Back to Journals » Risk Management and Healthcare Policy » Volume 16
Genetic Variations of AKT1 are Associated with Risk Screening for Non-Alcoholic Fatty Liver Disease
Authors Ding Y, Tang Z, Zhang R , Zhang M, Guan Q, Zhang L, Wang H, Chen Y, Zhang W, Wang J
Received 10 April 2023
Accepted for publication 12 July 2023
Published 26 July 2023 Volume 2023:16 Pages 1365—1376
DOI https://doi.org/10.2147/RMHP.S416592
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Haiyan Qu
Yajie Ding,1,* Zongzhe Tang,1,* Ru Zhang,1 Mengting Zhang,1 Qing Guan,1 Liuxin Zhang,2 Hongliang Wang,3 Yue Chen,1 Wei Zhang,4 Jie Wang1
1Department of Fundamental and Community Nursing, School of Nursing, Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China; 2The Nethersole School of Nursing, Faculty of Medicine, The Chinese University of Hong Kong, Shatin, New Territories, Hong Kong (SAR), People’s Republic of China; 3Department of General Practice, Ninghai Road Community Health Service Center, Nanjing, Jiangsu, People’s Republic of China; 4Department of Epidemiology, Shanghai Cancer Institute, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jie Wang, Department of Fundamental and Community Nursing, School of Nursing, Nanjing Medical University, 101 Longmian Avenue, Nanjing, Jiangsu, 211166, People’s Republic of China, Tel +86-25-86869557, Fax +86-25-86868501, Email [email protected]
Purpose: Protein kinase B (PKB/AKT) has shown a high profile in the research of metabolic diseases. This research sought to determine whether the AKT1 gene’s single nucleotide polymorphisms (SNPs) and the risk of developing non-alcoholic fatty liver disease (NAFLD) were related.
Patients and Methods: Recruited in this case–control study were 2693 subjects, including 815 with NAFLD and 1878 without NAFLD. Three SNPs of AKT1 (rs2494732, rs2494752 and rs1130233) were genotyped. To examine the correlation between SNPs and NAFLD susceptibility, logistic regression was performed.
Results: After adjusting for sex, age, triglyceride and glucose, AKT1 rs2494732-C (all P < 0.05 in co-dominant model, dominant model and additive model) and rs2494752-G (P < 0.05 in co-dominant model) were linked to a lower risk of NAFLD. The combined effect of both SNPs on NAFLD risk was statistically significant, showing a dose dependence (Ptrend = 0.010). Sex, body mass index, hypertension, hyperglycemia, hypertriglyceridemia, high-density lipoprotein-cholesterol, alanine aminotransferase, and beneficial alleles were all significant predictors of NAFLD risk (all P < 0.05). The prediction model achieved good discrimination, with an area under the receiver operating characteristic curve of 0.779. The Hosmer–Lemeshow test suggested an inadequate calibration of the model (χ2 = 21.073, P = 0.007).
Conclusion: AKT1 rs2494732 and rs2494752 may be related to Chinese NAFLD susceptibility. The prediction model combining both SNPs with clinical factors displays a strong ability to discriminate NAFLD patients. Both SNPs may be exploited to design new models for early screening of NAFLD high-risk population.
Keywords: NAFLD, susceptibility, AKT1 gene, polymorphism, risk screening
Graphical Abstract:
Introduction
The prevalence of nonalcoholic fatty liver disease (NAFLD) keeps rising globally, especially in adolescents and young adults.1,2 Characterized by intemperate amassing of triglycerides and cholesterol in hepatocytes, NAFLD is regarded as a hepatic sign of metabolic disorder.3 It causes nonalcoholic steatohepatitis, liver fibrosis, cirrhosis, and liver cancer,4 and also involves an increased risk of extrahepatic diseases, such as type 2 diabetes mellitus (T2DM),5 cardiovascular disease,6 and chronic kidney disease,7 impairing the health-related quality of life of patients.8 Therefore, early screening and prevention should be enhanced through tools designed with factors associated with its susceptibility.
The pathogenesis of NAFLD remains unsolved but may involve multiple factors, including insulin resistance, adipose tissue dysfunction, mitochondrial dysfunction, endoplasmic reticulum stress, inflammatory activation, intestinal microbiota, as well as genetic and epigenetics.9,10 Metabolic associated fatty liver disease (MAFLD), a more appropriate term, has been proposed to redefine NAFLD in an international consensus, emphasizing the association with metabolic comorbidities.11
During the progression of NAFLD, protein kinase B (PKB, also known as AKT) is overactivated to promote the maturation of proteins associated with lipid accumulation (a type of metabolic disorder).12 AKT, as a serine and threonine kinase, is activated by phosphoinositide 3-kinase (PI3K) through phosphorylation, thereby regulating a range of cellular processes, such as metabolism, cell survival, motility, lipid synthesis, protein synthesis and degradation.13 Implications of AKT in tumors, diabetes, obesity, neurological disorders, inflammation, and other diseases have been reported.14–16 AKT contains three isoforms (AKT1, AKT2, and AKT3), each with distinctive but overlapping functions.17 AKT1 is mainly involved in growth control, AKT2 in metabolic regulation, and AKT3 in brain development.18 In recent years, the role of AKT1 in metabolism has attracted mounting attention. AKT1 deletion protects mice from insulin resistance and obesity induced by diet, indicating a role of AKT1 in energy metabolism.17 In addition, studies have identified associations between AKT1 variants and metabolic-related diseases, such as T2DM,19 obesity,20 and metabolic syndrome.21 Considering that NAFLD is a metabolic stress-related disease, we speculate that AKT1 may regulate the susceptibility to NAFLD.
