Back to Journals » Journal of Inflammation Research » Volume 16
Fusing WGCNA and Machine Learning for Immune-Related Gene Prognostic Index in Lung Adenocarcinoma: Precision Prognosis, Tumor Microenvironment Profiling, and Biomarker Discovery
Authors He J, Luan T, Zhao G, Yang Y
Received 19 September 2023
Accepted for publication 3 November 2023
Published 16 November 2023 Volume 2023:16 Pages 5309—5326
DOI https://doi.org/10.2147/JIR.S436431
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Adam D Bachstetter
Jiaming He,1,2,* Tiankuo Luan,3,* Gang Zhao,4 Yingxue Yang5
1Laboratory of Stem Cells and Tissue Engineering, Department of Histology and Embryology, Chongqing Medical University, Chongqing, 400016, People’s Republic of China; 2Institute of Life Sciences, Chongqing Medical University, Chongqing, 400016, People’s Republic of China; 3Department of Anatomy, Chongqing Medical University, Chongqing, People’s Republic of China; 4Department of Gastroenterology, Wushan County People’s Hospital of Chongqing, Chongqing, 404700, People’s Republic of China; 5Department of Gastroenterology, the Second Affiliated Hospital of Chongqing Medical University, Chongqing, 400016, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yingxue Yang, Email [email protected]
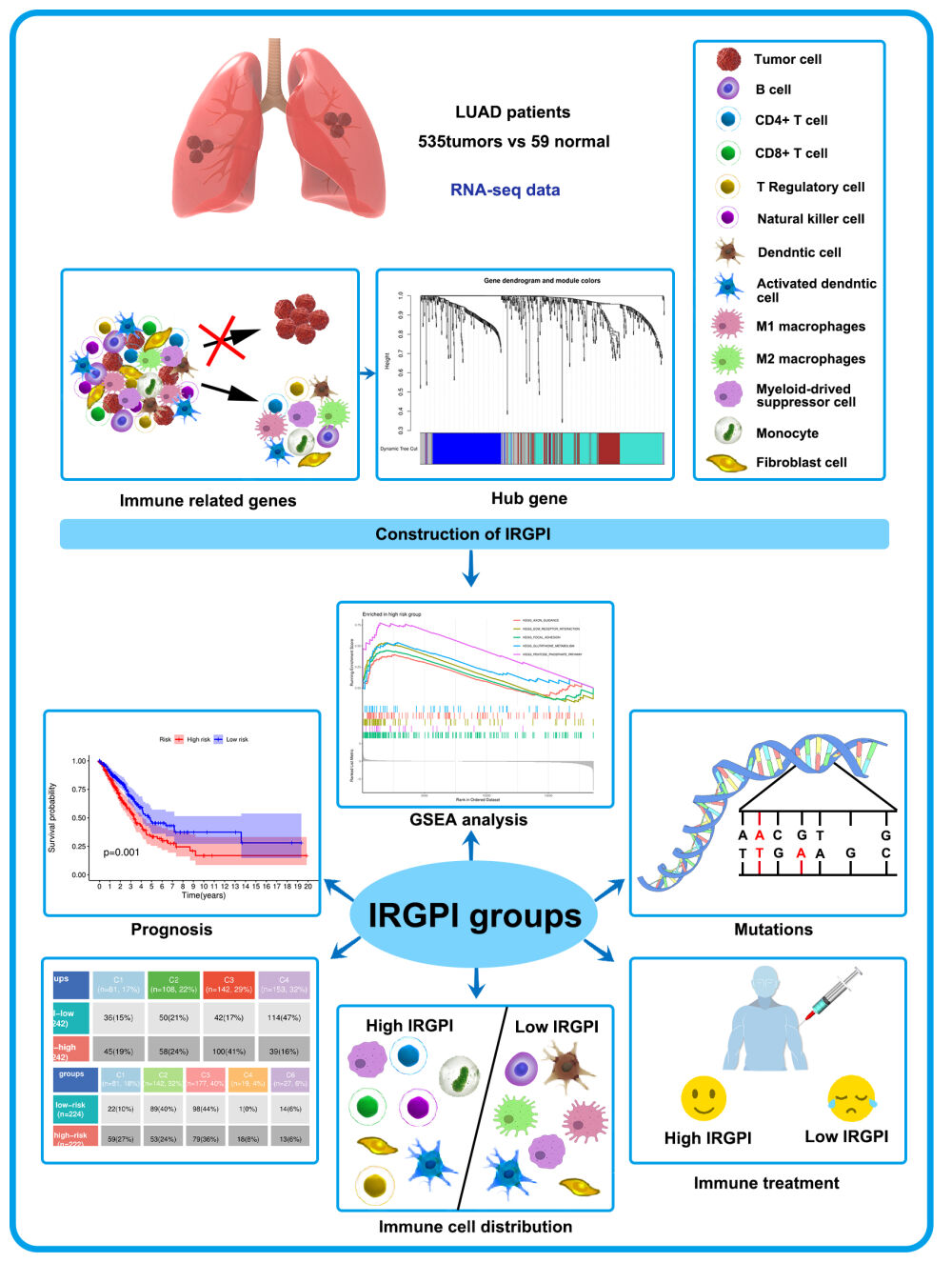
Background: The objective is to create an IRGPI (Immune-related genes prognostic index), which could predict the survival and effectiveness of immune checkpoint inhibitor (ICI) treatment for lung adenocarcinoma (LUAD).
Methods: By applying weighted gene co-expression network analysis (WGCNA), we ascertained 13 genes associated with immune functions. An IRGPI was constructed using four genes through multicox regression, and its validity was assessed in the GEO dataset. Next, we explored the immunological and molecular attributes and advantages of ICI treatment in subcategories delineated by IRGPI. The model genes were also validated by the random forest tree, and functional experiments were conducted to validate it.
