureus, which can catalyze the nucleophilic conversion of L-cysteine (L-Cys) or bacillus thiol (BSH) addition to antibiotics yields modified compounds without bactericidal properties.35 The biofilm produced by S. aureus can help the bacteria by escaping the host’s immune system and producing virulence factors that destroy immune cells; this is highly conducive to persistent infection by S. aureus.
The widespread infection of the Staphylococcus aureus has led us to study it. This study used NCTC8325 and N315 as control strains. The former, isolated from sepsis patients in the 1960s, is a widely used model strain, sensitive to all known antibiotics and highly virulent.36 The N315 is a less toxic methicillin-resistant Staphylococcus aureus (MRSA) strain.37 Next-generation sequencing (NGS) and WGS were performed on eight isolates. 21092239, X21111206, and 21112607 were selected for qRT-PCR to detect the expression of virulence factors; the phenotypic differences between strains were further explained from the gene perspective.
Methods
Bacterial Strains, Primers, and Growth Conditions
In this study, 221 clinical strains were collected from The First Affiliated Hospital Of USTC, from August 2021 to January 2022. According to the hemolytic activity of all clinical isolates, eight isolates with higher hemolytic activity were selected. The information for the clinical bacterial strains used in this study is listed in Table 1 and Table 2. The eight clinical isolates were derived from eight different patients. Primers used in this study are listed in Table 3. MLST primer sequences were obtained from the website (https://pubmlst.org/organisms/staphylococcus-aureus/primers). qRT-PCR primer is designed by ourselves, size of the amplicon hlgA 177bp, hlgB 139bp, hlgC 174bp, hla 166bp, clfA 73bp, clfB 63bp, psmα 23bp, psmβ 70bp, pta 121bp, spa. All S. aureus were grown at 37 °C and 220 rpm in Tryptic Soy Broth (TSB, BD, America) or Tryptic Soy Agar (TSA). All isolates were stored in 40% (v/v) glycerol broth at −80 °C until use. The TSB medium was supplemented with 2.5% NaCl in biofilm assay.
 |
Table 1 Clinical Strains Used in This Study |
 |
Table 2 MIC of Clinical Strains |
 |
Table 3 Primers Used in This Study |
Antibiotic Susceptibility Testing (AST)
AST was performed using a VITEK2 Compact System 3. AST was done on all 221 isolates of S. aureus. The antibiotics included: oxacillin (OXA), vancomycin (VAN), gentamicin (GEN), erythromycin (ERY), levofloxacin (LVX), moxifloxacin (MFX), clindamycin (CLI), paediatric compound sulfamethoxazole tablets (SXT), rifampin (RIF), linezolid (LNZ), and tigecycline (TGC).
Detection of Hemolytic Activity
The hemolytic activity of bacteria was detected by a quantitative method. Bacteria were cultured in TSB media and passaged twice. The overnight culture was diluted with blank TSB to OD600 = 3 (1.5×109 CFU/mL). The diluted bacterial liquid was collected by centrifugation at 12,000 rpm for 1 min, from which 100 μL of the supernatant was mixed with 900 μL of phosphate-buffered saline (PBS) buffer containing 3% sheep erythrocytes. The mixture was incubated at 37 °C for 20 min. After centrifugation at 12,000 rpm for 1 min, the supernatant’s absorbance (543 nm) was measured. The positive and negative controls consisted of 1 mL of ddH2O (double distilled H2O) containing 3% sheep erythrocytes and 1 mL of PBS buffer containing 3% sheep erythrocytes, respectively. The hemolysis rate of each sample was calculated considering a rate of 100% for the positive and 0% for the negative controls.
Multilocus Sequence Typing (MLST) of Isolates
S. aureus genomic DNA was extracted by the StarPrep Plasmid Miniprep Kit (Genstar, Beijing, China). The ST (Sequence Typing) of the eight isolates was then determined using the MLST protocol, MLST was performed as described previously and included internal fragments of the following seven housekeeping genes: arcC, carbamate kinase; aroE, shikimate dehydrogenase; glp, glycerol kinase; gmk, guanylate kinase; pta, phosphate acetyltransferase; tpi, triosephosphate isomerase; and yqiL, acetyl coenzyme A acetyltransferase.38 These genes were amplified by PCR from the genomic DNA using seven pairs of primers. The fragments’ nucleic acid sequences were determined by sequencing, and the allelic value and ST at each locus of each isolate was determined via http://pubmlst.org/.