As a genetic marker, single nucleotide polymorphisms (SNPs) have shown their value in etiological studies, due to their high stability across phenotypes of diseases.16 Therefore, this present work attempted to investigate the associations between functional SNPs of AKT1 gene and NAFLD susceptibility, and evaluate the ability of AKT1 polymorphisms combined with clinical variables in predicting NAFLD risk.
Materials and Methods
Study Participants and Conception
This case–control study involved 2693 individuals, including 815 in the NAFLD case group and 1878 in the control group, all recruited between April and October 2020 from a community in Nanjing, Jiangsu, China. Ethical approval was obtained from the Institutional Ethics Review Committee of Nanjing Medical University, with a project approval number of (2019) 740. Prior to the trial, written informed permission was obtained from each participant.
Included were individuals with Han nationality, ages over 18 years, post-fasting blood samples, sufficient comprehending and communicating skills to complete the questionnaire. Excluded were those with infection, autoimmune diseases, malignant tumors, other liver diseases (such as viral hepatitis, alcoholic liver disease, and drug-induced fatty hepatitis), excessive alcohol consumption (≥20 g/day in females and ≥30 g/day in males), willingness to undergo liver transplantation within one year, or advanced liver diseases with complications such as variceal bleeding or ascites.
The cases diagnosed with NAFLD were classified into the case group in accordance with the 2018 modification of the Chinese NAFLD guideline,22 while the subjects in the same community without NAFLD were assigned to the control group. Detailed diagnostic criteria for NAFLD include (1) no history of abusing alcohol excessively (<20 g/day in females and <30 g/day in males); (2) exclusion of conditions such drug-related liver disease, viral hepatitis, complete parenteral feeding, hepatomegaly, and autoimmune liver disease (all of which are causes of fatty liver); and (3) histological changes on liver biopsy. Given the difficulty in obtaining a histologic diagnosis of the liver, the working definition of NAFLD, provided that (1) and (2) were met, was (i) liver imaging findings that meet the diagnostic criteria for diffuse fatty liver with no other explanation; and/or (ii) metabolic syndrome-related components presenting with unexplained persistent elevations of serum alanine aminotransferase (ALT) and/or aspartate aminotransferase (AST) and γ-glutamyl transpeptidase (γ-GT) for more than 6 months.
Age (±3 years) was frequency-matched between the two groups. Sample size was estimated based on preliminary study and PASS 11.0 software, with two-sided α = 0.05, power = 80, odds ratio (OR) = 1.5, frequency of gene variation in the general population = 20%. The sample sizes in two groups met the requirements (at least 534 in each group).
Data and Specimen Collection
Demographic data were gathered from self-administered questionnaire surveys. Clinical data about medical history, anthropometric, biochemical and imaging information were obtained from electronic medical records.
Participants who were fasting had their venous blood drawn (at least 5 mL) and kept in tubes containing ethylene diamine tetraacetic acid (EDTA), an anticoagulant. Plasma and blood cells were separated from each blood sample within 2 hours and frozen at −80°C before genotyping assays.
SNP Selection Criteria
The following methods were used to select SNPs: (1) The genotype information of AKT1 (upstream and downstream extended by 2000bp each) in Han Chinese in Beijing (CHB) was downloaded from the 1000 Genomes Project database (http://www.1000genomes.org/) and imported into Haploview 4.2 software (Broad Institute, Cambridge, MA, USA). (2) Tagging SNPs (tagSNPs) of AKT1 gene were selected according to the correlation coefficient r2 ≥ 0.8, the minor allele frequency (MAF) > 0.05 and Hardy–Weinberg P-value cutoff = 0.05 in the software. At this point, 62 tagSNPs were generated. (3) The MAFs of tagSNPs in the Chinese population were available from NCBI dbSNP (http://www.ncbi.nlm.nih.gov/); the SNPs with MAF ≤ 0.05 were excluded, leaving 18 tagSNPs with high frequency. (4) Finally, the SNPs associated with other diseases (especially metabolic syndrome, type 2 diabetes, hypertension, hyperlipidemia, etc.) were selected through literature review. Based on the above four processes, three polymorphisms (rs2494732, rs2494752, and rs1130233) in the AKT1 gene were selected for further genotyping.