Results: The IRGPI relied on the genes CD79A, IL11, CTLA-4, and CD27. Individuals categorized as low-risk exhibited significantly improved overall survival in comparison to those classified as high-risk. Extensive findings indicated that the low-risk category exhibited associations with immune pathways, significant infiltration of CD8 T cells, M1 macrophages, and CD4 T cells, a reduced rate of gene mutations, and improved sensitivity to ICI therapy. Conversely, the higher-risk group displayed metabolic signals, elevated frequencies of TP53, KRAS, and KEAP1 mutations, escalated levels of NK cells, M0, and M2 macrophage infiltration, and a diminished response to ICI therapy. Additionally, our study unveiled that the downregulation of IL11 effectively impedes the proliferation and migration of lung carcinoma cells, while also inducing cell cycle arrest.
Conclusion: IRGPI is a biomarker with significant potential for predicting the effectiveness of ICI treatment in LUAD patients and is closely related to the microenvironment and clinicopathological characteristics.
Keywords: lung adenocarcinoma, immune-related genes, bioinformatics, prognostic index, IL11
Graphical Abstract:
Introduction
Therapies targeting programmed death 1 (PD1) and its ligand (PD-L1), as well as CTL-associated protein 4 (CTLA-4), referred to ICI, exhibit remarkable survival benefits compared to conventional treatments.1–4 Over the decades, the application of ICI has demonstrated promising results in treating LUAD patients.5–7 Nevertheless, a primary worry revolves around the inadequate patient response rate to ICI response rates.8–11 The effectiveness of ICI is influenced by multiple elements, including the tumor immune microenvironment (TME), whereas only a few indicators can forecast patient results.12 Exploring possible prognostic-related indicators for treatment advantages can enable personalized immunotherapy for patients with LUAD. Nevertheless, the evaluation of LUAD’s TME and its prognostic and therapeutic indicators remains incomplete.