Whole-Genome Sequencing, Assembly, and Annotation
The whole genome of S. aureus was sequenced using the Illumina HiSeq 4000 platform of the Beijing Genome Institute (BGI, Shenzhen, China). The program Pbdagcon (https://github.com/PacificBiosciences/pbdagcon) was used for self-correction. Draft genomic units, uncontroversial sets of fragments, were assembled against a high-quality corrected set of circular consensus sub-reads using a Celera Assembler. A single base correction was performed using GATK (https://www.broadinstitute.org/gatk/) and SOAP toolkits (SOAP2, SOAPsnp, SOAPindel) to improve genome sequence accuracy. De novo mixed assembly of short Illumina reads and long PacBio reads was performed using Unicycler v0.4.839 and annotated using Prokka 1.14.6.40
Genome Analysis and Comparative Genomics Analysis
Acquired antimicrobial resistance genes (ARGs) were identified using ABRicate version 1.0.1 (https://github.com/tseemann/abricate) by aligning genome sequences with the ResFinder and NCBI databases.41 The isolates’ virulence related genes were identified by aligning genome sequences with the VFDB database using Kleborate and ABRicate version 1.0.1.41,42 Comparative genomics and phylogenetic analysis of different isolates were performed using the HarvestTools kit (Parsnp, Gingr, and HarvestTools) and BacWGSTdb, and constructed using the Interactive Tree of Life (iTOL) v5 (http://itol.embl.de/) phylogenetic tree.43–45
Biofilm Assay
Biofilm is an important structure conducive to the survival of bacteria in the host. Therefore, the biofilm-forming ability of these eight strains was further tested. Biofilm assays were performed on eight highly virulent clinical strains under static based on the methods described previously.46 The twice-passaged overnight bacterial solution was diluted with blank TSB to OD600 = 0.05 (2.5×107 CFU/mL), and then aliquoted (200 μL per well) into sterile flat-bottom 96-well polystyrene plates (Costar, America). After incubation at 37 °C for 24 h, the upper bacterial liquid was carefully extracted with a pipette; the wells were washed twice with ddH2O to remove non-adherent cells and stained with an appropriate 100mL crystal violet for 20 minutes. After aspirating the staining solution and washing twice with ddH2O, the biofilm was dried by backing and dissolved with 30% glacial acetic acid. The absorbance at OD560 was measured with an enzyme-linked immunosorbent assay reader ELX800 (Bio-Tek, America), in a 3×3 scan model.
Growth Curve
The growth curve of S. aureus was determined using a microplate reader based on a previously described method.47 The overnight bacterial solution was transferred to blank TSB at an initial OD600 = 0.05 and aliquoted (200 μL per well) into sterile flat-bottom 96-well polystyrene plates. After incubation at 37 °C and 220 rpm, the absorbance value of OD600 was measured with ELX800 (Bio-Tek, America) in a 3×3 scan model every hour until the bacteria reached the plateau phase.
Galleria mellonella Infection Model
G. mellonella was used as the infection model to observe the isolates’ virulence more intuitively.48 The twice-passaged overnight bacterial solution was washed with sterile saline and then diluted to 1×108 CFU/mL with sterile saline.
The G. mellonella was kept in the dark and used within three days after shipment, after selection for individuals weighing between 250 and 300 mg and presenting a healthy milky white color. After randomly selecting a group of 20 individuals for injection, a 100 μL syringe was used to inject 10 μL of the diluted bacterial solution into the middle of the second abdominal foot of G. mellonella. After injection and infection, G. mellonella individuals were incubated at 37 °C; their survival was observed every 24 h for seven days. No dead larvae were observed in the negative control group, composed of individuals subjected to no injection or injection of 10 μL of sterile saline.