DNA Extraction and Genotyping
By using a magnetic bead technique based on blood genomic extraction kit (Pangu Genome Nanotechnology Co., Ltd.; Nanjing, China), genomic DNA was isolated from blood samples, and its concentration was set to 50 ng/mL.
Genotyping was performed in a 384-well plate on a Light Cycler 480 II Real-Time PCR System (Roche, Switzerland) using the TaqMan allelic discrimination method. Primer and probe information are detailed in Supplementary Table S1. A random selection of 10% of the samples was used in subsequent tests to confirm the quality control of the experimental data, and the concordance was 100%. The entire genotyping was performed in a blinded manner (all technicians were unaware of the participants’ information) and followed the manufacturer’s instructions. The success rate of genotyping was more than 96% for all SNPs.
In silico Analysis
Potential biological function of AKT1 gene polymorphisms in NAFLD susceptibility was explored using several online bioinformatic databases. (1) Genetic variation sites on chromosomes were determined through NCBI dbSNP (https://www.ncbi.nlm.nih.gov/snp). (2) The RegulomeDB scores of SNPs were obtained using the RegulomeDB database (http://www.regulomedb.org/), with a lower score implying a higher profile in transcriptional regulation. (3) The functions of SNPs were predicted in the SNP Function Prediction database (https://snpinfo.niehs.nih.gov/). (4) Whether genetic variation sites were associated with histone modifications was checked using the UCSC Genome Browser database (http://genome.ucsc.edu/). (5) The RNAfold WebServer (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi) projected the influence of genetic variation of positive SNPs on AKT1 mRNA secondary structure.
Statistical Analysis
Statistical analyses were conducted using SPSS 23.0, R software v3.4.3, and MedCalc 19.1. The Kolmogorov–Smirnov test and histogram were used to evaluate data normality. Descriptive analysis was conducted according to data type, including constituent ratios, mean ± standard deviation, and median (interquartile range). Baseline information for both groups was compared using Chi-square (χ2) test (for categorical variables), Student’s t-test (for normal continuous variables), and Mann–Whitney U-test (for non-normal continuous variables). A goodness-of-fit χ2-test was used to determine Hardy–Weinberg equilibrium. Logistic regression analysis based on four genetic models (co-dominant model, dominant model, recessive model and additive model) was performed to construct odds ratio (OR) and 95% confidence interval (CI) to investigate the correlations of SNPs with susceptibility to NAFLD, with age, sex, triglyceride, and glucose adjusted for potential confounding effects. The rules for implementing the four genetic models are detailed in Table 1. To account for the impact of multiple comparisons, the false discovery rate (FDR) is adjusted. Cochran–Armitage trend was tested to analyze the combined effects of statistically significant SNPs. We further performed subgroup analysis to explore the influence of confounding factors, and heterogeneity between subgroups was assessed by Q test. To identify NAFLD predictors and proceed to construct a predictive model, multivariate stepwise logistic regression and receiver-operating characteristic curve (ROC) were used. The discrimination and calibration of the prediction model were assessed by the area under the receiver operating characteristic curve (AUROC) and the Hosmer–Lemeshow test, respectively. In all analyses, a P-value < 0.05 in the two-tailed test was considered statistically significant.
 |
Table 1 Genotype Distributions of Three AKT1 SNPs and Their Associations with NAFLD Risk |
Results
Basic Characteristics of Study Subjects
Table 2 summarizes the demographic and clinical features of the NAFLD cases and the controls. Between the two groups, there was no discernible age difference (P > 0.05). Sex, body mass index (BMI), systolic blood pressure (SBP), diastolic blood pressure (DBP), alanine aminotransferase (ALT), aspartate aminotransferase (AST), γ-glutamyl transpeptidase (γ-GT), total cholesterol (TC), triglyceride (TG), high-density lipoprotein-cholesterol (HDL-C), low-density lipoprotein-cholesterol (LDL-C), and glucose (GLU) were the variables where there were significant differences (all P < 0.05). Besides, all SNPs in the control group’s genotype frequencies were in Hardy–Weinberg equilibrium (all P > 0.05) (Supplementary Table S1), indicating that the selected samples were representative of the population.
 |
Table 2 Distributions of Demographic and Clinical Characteristics in NAFLD Case and Control Groups |
Associations Between AKT1 SNPs and NAFLD Susceptibility
The genotype information for the three SNPs in both groups is displayed in Table 1. After adjusting for sex, age, TG, and GLU, logistic regression analyses revealed that AKT1 variants rs2494732-C (TC genotype vs TT genotype: adjusted OR = 0.784, 95% CI = 0.648–0.947, P = 0.012; dominant model: adjusted OR = 0.775, 95% CI = 0.647–0.928, P = 0.006; additive model: adjusted OR = 0.823, 95% CI = 0.714–0.949, P = 0.007) and rs2494752-G (AG genotype vs AA genotype: adjusted OR = 0.826, 95% CI = 0.684–0.999, P = 0.048) were significantly correlated with low NAFLD risk in different models. After false discovery rate correction for multiple comparisons (Supplementary Table S2), their associations were still significant with all PFDR≤0.25.23 AKT1 1130233 and NAFLD susceptibility did not, however, appear to be significantly correlated with any of the models (all P > 0.05).