We are committed to constructing a predictive model that can predict the response of LUAD patients to ICI treatment. By analyzing sequencing results of LUAD patients, we identified immune-related genes and employed WGCNA to construct an IRGPI to screen immune-related hub genes linked to patient prognosis. Subsequently, we delineated the immunological and molecular characteristics of IRGPI and assessed its prognostic implications in individuals receiving immunotherapy. Additionally, we compared alternative biomarkers, namely tumor inflammation signature (TIS) and tumor immune dysfunction and exclusion (TIDE). The creation of the IRGPI aimed to establish a robust prognostic predictor for patients afflicted by LUAD undergoing immunotherapeutic interventions (Graphical abstract).
Materials and Methods
Collection of Patient Data
The Cancer Genome Atlas (TCGA) provided RNA-seq data, mutation gene data, survival information for 535 LUAD samples and 59 normal samples, and associated clinicopathological details. The datasets were obtained from the online repository available at (https://portal.gdc.cancer.gov/). Survival data and RNA-seq profiles for 442 LUAD samples (GSE72094) were sourced from the GEO database accessible at https://www.ncbi.nlm.nih.gov/geo/. The lists of immune-related genes were acquired from the ImmPort (https://www.immport.org/shared/home) and InnateDB (https://www.innateDBdb.com/) databases.
Identification of Immune-Related Genes (IRGs)
After obtaining the RNA-seq data, we utilized the “limma” package in R to identify different expressed genes (DEGs) with a significance level of P <0.05 and an absolute log2 fold change (|log2FC|) exceeding 1. Subsequently, we successfully pinpointed a set of DEGs associated with the immune system by intersecting these DEGs with the IRGs retrieved from ImmPort and InnateDB databases; we. Then, we employed the “clusterProfiler” R package to perform enrichment analyses involving Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
After constructing co-expression survival-related modules using WGCNA, potential prognostic markers in LUAD were discovered.13 Thirteen immune-related hub genes were significantly linked to survival (analyzed using the “survival” package) and subsequently subjected to further examination (P <0.05, Log rank test).
The IRGPI Was Constructed and Validated
Thirteen immune-related core genes were identified through Multivariate Cox regression analyses and subsequently used to construct the IRGPI. By adding the products of coefficients and expression levels of genes, including Gene 1, Gene 2, and so on, up to Gene n, we calculated the risk score referred to as IRGPI. The predictive capability of IRGPI was assessed using Kaplan-Meier survival curves. By employing univariate and multivariate Cox regression analyses, the potential prognostic value of IRGPI was comprehensively evaluated. Furthermore, the accuracy and sensitivity of IRGPI were assessed using R through the validation of receiver operating characteristic (ROC) curve analysis.
Analysis of Gene Sets Enrichment
Utilizing the “clusterProfiler” R package, we conducted a Gene Set Enrichment Analysis (GSEA) on the IRGPI score by dividing it into high and low groups based on the median value.
Analysis of Gene Mutations Burden
The R software package “Maftools”14 was selected for the analysis of somatic mutation profiles between the two IRGPI subgroups. Subsequent analyses explored the correlation between the overall tumor mutation burden (TMB) and IRGPI.
Analysis of Immune Infiltration and the Effectiveness of ICI Treatment
To determine the immune characteristics of 535 LUAD samples, we applied the CIBERSORT algorithm to assess the proportions of 22 distinct immune cell categories.15 We utilized TIMER2.0 for a more comprehensive analysis of immune cell infiltration proportions and visualized the results using the “ggplot2” package in the R programming language.16 The infiltration estimation profile for all TCGA tumors is also available for download from TIMER2.0.
Subsequently, we conducted ssGSEA analysis to obtain scores for specific gene features and compared the biological characteristic changes among different IRGPI groups. Correlation investigations were undertaken to explore the link between the IRGPI score and the expression of PD-L1.