Total RNA Extraction, cDNA Generation, and Real-Time Quantitative Reverse Transcription-PCR
The twice-passaged overnight bacterial culture was transferred to blank TSB at an initial OD600 = 0.05, shaken at 37 °C until reaching OD600 = 2.5 in the middle stage of exponential growth, and then collected. The supernatant was discarded by centrifugation to collect the bacterial cells. To resuspend the cells, 900 μL of RNAiso Plus (TaKaRa, Japan) was added and then transferred to a crushing tube containing 0.6 g of zirconia-silica beads with a diameter of 0.1 mm. Further fragmentation was performed twice with a FastPrep-24 automated system (MP Biomedicals Solon, America) with an interval of 5 min. Residual DNA was removed with RNase-free recombinant DNase I (TaKaRa, Japan, 5 U/µL). For reverse transcription, using random primers, cDNA was synthesized with the PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, Japan). Quantitative reverse transcription-PCR (qRT-PCR) was performed using the StepOne Real-Time PCR System (Applied Biosystems, America) using SYBR Premix Ex Taq (TaKaRa, Japan). The amount of cDNA was measured by the 2−ΔΔCt method, using pta as the reference gene and the corresponding control samples as running calibrators. Table 3 lists the primers used in this study. All qRT-PCR assays were repeated at least three times.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5. Data were analyzed using unpaired t-tests to compare two different conditions, and ANOVA was performed for more conditions. All experiments were performed in biological triplicates.
Results
Plasmids, Antibiotic Resistance Related Genes, Virulence Related Genes, and Multilocus Sequence Typing (MLST) Results of Eight Isolates
MLST of the isolates was determined by comparing the sequence numbers of the seven housekeeping genes (Table 4). The isolates 21101007, 21092239, 21112741, and 21083150 were ST6697; 21112543 and 21112607 were ST59; 21122023 was ST630, and X21111206 was ST188. The four ST types belong to four different clonal complexes (CC), ST6697 belongs to CC181, ST59 belongs to CC59, ST630 belongs to CC8, ST188 belongs to CC1. By analyzing the sequencing results, we calculated the single nucleotide polymorphisms (SNP) distances of the eight strains (Figure S1), roughly observing their genetic relationships (Figure S2). NGS was performed on the eight clinical strains with higher hemolysis. Sequencing results showed antibiotic resistance, virulence, and plasmids (Figure 1). Whole-genome sequencing showed that, from the eight S. aureus isolates, 21112741, 21101007, and 21083150 contained four plasmids (pN315, pNE131, pSJH901, and pSJH101), 21112607 and X1206 contained three plasmids (pN315, pSJH901, and pSJH101), 21122023 contained two plasmids (SAP106B and MSSA476) and 2092239 and 21112543 did not contain any of these six plasmids.
 |
Table 4 Serial Number of Seven Housekeeper Genes, ST and CC |
The eight strains contained eight antibiotic resistance related genes, namely blaR1, blaI, blaZ, mecA, erm(C), erm(T), tet(48), and fosB-Saur (Figure 1). According to the results of methicillin susceptibility tests, 21112741, 21101007, 21083150, 21112607, 21112543, and 21122023 are MRSA, X21111206 and 21092239 are MSSA. From WGS, we observed that the genes blaR1, blaI, and blaZ were present in five strains (21112741, 21101007, 21083150, 21112607, and X21111206); erm(C) existed in 21112741, 21101007, and 21083150; erm(T) only existed in 210922239; fosB-Saur was only present in 21122023; mecA was present in six strains (21112741, 21101007, 21083150, 21112607, 21112543, and 21122023); and tet(48) existed in all eight strains. According to the WGS results, a total of 72 virulence related genes were analyzed through gene sequence comparison in the eight strains; the strain with the most virulence related genes (67) was 21112543 and 21112607, followed by X21111206 with 66, 21112741 with 61, 210922239, 21101007 and 21083150 with 60, and 21122023 with 54. The virulence related genes missing from eight isolates shown by WGS are listed in Table 5
 |
Table 5 WGS Showed the Deletion of Virulence Related Genes in Eight Isolates |
Phenotypic Characterization of the Eight Isolates
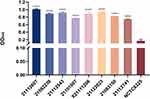
Phenotypic experiments were performed on these eight highly hemolytic strains; their biofilm-forming ability, lethal ability to G. mellonella, and growth curves were measured. First, the eight clinical isolates (21112607, X21111206, 21092239, 21122023, 21112543, 21112741, 21101007, and 21083150) presented significantly higher hemolytic activity than NCTC8325 (Figure 2; Figure S3). Regarding growth rates, 21112607 and 21112543 presented relatively slow growth rates, with no difference from NCTC8325. The growth rates of the other six isolates were faster, with clear differences compared to NCTC8325, generally, the difference in growth rate between strains is more significant after 4 hours (Figure 3).