As shown in Table 3, beneficial alleles (rs2494732-C and rs2494752-G) were counted to evaluate the combined effects of both SNPs on NAFLD susceptibility. A lower incidence of NAFLD was observed in the subjects with more beneficial alleles. Those with beneficial alleles (“1–2” or “3–4”) showed a significantly lower risk of NAFLD, compared to those carrying none of these beneficial alleles (adjusted OR = 0.820, 95% CI = 0.677–0.993, P = 0.042; adjusted OR = 0.683, 95% CI = 0.488–0.957, P = 0.027, respectively). It appears that the combination allele affects NAFLD susceptibility in a dose-dependent way (Ptrend = 0.010) as more beneficial alleles were linked to a reduced incidence of NAFLD.
 |
Table 3 Combined Effect of AKT1 rs2494732-C and rs2494752-G on NAFLD Risk |
The combined effect of rs2494732-C and rs2494752-G was further evaluated in subgroups stratified according to sex, age, BMI, BP, GLU, TG, HDL-C, ALT, AST, and γ-GT using additive models (Supplementary Table S3). The stratification was based on the health industry standard of the People’s Republic of China (WS/T 404.1–2012), clinical guidelines and the population characteristics in this study.22,24–26 We found that the combined protective effect of two SNPs was more pronounced in the subgroups of males, age <40 years, BMI <24 kg/m2, SBP <140 and DBP <90 mmHg, GLU ≥5.6 mmol/L, TG <1.7 mmol/L, HDL-C >1.04 mmol/L, ALT ≥40 U/L, AST <40 U/L, and γ-GT <50 U/L (all adjusted P < 0.05). Except for age (P = 0.022), the effects in the remaining subgroups showed no significant heterogeneity (all P > 0.05).
Functional Prediction of Positive SNPs
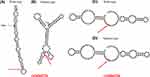
The Regulome DB score for both AKT1-rs2494732 and rs2494752 was 4. The SNP function prediction database predicted rs2494752 as a transcription factor binding site. UCSC database predictions showed that both rs2494732 and rs2494752 were enriched close to the H3K4Me1 marker (Figure 1). The impact of rs2494732-C and rs2494752-G on AKT1 mRNA secondary structure was predicted by RNAfold WebServer (Figure 2). The arrows indicated the position of the variation. The T and C alleles of rs2494732 were calculated to have minimum free energy (MFE) values of −38.6 and −39.1 kcal/mol, respectively. The MFE value for rs2494752 was calculated to be −24.1 kcal/mol for both the A and G alleles. The results of the linkage disequilibrium (LD) test with Haploview software showed no strong LD among the three candidate SNPs (Supplementary Figure S1), implying that the haplotype analysis was not necessary.
 |
Figure 1 Functional prediction of positive single nucleotide polymorphisms in AKT1. The red dotted line indicates the position of AKT1 rs2494732 and rs2494752 (available at http://genome.ucsc.edu/). |
Independent Predictors of NAFLD
A stepwise regression model was conducted incorporating sex, age, BMI, hypertension, hyperglycemia, hypertriglyceridemia, HDL-C, ALT, AST, γ-GT, and combined beneficial alleles (rs2494732-C and rs2494752-G). These variables showed no evident multi-collinearity (Supplementary Table S4). As outlined in Table 4, sex (OR = 0.413, 95% CI = 0.314–0.542, P < 0.001), BMI (OR = 3.207, 95% CI = 2.630–3.911, P < 0.001), hypertension (OR = 1.477, 95% CI = 1.198–1.821, P < 0.001), hyperglycemia (OR = 1.242, 95% CI = 1.021–1.510, P=0.030), hypertriglyceridemia (OR = 1.932, 95% CI = 1.567–2.383, P < 0.001), low HDL-C (OR = 1.428, 95% CI = 1.119–1.822, P = 0.004), ALT (OR = 1.762, 95% CI = 1.374–2.260, P < 0.001), and beneficial alleles (OR = 0.846, 95% CI = 0.723–0.991, P = 0.038) were independent predictors of NAFLD.
 |
Table 4 Multivariate Stepwise Logistic Regression Analysis for Independent Factors Influencing NAFLD Risk |
By combining these 8 variables, a NAFLD risk prediction model was constructed. As shown in Figure 3, the AUROC of the prediction model was 0.779 (95% CI = 0.763–0.795). At a cutoff value of −1.098, the sensitivity and specificity of this model were 79.7% (76.7–82.5%) and 63.7% (61.4–65.9%), respectively. The calculated positive and negative predictive values were 48.8% and 87.9%, respectively. The NAFLD risks estimated by the model and those observed in the real setting differed statistically significantly, according to the Hosmer–Lemeshow test (χ2 = 21.073, P = 0.007).