In addition, we executed time-dependent ROC curve assessments to derive the area under the curve (AUC), and we contrasted the predictive importance among IRGPI, TIDE, and TIS by employing the timeROC package within the R environment. The TIDE score was computed online via the following web address (https://tide.dfci.harvard.edu/), and the TIS score was determined as the average logarithmically scaled normalized expression of the 18 characteristic genes.17,18
Cell Culture
The Chongqing Key Laboratory of Stem Cells and Tissue Engineering provided the LUAD cell lines (A549 and H1299). The cells were cultured at 37°C in a humidified environment with 5% CO2 using RPMI-1640 medium (Gibco BRL, USA) supplemented with 10% fetal bovine serum, and 1% penicillin-streptomycin solution (C100C5, NCM Biotech, Suzhou, China).
Transient Transfection
Tsingke (Shanghai, China) designed and synthesized four shRNAs, including three IL-11 shRNAs and one negative control shRNA. All shRNAs were constructed in plko.1-copGFP-puro vector. The shRNA sequences are listed in Table S1. And, shRNAs were introduced into the cells by Lipo 3000 (Invitrogen, Carlsbad, CA, USA) following the instructions provided by the manufacturer.
Cell Viability Assay and 5-Ethynyl-2′-Deoxyuridine (EdU) Assay
To assess cell viability, the Cell counting Kit-8 (CCK8, NCM Biotech, Suzhou, China) was employed. Following various procedures, the cells were placed in 96-well plates with a concentration of 2.5×103 cells per well for 24, 48, and 72 hours, correspondingly. Next, 10 microliters of CCK8 were added following the provided guidelines. The measurement of absorbance was observed at a wavelength of 450 nm.
The EdU cell proliferation kit (Cellorlab, CX004, Shanghai, China) was also employed to examine cell proliferation by the manufacturer. Images were captured and analyzed using a fluorescence microscope (Leica, Munich, Germany).
Scratch Assay
The cells were introduced into the 6-well dish. Once the cells reached full growth, a sterilized 200 μL head was used to create a straight scratch line. Following 24 hours, a random selection was made from each group to observe and capture photographs of the field of view.
Extraction of RNA and Performing Real-Time PCR
The Trizol reagent (15596018, Thermo) was used to extract the entire cellular RNA. Reverse transcription was performed following the protocol of ABScript cDNA First-Strand Synthesis Kit (RK20400, Wuhan, Abclonal). Genious 2x SYBR Green Fast qPCR Mix (RK21204, Wuhan, Abclonal) was used for conducting real-time PCR.
The primers are listed in Table S1. The relative expression levels were quantified using the 2−ΔΔCt method.
Random Forest and GSEA Screening and Exploring Important Genes
The effect of distinct gene expression on patient OS was evaluated using the random forest algorithm provided by the R package “randomForest”. This analysis also yielded variable importance values for each gene.19
To investigate the mechanism through which IL11 regulates the progression of LUAD, a co-expression network related to IL11 was constructed within the TCGA-LUAD dataset. This was achieved by utilizing a functional module derived from the WEB-based gene set analysis toolkit of the LinkedOmics database.
Cell Cycle Analysis
The cells were enzymatically digested following transfection using a 0.1% trypsin solution. Next, the cells were spun at a temperature of 4°C at 500 rpm for 3 mins. Then, the cells were rinsed with PBS on two occasions. The pellets were immersed in ice-cold 70% ethanol and treated with propidium iodide for staining. The CELL Quest kit (BD Biosciences, San Jose, CA, USA) was utilized for data analysis.
Statistical Analysis
An independent t-test was employed to compare continuous variables between the two groups. For categorical data analysis, the chi-square test was utilized. The Wilcoxon signed-rank test was also applied to assess immune infiltration differences among the subgroups. The comparison of TIDE scores between groups was conducted using the Wilcoxon test. Values of P <0.05 were used to determine statistical significance. P values less than 0.05 were represented by *P values less t0.01 were represented by **and P values less than 0.001 were represented by ***. The abbreviation “ns” was used to indicate results that were not statistically significant.