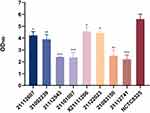
Four of the eight isolates (21112607, X221111206, 21092239, and 21122023) presented relatively strong biofilm-forming abilities, and the remaining four strains presented weaker biofilm-forming abilities. All eight isolates presented lower biofilm-forming abilities than the highly virulent strain NCTC8325 (Figure 4).
Although bacteria virulence can partially be assessed through the hemolytic activity and biofilm-forming ability of bacteria, virulence factors are diverse and complex. For a more comprehensive and intuitive judgment of bacteria virulence, G. mellonella was used as an animal infection model. By calculating the mortality rate of G. mellonella exposed to S. aureus, we can clearly illustrate the virulence of the isolates. The experimental results showed that, among the eight isolates, 21112607, X21111206, and 21092239 presented significantly higher lethality in G. mellonella than the highly virulent strain NCTC8325, selected as a positive control (Figure 5), indicating a higher virulence. The mortality of the remaining five clinical isolates was not significantly different than NCTC8325.
Virulence Factor Expression
The three isolates found to have significantly higher virulence than NCTC8325 in the infection experiment were subjected to Real-Time Quantitative Reverse Transcription-PCR (qRT-PCR) to detect the expression of virulence genes (Figure 6). We selected 10 representative virulence genes, namely psmα, psmβ, hlgA, hlgB, hlgC, hla, clfA, clfB, spa, and sak, each with respective functions during an S. aureus infection. Compared with the positive control NCTC8325, we could observe that the expression levels of the genes clfA, clfB, and spa were significantly higher in N315. The psmα, psmβ, and spa genes were significantly higher in X21111206 than in NCTC8325. In the 21112607 isolate, the expression levels of all genes, except for hla and sak, were higher than in NCTC8325; psmα, psmβ, hlgA, clfB, hlgC, and spa were higher in 21092239 than in NCTC8325. The expression level of hlgC was highest in 21092239; NGS results showed that the sak gene was deleted in this isolate, and the gene was not detected when the gene expression level was detected, since this isolate does not contain the sak gene, and the expression levels of the remaining three genes (hlgB, clfA and hla) in this isolate were all very low. The expression level of virulence related genes of the three highly virulent isolates showed that the expression level of psms in the three strains was very high, which was significantly different from that of the highly virulent strain NCTC8325. Therefore, psms may have a close relationship with the high virulence of the strain.