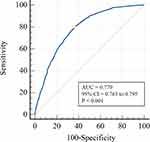 |
Figure 3 The receiver-operating characteristic curve for the risk prediction model of nonalcoholic fatty liver disease. Abbreviations: AUC, area under the curve; 95% CI, 95% confidence interval. |
Discussion
This research investigated the links between AKT1 SNPs and the risk of NAFLD in a Chinese Han cohort. Our findings demonstrated that AKT1 rs2494732-C and rs2494752-G were beneficial alleles related to low NAFLD risk, but no association of rs1130233 with NAFLD risk was found. Furthermore, the combination of beneficial alleles and clinical factors showed an ideal predictive ability for the risk of NAFLD.
The AKT gene is located in the chromosome 14q32.33 region. As a downstream effector of the PI3K pathway, it can be activated to regulate the function of multiple substrates in various physiological processes, such as metabolism, cell survival, motility, transcription and cell cycle.27 Genetic variants of AKT are associated with the development of cancer, neurological disorders, obesity, diabetes and other metabolic diseases.15 Among the three isoforms of AKT, AKT1 is the most widely expressed and associated with growth and adipogenesis.28,29 McKenzie et al20 found that AKT1 variants might influence obesity-associated metabolic phenotypes in elderly Caucasians. The results of genotyping experiments by Eshaghi et al21 also suggested an association between AKT1 polymorphisms and the components of metabolic syndrome. All these findings verify the implication of AKT1 gene in metabolism disorders.
NAFLD is a metabolic condition that is intimately linked to metabolic syndrome,11 T2DM,5 and obesity,30 and its association with AKT1 variants has not been explored. This study is the first to show a connection between AKT1 polymorphisms and the likelihood of developing NAFLD. Additionally, we discovered that people with both rs2494732-C and rs2494752-G had a dose-dependently decreased risk of developing NAFLD. These findings offer fresh arguments for decriminalizing NAFLD.
rs2494732 belongs to an intron polymorphic locus of AKT1 gene, and studies have focused on its association with schizophrenia,31 psychosis in cannabis users,32 and cancers, such as head and neck squamous cell carcinoma33 and non-small cell lung cancer.34 For the first time, we discovered in the current investigation, those who carried the rs2494732-C allele had a decreased chance of developing NAFLD. Liemburg et al explored the association of AKT1 with BMI in cannabis users, finding that the rs2494732 polymorphism was associated with BMI, glycosylated hemoglobin (HBA1c) level and total metabolic risk.35 BMI, a known risk predictor for NAFLD, has been incorporated into several NAFLD risk assessment models, such as the ZJU index,36 fatty liver index (FLI),37 and hepatic steatosis index (HIS).38 In addition, HBA1c, an index of insulin resistance and type 2 diabetes, is usually increased during the development of NAFLD.39 More importantly, we noticed that the rs2494732-C variant might affect the expression level of AKT1 by changing its RNA secondary structure (Figure 2). All these evidences provide clues to explain an association between rs2494732 and NAFLD was found in this study.
Similarly, we also identified rs2494752-G of AKT1 as a protective genotype against NAFLD. In the previous studies, rs2494752 was found to be involved in various cancers, including liver cancer,40 esophageal squamous cell carcinoma,41 gastric cancer,42 breast cancer,43 and non-small cell lung cancer.44 No studies have been conducted to analyze the role of rs2494752 polymorphism in NAFLD, but our data from the UCSC database indicated that the locus was located in the promoter region of AKT1 gene and close to the peak of H3k4me1 level. It also served as a transcription factor binding site, as shown by the database predicting SNP functions. These findings imply that rs2494752 variant may influence cis-regulatory modules in transcription and translation to regulate the protein level and biological effects of AKT, thus reducing the risk of NAFLD. Further genetic and epigenetic studies are needed to determine the role of this polymorphism in metabolism. No association of rs1130233 polymorphism with NAFLD susceptibility was found in this study, although Eshaghi et al21 revealed that rs1130233 was connected to key metabolic syndrome markers as hs-CRP and BMI. Genotyping of Iranians showed that AKT1 rs1130233 was not linked to an increased risk of cardiovascular disease or the metabolic syndrome,45 which is similar to our findings. Therefore, whether rs1130233 is associated with susceptibility to NAFLD remains to be explored.