Results
Identification of DIRGs
In the analysis of differential expression (535 tumors compared to 59 normal samples), a total of 8109 DEGs were acquired from tumor samples compared to normal samples (Figure S1A). By overlapping these genes with IRGs sourced from ImmPort and InnateDB databases, a collective count of 681 genes associated with the immune system and displaying differential expression (DIRGs) was pinpointed. Within this set, 423 genes demonstrated upregulation, whereas 258 genes exhibited downregulation (Figure S1B). The DIRGs contained 1680 GO terms and 59 KEGG pathways (see Table S2), and Figure S1C and S1D display the top 20 enrichments for GO and KEGG results.
The candidate genes (totaling 681) underwent WGCNA analysis to identify key genes linked with immune processes. Achieving a correlation coefficient exceeding 0.9, the scale-free network’s ideal soft-thresholding exponent was pinpointed as 4, as indicated in Figure S2. The identification of four distinct modules followed this. Utilizing the best-suited soft-thresholding exponent and average linkage hierarchical clustering, four modules were distinguished (Figure S3A and S3B). In terms of the Pearson correlation coefficient measuring the relationship between each module and sample characteristics, it was observed that the blue module exhibited a robust correlation with LUAD tumors. Consequently, the genes within this module were utilized for further analysis. Figure S3C and S3D display separate GO terms and KEGG enrichments for the genes of the modules (additional information in Table S3). The link between the OS of patients with LUAD and the expression levels of 13 hub genes related to immunity was assessed through Kaplan-Meier analysis, illustrated in Figure S4.
Next, we investigated the attributes of the 13 hub genes. According to Figure S5, a portion of the 13 hub genes related to the immune system exhibited missense and nonsense mutations, with IGHM, TLR10, and IRF4 showing a mutation rate of 2%.
IRGPI Predicts Survival of LUAD Patients
Univariate Cox regression analyses were performed to identify the independent genes associated (Figure 1A) with prognosis regarding OS. The OS of LUAD patients was significantly influenced by 13 genes (CD79A, CD19, IGHD, IGHM, IGHV7-81, IGKV3D-15, IL11, TNFRSF13C, TNFRSF17, CTLA-4, IRF4, CD27, and TLR10), as depicted in Figure S4. By employing multifactorial Cox regression assessments on datasets from both TCGA and GEO, we created a prognostic index called IRGPI. The IRGPI risk score was calculated by multiplying the expression of a specific gene in the sample by its weight in the multivariate Cox model and then summing it. This index was calculated using the following formula: IRGPI = (−0.330) * CD79A expression levels + 0.356 * IL11 expression levels + (−0.213) * CTLA-4 expression level + 0.339 * CD27 expression levels, after merging survival states and times.
Patients with LUAD were categorized into low- and high-risk groups by utilizing the median risk score value of IRGPI. Compared to patients in the high-risk group, patients in the low-risk group demonstrated superior overall survival (OS, P = 0.001, based on the Log rank test) (Figure 1B). Subsequently, IRGPI was validated using the LUAD dataset GSE72094 (n = 442) to confirm its role. The findings from the GEO cohort were in line with the TCGA dataset (P = 0.001, Log rank test) (Figure 1C).
Moreover, the reliability of IRGPI was evaluated of time-dependent ROC curves (Figure 1D and E). In both cohorts, the AUC exceeded 0.6 for survival at 1-, 3-, and 5-year intervals. Furthermore, IRGPI demonstrated a strong ability to predict survival in both the TCGA and GEO groups accurately.
IRGPI is a Dependent Prognostic Indicator
In Table S4, an inclusive presentation of clinicopathological characteristics about LUAD patients from the TCGA dataset was showcased, predominantly falling within the two IRGPI subgroups. Figure 1F showed that the prognosis of LUAD was significantly correlated with tumor and TNM stages according to the results of univariate Cox analyses. According to the results of multivariate Cox analyses, IRGPI was identified as a prognostic factor that acts independently (Figure 1G).