Discussion
S. aureus is a very successful pathogen since it secretes multiple virulence factors that destroy different cells of the host, besides producing biofilms that increase tolerance to antibiotics and immune defenses.49–51 Heavy use of antibiotics has also led to the development of resistance genes in the Staphylococcus aureus. All eight clinical isolates contained antibiotic resistance related genes with their own types of antibiotics. Here, we first selected eight clinical isolates with the highest hemolytic activity from all isolates’ hemolytic results, including four sequence type (ST) types: X21111206 was ST188, 21101007, 21112741, 21083150, and 21092239 were ST6697, 21122023 was ST630, and 21112543 and 21112607 were ST59. The WGS showed that vwb virulence related genes were missing: namely, 21092239, 21112741, 21101007, 21083150, 21112607 and 21112543 did not contain vwb. The von Willebrand factor-binding protein (vWbp) is a coagulase secreted by S. aureus and a type of MSCRAMM. The adhesins can help S. aureus to adhere to and colonize the host and help in biofilm formation. Additionally, 21122023 lacked the icaB and icaC genes. The ica operon encodes four genes, icaA, icaD, icaB, and icaC, and the icaADBC gene cluster is responsible for the production of poly-N-acetylglucosamine (PNAG), a major component of biofilms.52 Although X21111206 did not lack the adhesin gene in WGS, the qRT-PCR results showed a low clfA gene expression. Deleting some genes that help bacteria form biofilms, as well as the decrease in gene expression, It may be the reason for the decline of biofilm forming ability. Specifically, the biofilm formation ability of these eight isolates was lower than of NCTC8325. Moreover, the large expression of most hemolysin genes may also be the reason for the strong hemolysis ability, especially there are many non-hemolysin toxins that can destroy cells, such as PSMs. However, neither the hemolytic nor biofilm-forming ability can fully represent the virulence of the strains; therefore, the applied G. mellonella infection model showed X21111206, 21092239, and 21112607 as the three isolates showing higher virulence. The qRT-PCR results of these three isolates revealed a very important virulence factor, psm, which plays an important role in various infectious diseases caused by S. aureus,53 and PSM can be directly regulated by the quorum sensing system Agr.54 PSM can promote the release of lipoproteins, which in turn stimulate TLR2 to initiate an immune response, leading to sepsis.55 This virulence factor also increases biofilm stability through its aggregation to form fibrillar amyloid structures that help stabilize S. aureus biofilms.56 Moreover, psmα can act on keratinocytes and promote IL-17-mediated epidermal skin inflammation.57 It also induces the death of human osteoblasts, inhibiting bone remodeling, and causing bone destruction, an important virulence factor in the pathogenesis of osteomyelitis. PSM can also induce the production of IL-8, promoting osteoclast differentiation and increasing osteoclast activity. This may be one of the mechanisms by which PSM indirectly leads to bone destruction.58
Notably, previous studies have shown that, except for the two toxins α-hemolysin and PVL, there are not that many toxins that can currently become vaccine candidates for multiple pathogenic staphylococci.59 As found in this study, PSM plays an important role in the pathogenesis of staphylococcal infectious diseases and may provide strong evidence that PSMs could be targeted as anti-Staphylococcus aureus therapeutic agents.
Conclusions
In this study, we screened eight clinical strains of S. aureus for their hemolytic activity and explored their phenotypes and genomes. Using the G. mellonella infection model, three clinical strains (21092239, X21111206, and 21112607) had significantly higher virulence than the positive control NCTC8325. For these three strains, the expression number of virulence genes was further detected. We found that psms, an important virulence factor, was highly expressed in these three highly virulent strains. This further demonstrates the feasibility of PSM as a new target for clinical therapy.
Abbreviations
SSSS, staphylococcal scalded skin syndrome; WGS, Whole genome sequencing; PSM, phenol-soluble modulin; SpA, Staphylococcus protein A; IG, immunoglobulin; Sak, staphylokinase; ClfA, clumping factor A; ClfB, clumping factor B; FNBP, fibronectin-binding protein; fnbA/ fnbB, fibronectin adhesin genes; PNAG, poly-N-acetylglucosamine; MSCRAMM, microbial surface component-recognizing adhesion matrix molecule; TLR, toll-like receptor; IL, Interleukin; PVL, Panton-Valentine leukocidin; ET, exfoliative toxin; BSH, bacillus thiol; NGS, Next-generation sequencing; MLST, Multilocus Sequence Typing; ST, sequence type; arc, carbamate kinase; aroE, shikimate dehydrogenase; glp, glycerol kinase; gmk, guanylate kinase; pta, phosphate acetyltransferase; tpi, triosephosphate isomerase; yqiL, acetyl coenzyme A acetyltransferase; PCR, Polymerase chain reaction; vWbp, von Willebrand factor-binding protein; qRT-PCR, Quantitative Real-Time Reverse Transcription-PCR.
Data Sharing Statement
The whole genome sequences generated in the current study are available in the NCBI database (BioProject: PRJNA824222).
Ethics Approval and Consent to Participate
All isolates in this study were collected during the bacteriological analysis in the clinical microbiology laboratory of a public hospital, and patients were treated anonymously and therefore did not require ethical approval.