Stratified analysis revealed that in the majority of subgroups, the combined effects of rs2494732 and rs2494752 were statistically significant, meanwhile exhibiting no heterogeneity between subgroups (except for that in the age subgroup), suggesting that these variables did not alter this effect. Although age may confound the results, we have adjusted it as a covariate in our analysis. Further multivariate stepwise logistic regression results showed that female and beneficial alleles were independent protective factors for NAFLD. A meta-analysis of the burden of NAFLD in China from 2008 to 2018 suggested that the prevalence of NAFLD was higher in males.46 This may be related to the greater levels of serum TG, glucose, and body fat, as well as higher prevalence of hypertension and liver enzyme abnormalities in males.47 Our results also showed that BMI, hypertension, hyperglycemia, hypertriglyceridemia, low HDL-C, and high ALT were independent risk factors for NAFLD. Variables other than ALT are risk factors proposed by the diagnostic criteria for metabolic syndrome.48 ALT elevates mildly in NAFLD, making it an indicator of the biochemistry in the liver.49 What’s more, the importance of these variables in screening NAFLD has also been written into the NAFLD guideline.22 The AUROC of the prediction model implied a good discriminative power. Disappointingly, the results of Hosmer–Lemeshow test showed poor calibration of the predictive model. More studies are needed to establish NAFLD-predicting models with strong abilities of discrimination and calibration. Our predictive models combining genetic and clinical factors may be used in early screening for high-risk NAFLD populations. However, their costs and benefits should be analyzed in future studies.
Some limitations deserve our attention. First, this study was a single-center study and the sample size might not be large enough. We performed frequency matching based on age. Considering the sample size, we did not match sex in two groups. Consequently, we adjusted for sex as a covariate, and performed multivariate and stratified analyses to control the effect of confounding factors. Second, only three SNPs of AKT1 were selected. There is a need to investigate the combined effect of multiple genes on NAFLD susceptibility. In addition, bioinformatic analysis has limitations in predicting the biological functions of SNPs. Further functional studies are needed. Finally, we did not include behavioral factors (such as diet and exercise) in our predictive models. In future, the combined effects of multiple genetic and behavioral factors on NAFLD susceptibility need to be explored in a prospective context and in large multicenter samples of different ethnicities.
Conclusion
In conclusion, this study revealed for the first time that AKT1 (rs2494732 and rs2494752) variants are associated with NAFLD susceptibility in the Chinese Han population. These beneficial SNPs and clinical variables work well together to predict NAFLD susceptibility. Our findings offer hints for further functional research as well as markers for screening high-risk NAFLD populations.
Data Sharing Statement
The original contributions presented in the study were included in the manuscript/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Ethics Review Committee of Nanjing Medical University [NO. (2019) 740], complying with the Declaration of Helsinki. The patients/participants provided their written informed consent to participate in this study.
Acknowledgments
We thank Dr. Yongke Cao for providing English language polishing support and all members who participated in this work.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Six Talent Peaks Project in Jiangsu Province, China (2019, WSN-049), “333 Project” of Jiangsu Province, and Connotation Construction Project of Nanjing Medical University for Priority Academic of Nursing Science (General Office, the People’s Government of Jiangsu Province <2018> No.87).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Xue R, Yang RX, Fan JG. Epidemiological trends and clinical characteristic of NAFLD/MAFLD in Asia. J Dig Dis. 2022;23(7):354–357. doi:10.1111/1751-2980.13117