Molecular Characteristics of Different IRGPI Subgroups
According to GSEA analysis, it was ascertained that the high-risk cluster displayed notable enhancement in terms of axon guidance and metabolic-related pathways (Figure 2A). Conversely, the low-risk group showed significant enrichment in pathways associated with immune response and cell receptors (Figure 2B and Table S5) (P <0.05). Table S6 listed the genes that showed differential expression between the two subgroups.
Afterward, we examined genetic mutations to clarify the immunological characteristics of the IRGPI subcategories. The most frequent type of mutation was missense variations, followed by multi-hit and nonsense mutations. Figure 2C and D revealed that the IRGPI subgroups exhibited the most elevated mutation rates among the top 20 genes. In both groups, the mutation frequencies of TP53, MUC16, TTN, RYR2, LRP1B, CSMD3, ZFHX4, KRAS, USH2A, SPTA1, and FLG exceeded 20%.
Next, we investigate the correlation between IRGPI score and the expression of PD-L1 (also named CD274), as well as TMB. Consequently, the correlation between PD-L1 and the IRGPI score was significant, whereas the association between the IRGPI score and TMB was minor (r = 0.05, P = 0.27), as depicted in Figure S6.
The IRGPI Forecasted the Infiltration of Immune Cells into the Microenvironment of LUAD
To examine the makeup of immune cells in different IRGPI subcategories, we employed the Wilcoxon test to compare the distribution of immune cells among various IRGPI subgroups. In the high-risk subgroup, there was a higher presence of monocytes, M0 and M2 macrophages, activated mast cells, neutrophils, and activated dendritic cells. Conversely, the low-risk subgroup had abundant naïve B cells, plasma cells, CD8 T cells, resting memory CD4 T cells, activated memory CD4 T cells, Tregs, and M1 macrophages (Figure 3A). Figure 3B displays the clinicopathological characteristics of various IRGPI subgroups associated with the immune landscape’s attributes. Using the multi-platform algorithm, we utilized a bubble chart to display the findings of immune infiltration in Figure 3C and Table S7.
Next, we utilized specific gene patterns to characterize the immune and molecular activities among distinct IRGPI subcategories. Consequently, the high-risk subgroup exhibited an increased presence of immunosuppressive cells and metabolism signals. In contrast, the low-risk subgroup displayed a higher abundance of CD8 T cells and MHC class I molecules (Figure 2A, B and 3D).
We additionally examined if the predictive significance of IRGPI arose from enhanced immune regulation or reduced malignant tumor progression. According to Figure S7, it is evident that individuals with a favorable outlook exhibit higher levels of resting dendritic cells, naïve B cells, resting mast cells, monocytes, and plasma cells. Conversely, those with a poorer prognosis display increased levels of activated dendritic cells, M0 macrophages, resting NK cells, and activated memory CD4 T cells. Hence, we proposed that the correlation between IRGPI and immune cell infiltrations indicates its ability to represent the tumor microenvironment (TME) state.
Relationship Between IRGPI Groups Based on Other Immune and Molecular Subtypes
We then evaluated the accuracy of our model in other reported immune subtype studies20 which contained Cluster (C1-C4). Figure 4A demonstrates that the distribution of C1 and C2 subtypes was nearly equal in both groups, while the high-risk subgroup had a higher proportion of C3 compared to the low-risk subgroup (P <0.001, chi-square test). Next, an in-depth immunogenomic analysis21 was used to categorize 446 immune samples further. According to the data in Figure 4B, the low-risk subgroup had a higher prevalence of C2 and C3 subtypes, whereas the high-risk subgroup had a more significant proportion of C4 subtypes (P <0.001, chi-square test).