Acknowledgments
We thank Yuanyuan Dai and Huaiwei Lu for providing us with clinical strains.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was supported by the National Key Research and Development Program of China (2021YFC2300300), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29020000), grants from National Natural Science Foundation of China (32070132) and the Fundamental Research Funds for the Central Universities (YD9100002013).
Disclosure
The authors declare that they have no competing interests for this work.
References
1. Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28(3):603–661. doi:10.1128/CMR.00134-14
2. Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10(3):505–520. doi:10.1128/CMR.10.3.505
3. Dziewanowska K, Patti JM, Deobald CF, Bayles KW, Trumble WR, Bohach GA. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infect Immun. 1999;67(9):4673–4678. doi:10.1128/IAI.67.9.4673-4678.1999
4. Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000;13(1):16–34. doi:10.1128/CMR.13.1.16
5. Tasneem U, Mehmood K, Majid M, Ullah SR, Andleeb S. Methicillin resistant Staphylococcus aureus: a brief review of virulence and resistance. J Pak Med Assoc. 2022;72(3):509–515. doi:10.47391/JPMA.0504
6. Li M, Diep BA, Villaruz AE, et al. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2009;106(14):5883–5888. doi:10.1073/pnas.0900743106
7. Li M, Cheung GY, Hu J, et al. Comparative analysis of virulence and toxin expression of global community-associated methicillin-resistant Staphylococcus aureus strains. J Infect Dis. 2010;202(12):1866–1876. doi:10.1086/657419
8. Lina G, Bohach GA, Nair SP, et al. Standard nomenclature for the superantigens expressed by staphylococcus. J Infect Dis. 2004;189(12):2334–2336. doi:10.1086/420852
9. Wang R, Braughton KR, Kretschmer D, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13(12):1510–1514. doi:10.1038/nm1656
10. Lamand V, Dauwalder O, Tristan A, et al. Epidemiological data of staphylococcal scalded skin syndrome in France from 1997 to 2007 and microbiological characteristics of Staphylococcus aureus associated strains. Clin Microbiol Infect. 2012;18(12):E514–521. doi:10.1111/1469-0691.12053
11. Gustafsson E, Oscarsson J. Maximal transcription of aur (aureolysin) and sspA (serine protease) in Staphylococcus aureus requires staphylococcal accessory regulator R (sarR) activity. Fems Microbiol Lett. 2008;284(2):158–164. doi:10.1111/j.1574-6968.2008.01198.x
12. Speziale P, Pietrocola G, Foster TJ, Geoghegan JA. Protein-based biofilm matrices in Staphylococci. Front Cell Infect Microbiol. 2014;4:171. doi:10.3389/fcimb.2014.00171
13. Peacock SJ, Moore CE, Justice A, et al. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect Immun. 2002;70(9):4987–4996. doi:10.1128/IAI.70.9.4987-4996.2002
14. Perkins DN, Vaudaux P, Vaudaux P, Höök M, Foster TJ, Foster TJ. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol Microbiol. 1998;30(2):245–257. doi:10.1046/j.1365-2958.1998.01050.x
15. Cooper LZ, Madoff MA, Weinstein L. Heat stability and species range of purified staphylococcal alpha-toxin. J Bacteriol. 1966;91(5):1686–1692. doi:10.1128/jb.91.5.1686-1692.1966
16. Grimminger F, Rose F, Sibelius U, et al. Human endothelial cell activation and mediator release in response to the bacterial exotoxins Escherichia coli hemolysin and staphylococcal alpha-toxin. J Immunol. 1997;159(4):1909–1916. doi:10.4049/jimmunol.159.4.1909
17. Inoshima I, Inoshima N, Wilke GA, et al. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat Med. 2011;17(10):1310–U1196. doi:10.1038/nm.2451
18. Nygaard TK, Pallister KB, DuMont AL, et al. Alpha-toxin induces programmed cell death of human T cells, B cells, and monocytes during USA300 infection. PLoS One. 2012;7(5):e36532. doi:10.1371/journal.pone.0036532
19. Gouaux JE, Braha O, Hobaugh MR, et al. Subunit stoichiometry of staphylococcal alpha-hemolysin in crystals and on membranes: a heptameric transmembrane pore. Proce National Acad Sci. 1994;91(26):12828–12831. doi:10.1073/pnas.91.26.12828
20. Prévost G, Mourey L, Colin DA, Menestrina GJ. Staphylococcal pore-forming toxins. Pore-Forming Toxins. 2001;257:53–83.