2. Paik JM, Kabbara K, Eberly KE, Younossi Y, Henry L, Younossi ZM. Global burden of NAFLD and chronic liver disease among adolescents and young adults. Hepatology. 2022;75(5):1204–1217. doi:10.1002/hep.32228
3. Powell EE, Wong VW, Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397(10290):2212–2224. doi:10.1016/s0140-6736(20)32511-3
4. Cotter TG, Rinella M. Nonalcoholic fatty liver disease 2020: the state of the disease. Gastroenterology. 2020;158(7):1851–1864. doi:10.1053/j.gastro.2020.01.052
5. Tilg H, Moschen AR, Roden M. NAFLD and diabetes mellitus. Nat Rev Gastroenterol Hepatol. 2017;14(1):32–42. doi:10.1038/nrgastro.2016.147
6. Targher G, Byrne CD, Tilg H. NAFLD and increased risk of cardiovascular disease: clinical associations, pathophysiological mechanisms and pharmacological implications. Gut. 2020;69(9):1691–1705. doi:10.1136/gutjnl-2020-320622
7. Marcuccilli M, Chonchol M. NAFLD and chronic kidney disease. Int J Mol Sci. 2016;17(4):562. doi:10.3390/ijms17040562
8. Huang R, Fan JG, Shi JP, et al. Health-related quality of life in Chinese population with non-alcoholic fatty liver disease: a national multicenter survey. Health Qual Life Outcomes. 2021;19(1):140. doi:10.1186/s12955-021-01778-w
9. Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24(7):908–922. doi:10.1038/s41591-018-0104-9
10. Meroni M, Longo M, Rustichelli A, Dongiovanni P. Nutrition and genetics in NAFLD: the perfect binomium. Int J Mol Sci. 2020;21(8):2986. doi:10.3390/ijms21082986
11. Eslam M, Sanyal AJ, George J. MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158(7):1999–2014.e1. doi:10.1053/j.gastro.2019.11.312
12. Jeong SH, Kim HB, Kim MC, et al. Hippo-mediated suppression of IRS2/AKT signaling prevents hepatic steatosis and liver cancer. J Clin Invest. 2018;128(3):1010–1025. doi:10.1172/jci95802
13. Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169(3):381–405. doi:10.1016/j.cell.2017.04.001
14. Di Lorenzo A, Fernández-Hernando C, Cirino G, Sessa WC. Akt1 is critical for acute inflammation and histamine-mediated vascular leakage. Proc Natl Acad Sci U S A. 2009;106(34):14552–14557. doi:10.1073/pnas.0904073106
15. Cohen MM Jr. The AKT genes and their roles in various disorders. Am J Med Genet A. 2013;161(12):2931–2937. doi:10.1002/ajmg.a.36101
16. Avan A, Avan A, Le Large TY, et al. AKT1 and SELP polymorphisms predict the risk of developing cachexia in pancreatic cancer patients. PLoS One. 2014;9(9):e108057. doi:10.1371/journal.pone.0108057
17. Wan M, Easton RM, Gleason CE, et al. Loss of Akt1 in mice increases energy expenditure and protects against diet-induced obesity. Mol Cell Biol. 2012;32(1):96–106. doi:10.1128/mcb.05806-11
18. Heljić M, Brazil DP. Protein kinase B/Akt regulation in diabetic kidney disease. Front Biosci. 2011;3(1):98–104. doi:10.2741/s135
19. Yin X, Xu Z, Zhang Z, et al. Association of PI3K/AKT/mTOR pathway genetic variants with type 2 diabetes mellitus in Chinese. Diabetes Res Clin Pract. 2017;128:127–135. doi:10.1016/j.diabres.2017.04.002
20. McKenzie JA, Witkowski S, Ludlow AT, Roth SM, Hagberg JM. AKT1 G205T genotype influences obesity-related metabolic phenotypes and their responses to aerobic exercise training in older Caucasians. Exp Physiol. 2011;96(3):338–347. doi:10.1113/expphysiol.2010.055400
21. Eshaghi FS, Ghazizadeh H, Kazami-Nooreini S, et al. Association of a genetic variant in AKT1 gene with features of the metabolic syndrome. Genes Dis. 2019;6(3):290–295. doi:10.1016/j.gendis.2019.03.002
22. National Workshop on Fatty Liver and Alcoholic Liver Disease, Chinese Society of Hepatology, Chinese Medical Association; Fatty Liver Expert Committee, Chinese Medical Doctor Association. Guidelines of prevention and treatment for nonalcoholic fatty liver disease: a 2018 update. Zhonghua Gan Zang Bing Za Zhi. 2018;26(3):195–203. doi:10.3760/cma.j.issn.1007-3418.2018.03.008
23. Woods NT, Monteiro AN, Thompson ZJ, et al. Interleukin polymorphisms associated with overall survival, disease-free survival, and recurrence in non-small cell lung cancer patients. Mol Carcinog. 2015;54(1):E172–184. doi:10.1002/mc.22275
24. Chinese obesity working group. Prevention and control guide on overweight and obesity in Chinese adults (excerpt). Acta Nutrimenta Sinica. 2004;2004(1):1–4.