Consistently reported in extensive genomic profiling studies of LUAD, three different molecular subtypes (n=221) are the proximal-inflammatory (PI), terminal respiratory unit (TRU), and proximal-proliferative (PP).22 Nevertheless, the subtype was nearly evenly distributed among both groups (Figure S8) (P = 0.804, chi-square test).
IRGPI Predicts the Benefit of Immunotherapy
Subsequently, TIDE was employed to evaluate immunotherapy’s potential clinical benefits in different IRGPI subcategories. A high level of TIDE indicates a greater likelihood of immune evasion, which implies that patients undergoing ICI therapy may experience poorer outcomes. However, our findings indicate that the low-risk subgroup displayed a greater TIDE score than the high-risk subgroup, contradicting their OS duration (Figure 5A). Moreover, the subgroup at a greater risk demonstrated a significant microsatellite instability (MSI) score and a substantial T cell exclusions. In contrast, the subgroup at a lower risk displayed a notable T cell dysfunction score. Therefore, the decreased MSI and reduced T-cell exclusion score in low-risk patients might contribute to favorable results. Subsequently, it was discovered that the IRGPI exhibited superior AUCs compared to TIDE and TIS (Figure S9). Therefore, we proposed that the prognostic significance of IRGPI was similar to that of the 18-gene T cell-inflamed signature (TIS) and TIDE for OS in both groups. Furthermore, we evaluated the prognostic importance of IRGPI within the urothelial carcinoma (UC) cohort subjected to treatment involving anti-PD-L1 therapy.23 Based on the findings in Figures 5B and C, it is evident that individuals classified as low-risk experienced more favorable OS results compared to those in the high-risk category.
The Function Role of IL11 in Lung Cancer Cells
Additionally, we also aim to investigate the biological roles of the IRGPI genes. In the TCGA cohort (Figure 6A and B), we utilize the random forest algorithm to compute the impact weights of the model genes. Interestingly, IL11 ranks the highest. Furthermore, the predictive models demonstrated the significance of IL11 expression in the prognosis of LUAD patients, with its highest coefficient score. Consequently, we initiated an investigation into the biological role of this in lung cancer cells H1299 and A549. We designed three different knockdown sequences and performed RT-PCR verification. Our subsequent experiments proved that IL11#1 had the highest efficiency (Figure 6C) among them. The proliferation and migration of lung cancer cells in CCK8, EdU, and wound-healing analysis (Figure 6D–H) were influenced by IL11. Subsequently, utilizing the GSEA examination, we discovered that IL11 played a role in influencing the cell cycle of individuals with LUAD (Figure 6I and J). Then, flow cytometry was conducted and revealed that the inhibition of IL11 can impede the progression of lung cancer cells into the G0/G1 phase (Figure 6K–L). Hence, we thought IL11 could be a promising target for treating LUAD.
Discussion
In numerous clinical studies, ICI treatment has proven to be efficacious for patients diagnosed with advanced and metastatic LUAD.5–7 By implementing this approach, the utilization of ICI will continue to be a fundamental aspect of cancer therapy.24 Since the overall success rate of ICI treatment remains minimal,8–11 it is imperative to carefully choose patients based on pertinent biomarkers to maximize the benefits of these therapies. Despite extensive evaluation of various prognostic markers, a validated biomarker for predicting immunotherapeutic responses and OS in LUAD remains elusive. Identifying a prognostic biomarker for immunotherapy in LUAD patients based on clinicopathological factors is crucial.