21. Spaan AN, Vrieling M, Wallet P, et al. The staphylococcal toxins gamma-haemolysin AB and CB differentially target phagocytes by employing specific chemokine receptors. Nat Commun. 2014;5:5438. doi:10.1038/ncomms6438
22. Spaan AN, Reyes-Robles T, Badiou C, et al. Staphylococcus aureus targets the Duffy antigen receptor for chemokines (DARC) to lyse erythrocytes. Cell Host Microbe. 2015;18(3):363–370. doi:10.1016/j.chom.2015.08.001
23. Cheung GY, Duong AC, Otto M. Direct and synergistic hemolysis caused by Staphylococcus phenol-soluble modulins: implications for diagnosis and pathogenesis. Microbes Infect. 2012;14(4):380–386. doi:10.1016/j.micinf.2011.11.013
24. Rasigade JP, Trouillet-Assant S, Ferry T, et al. PSMs of hypervirulent Staphylococcus aureus act as intracellular toxins that kill infected osteoblasts. PLoS One. 2013;8(5):e63176. doi:10.1371/journal.pone.0063176
25. Surewaard BG, de Haas CJ, Vervoort F, et al. Staphylococcal alpha-phenol soluble modulins contribute to neutrophil lysis after phagocytosis. Cell Microbiol. 2013;15(8):1427–1437. doi:10.1111/cmi.12130
26. Periasamy S, Joo HS, Duong AC, et al. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci U S A. 2012;109(4):1281–1286. doi:10.1073/pnas.1115006109
27. Falugi F, Kim HK, Missiakas DM, Schneewind O. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. Mbio. 2013;4(5):e00575–00513. doi:10.1128/mBio.00575-13
28. Grella DK, Castellino FJ. Activation of human plasminogen by staphylokinase. direct evidence that preformed plasmin is necessary for activation to occur. Blood. 1997;89(5):1585–1589. doi:10.1182/blood.V89.5.1585
29. Lijnen HR, Van Hoef B, Collen D. Interaction of staphylokinase with different molecular forms of plasminogen. Eur J Biochem. 1993;211(1–2):91–97. doi:10.1111/j.1432-1033.1993.tb19873.x
30. Kwiecinski J, Peetermans M, Liesenborghs L, et al. Staphylokinase control of staphylococcus aureus biofilm formation and detachment through host plasminogen activation. J Infect Dis. 2016;213(1):139–148. doi:10.1093/infdis/jiv360
31. Pence MA, Haste NM, Meharena HS, et al. Beta-lactamase repressor BlaI modulates staphylococcus aureus cathelicidin antimicrobial peptide resistance and virulence. PLoS One. 2015;10(8):e0136605. doi:10.1371/journal.pone.0136605
32. Peacock SJ, Paterson GK. Mechanisms of methicillin resistance in staphylococcus aureus. Annu Rev Biochem. 2015;84:577–601. doi:10.1146/annurev-biochem-060614-034516
33. Farrell DJ, Morrissey I, Bakker S, Morris L, Buckridge S, Felmingham D. Molecular epidemiology of multiresistant Streptococcus pneumoniae with both erm(B)- and mef(A)-mediated macrolide resistance. J Clin Microbiol. 2004;42(2):764–768. doi:10.1128/JCM.42.2.764-768.2004
34. Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65(2):232–260. doi:10.1128/MMBR.65.2.232-260.2001
35. Thompson MK, Keithly ME, Goodman MC, et al. Structure and function of the genomically encoded fosfomycin resistance enzyme, FosB, from Staphylococcus aureus. Biochemistry. 2014;53(4):755–765. doi:10.1021/bi4015852
36. Novick RP, Richmond MH. Nature and interactions of the genetic elements governing penicillinase synthesis in staphylococcus aureus. J Bacteriol. 1965;90:467–480. doi:10.1128/jb.90.2.467-480.1965
37. Kuroda M, Ohta T, Uchiyama I, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357(9264):1225–1240. doi:10.1016/S0140-6736(00)04403-2
38. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1015. doi:10.1128/JCM.38.3.1008-1015.2000
39. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi:10.1371/journal.pcbi.1005595
40. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi:10.1093/bioinformatics/btu153
41. Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–2644. doi:10.1093/jac/dks261
42. Wyres KL, Wick RR, Gorrie C, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2(12):e000102. doi:10.1099/mgen.0.000102
43. Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):524. doi:10.1186/s13059-014-0524-x
44. Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi:10.1093/nar/gkz239
45. Feng Y, Zou S, Chen H, Yu Y, Ruan Z. BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 2021;49(D1):D644–D650. doi:10.1093/nar/gkaa821
46. Kim YG, Lee JH, Kim SI, Baek KH, Lee J. Cinnamon bark oil and its components inhibit biofilm formation and toxin production. Int J Food Microbiol. 2015;195:30–39. doi:10.1016/j.ijfoodmicro.2014.11.028
47. Krishnamurthi VR, Niyonshuti II, Chen J, Wang Y. A new analysis method for evaluating bacterial growth with microplate readers. PLoS One. 2021;16(1):e0245205. doi:10.1371/journal.pone.0245205
48. Menard G, Rouillon A, Ghukasyan G, Emily M, Felden B, Donnio PY. Galleria mellonella larvae as an infection model to investigate sRNA-mediated pathogenesis in staphylococcus aureus. Front Cell Infect Microbiol. 2021;11:631710. doi:10.3389/fcimb.2021.631710
49. Beenken KE, Dunman PM, McAleese F, et al. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186(14):4665–4684. doi:10.1128/JB.186.14.4665-4684.2004
50. Fitzpatrick F, Humphreys H, O’Gara JP. The genetics of staphylococcal biofilm formation—will a greater understanding of pathogenesis lead to better management of device-related infection? Clin Microbiol Infec. 2005;11(12):967–973. doi:10.1111/j.1469-0691.2005.01274.x
51. Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi:10.1007/978-3-540-75418-3_10
52. Cue D, Lei MG, Luong TT, et al. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J Bacteriol. 2009;191(20):6363–6373. doi:10.1128/JB.00913-09
53. Sayed-Zaki ME, El-Sabbagh AM, Hammam H. Molecular study of mec-phenol soluble modulin gene in methicillin resistant staphylococcus aureus. Clin Lab. 2021;67:9. doi:10.7754/Clin.Lab.2020.200947
54. Yu D, Zhao L, Xue T, Sun B. Staphylococcus aureus autoinducer-2 quorum sensing decreases biofilm formation in an icaR-dependent manner. BMC Microbiol. 2012;12:288. doi:10.1186/1471-2180-12-288
55. Hanzelmann D, Joo HS, Franz-Wachtel M, et al. Toll-like receptor 2 activation depends on lipopeptide shedding by bacterial surfactants. Nat Commun. 2016;7:12304. doi:10.1038/ncomms12304
56. Le KY, Villaruz AE, Zheng Y, et al. Role of phenol-soluble modulins in staphylococcus epidermidis biofilm formation and infection of indwelling medical devices. J Mol Biol. 2019;431(16):3015–3027. doi:10.1016/j.jmb.2019.03.030
57. Nakagawa S, Matsumoto M, Katayama Y, et al. Staphylococcus aureus virulent PSMα peptides induce keratinocyte alarmin release to orchestrate IL-17-dependent skin inflammation. Cell Host Microbe. 2017;22(5):667–677.e665. doi:10.1016/j.chom.2017.10.008
58. Davido B, Saleh-Mghir A, Laurent F, et al. Phenol-soluble modulins contribute to early sepsis dissemination not late local USA300-osteomyelitis severity in rabbits. PLoS One. 2016;11(6):e0157133. doi:10.1371/journal.pone.0157133
59. Karauzum H, Venkatasubramaniam A, Adhikari RP, et al. IBT-V02: a multicomponent toxoid vaccine protects against primary and secondary skin infections caused by staphylococcus aureus. Front Immunol. 2021;12:624310. doi:10.3389/fimmu.2021.624310
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.