25. Joint Committee for Guideline Revision. 2016 Chinese guideline for the management of dyslipidemia in adults. Zhonghua Xin Xue Guan Bing Za Zhi. 2016;44(10):833–853. doi:10.3760/cma.j.issn.0253-3758.2016.10.005
26. Joint Committee for Guideline Revision. 2018 Chinese guidelines for prevention and treatment of hypertension-A report of the revision committee of Chinese guidelines for prevention and treatment of hypertension. J Geriatr Cardiol. 2019;16(3):182–241. doi:10.11909/j.issn.1671-5411.2019.03.014
27. Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36(2):131–137. doi:10.1038/ng1296
28. Chen WS, Xu PZ, Gottlob K, et al. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 2001;15(17):2203–2208. doi:10.1101/gad.913901
29. Chu CY, Chen CF, Rajendran RS, et al. Overexpression of Akt1 enhances adipogenesis and leads to lipoma formation in zebrafish. PLoS One. 2012;7(5):e36474. doi:10.1371/journal.pone.0036474
30. Polyzos SA, Kountouras J, Mantzoros CS. Obesity and nonalcoholic fatty liver disease: from pathophysiology to therapeutics. Metabolism. 2019;92:82–97. doi:10.1016/j.metabol.2018.11.014
31. Karege F, Méary A, Perroud N, et al. Genetic overlap between schizophrenia and bipolar disorder: a study with AKT1 gene variants and clinical phenotypes. Schizophr Res. 2012;135(1–3):8–14. doi:10.1016/j.schres.2011.12.015
32. Morgan CJ, Freeman TP, Powell J, Curran HV. AKT1 genotype moderates the acute psychotomimetic effects of naturalistically smoked cannabis in young cannabis smokers. Transl Psychiatry. 2016;6(2):e738. doi:10.1038/tp.2015.219
33. Li Y, Zhu L, Yao H, et al. Association of inflammation-related gene polymorphisms with susceptibility and radiotherapy sensitivity in head and neck squamous cell carcinoma patients in Northeast China. Front Oncol. 2021;11:651632. doi:10.3389/fonc.2021.651632
34. Jin Y, Yuan Y, Yi M, Han H, Liu B, Li Q. Phosphorylated-Akt overexpression is associated with a higher risk of brain metastasis in patients with non-small cell lung cancer. Biochem Biophys Rep. 2019;18:100625. doi:10.1016/j.bbrep.2019.100625
35. Liemburg EJ, Bruins J, van Beveren N, Islam MA, Alizadeh BZ. Cannabis and a lower BMI in psychosis: what is the role of AKT1? Schizophr Res. 2016;176(2–3):95–99. doi:10.1016/j.schres.2016.08.014
36. Li L, You W, Ren W. The ZJU index is a powerful index for identifying NAFLD in the general Chinese population. Acta Diabetol. 2017;54(10):905–911. doi:10.1007/s00592-017-1024-8
37. Chung TH, Kim JK, Kim JH, Lee YJ. Fatty liver index as a simple and useful predictor for 10-year cardiovascular disease risks determined by Framingham risk score in the general Korean population. J Gastrointestin Liver Dis. 2021;30(2):221–226. doi:10.15403/jgld-3404
38. Lee JH, Kim D, Kim HJ, et al. Hepatic steatosis index: a simple screening tool reflecting nonalcoholic fatty liver disease. Dig Liver Dis. 2010;42(7):503–508. doi:10.1016/j.dld.2009.08.002
39. Chen C, Zhu Z, Mao Y, et al. HbA1c may contribute to the development of non-alcoholic fatty liver disease even at normal-range levels. Biosci Rep. 2020;40(1). doi:10.1042/bsr20193996
40. Wang Z, Fu H, Li W. Association between AKT rs2494752 single nucleotide polymorphism and the risk of metastasis in hepatocellular carcinoma. Oncol Lett. 2018;16(3):3699–3705. doi:10.3892/ol.2018.9060
41. Zhu J, Wang M, He J, et al. Polymorphisms in the AKT1 and AKT2 genes and oesophageal squamous cell carcinoma risk in an Eastern Chinese population. J Cell Mol Med. 2016;20(4):666–677. doi:10.1111/jcmm.12750
42. Wang MY, He J, Zhu ML, et al. A functional polymorphism (rs2494752) in the AKT1 promoter region and gastric adenocarcinoma risk in an Eastern Chinese population. Sci Rep. 2016;6:20008. doi:10.1038/srep20008
43. Li X, Zhang R, Liu Z, Li S, Xu H. The genetic variants in the PTEN/PI3K/AKT pathway predict susceptibility and CE(A)F chemotherapy response to breast cancer and clinical outcomes. Oncotarget. 2017;8(12):20252–20265. doi:10.18632/oncotarget.15690
44. Xu JL, Wang ZW, Hu LM, et al. Genetic variants in the PI3K/PTEN/AKT/mTOR pathway predict platinum-based chemotherapy response of advanced non-small cell lung cancers in a Chinese population. Asian Pac J Cancer Prev. 2012;13(5):2157–2162. doi:10.7314/apjcp.2012.13.5.2157
45. Kermanshahi F, Ghazizadeh H, Hussein NA, et al. Association of a genetic variant in the AKT gene locus and cardiovascular risk factors. Cell Mol Biol. 2020;66(3):57–64. doi:10.14715/cmb/2020.66.3.9
46. Zhou F, Zhou J, Wang W, et al. Unexpected rapid increase in the burden of NAFLD in China from 2008 to 2018: a systematic review and meta-analysis. Hepatology. 2019;70(4):1119–1133. doi:10.1002/hep.30702
47. Hu XY, Li Y, Li LQ, et al. Risk factors and biomarkers of non-alcoholic fatty liver disease: an observational cross-sectional population survey. BMJ Open. 2018;8(4):e019974. doi:10.1136/bmjopen-2017-019974
48. Chinese Diabetes Society: Metabolic Syndrome Cooperative Study Group. The suggestion about metabolic syndrome from Chinese diabetes society. Chin J Diabetes Mellitus. 2004;12(3):156–161.
49. Liu Z, Que S, Xu J, Peng T. Alanine aminotransferase-old biomarker and new concept: a review. Int J Med Sci. 2014;11(9):925–935. doi:10.7150/ijms.8951
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.