According to a recent investigation, effectively foretelling the responsiveness to ICI treatment is likely to demand the amalgamation of TME in conjunction with establishing and validating markers linked to the immune system.25 WGCNA is a virtual method that can be employed to discover potential markers or targets related to the immune system. Using WGCNA on LUAD immune gene datasets, impacting LUAD patients’ OSact the OS of LUAD patients. Then, our IRGPI was constructed by multiple Cox regression analysis. IRGPI was calculated by multiplying the expression data of certain genes for each sample by their weights in the Cox model and then summing them up. Therefore, we developed the IRGPI by considering four genes (CD79A, IL11, CTLA-4, and CD27), which were found to be significant prognostic factors for OS independently. In both TCGA and GEO cohorts, the IRGPI demonstrated its validity as a prognostic immune-related biomarker for LUAD, exhibiting improved survival in low-risk subgroups and decreased survival in high-risk subgroups.
The IRGPI consisted of four genes, namely CD79A, IL11, CTLA-4, and CD27. CD79A is a B cell receptor.CD79A proteins are expressed throughout the entire process of B-cell development,26 while the majority of T-cell neoplasms lack CD79A protein expression.27 According to a recent investigation, the heightened presence of CD79A+ B cells within the stromal environment is a favorable predictive factor for individuals undergoing surgery due to colorectal liver metastasis.28 Nevertheless, the precise regulatory mechanism of CD79A in LUAD remains uncertain. IL11 belongs to the glycoprotein (GP) 130 cytokine family, which also has IL6 and IL27.29 These cytokines have a common receptor (GP130) plus additional specific -receptors.30 The IL11R (IL11-receptor) has been detected in lymphocytes, macrophages, endothelial cells, B-cells, and hematopoietic cells.IL11 is rare in the serum of healthy people.31 However, the amplification of IL11 and IL11R has been detected in lung, colorectal, gastric, breast, and prostate tumors, as well as in osteosarcoma. This suggests that IL11 signaling is crucial in the connection between cancer and inflammation. IL11 acts as an anti-inflammatory cytokine and directly affects macrophages and other effector cells at sites of inflammation.32,33 In a recent study, the potential of IL11 as a diagnostic biomarker for lung adenocarcinoma in BALF specimens is described.34 Notably, CTLA-4, also called CD152, has been incorporated into the IRGPI gene. The site has received approval as an ICI therapy location for patients with LUAD.35 T cells express the CTLA-4 molecule, a CD28 homolog.36,37 In the initiation stage of T cell activation,38 CTLA-4 substantially regulates T cell functions. Increased levels of CTLA-4 on both CD8+ and CD4+ T cells hinder the activation signals caused by the binding of CD28-CD80 or -CD82 and the binding of T cell receptor-MHC.39,40 CD27, a member of the superfamily of tumor necrosis factor receptors, is a molecule that acts as a co-stimulator and is present on T lymphocytes even in the absence of stimulation. By interacting with its CD70 receptor, it has the potential to enhance the production of plasma cells, the growth of B cells, the secretion of immunoglobulins, the formation of B-cell memory, and the initiation of cytolytic activities in natural killer (NK) cells. Furthermore, the interaction between CD27 and CD70 can also facilitate the activation, survival, and proliferation of T-cells, as well as the development of effector functions and T-cell memory, through the stimulation of concomitant antigen-associated T-cell receptors (TCR).41 The IRGPI calculation formula included negative coefficients for CD79A and CTLA-4 and positive coefficients for IL11 and CD27. Hence, a detrimental correlation existed between IRGPI and CD79A, as well as CTLA-4, whereas a favorable correlation was observed between IRGPI and IL11, along with CD27. Additionally, we employed the random forest technique, which aligns with the top IL11 coefficient in the IRGPI model, and the IL11 score obtained is also the highest. Furthermore, IL11 is involved in the cellular cycle of LUAD and impacts the advancement of lung tumor cells, further confirming the significance of the model. To summarize, IRGPI served as a marker linked to ongoitumor growth inhibitionibition of tumor growth.
To clarify the immunological characteristics of IRGPI subcategories, we assessed their genetic mutations. Missense mutations were highly prevalent, followed by multiple and nonsense mutations. We focused on genes which with significant variations of mutations between subgroups, such as ZFHX4 (32% vs 25%), XIRP2 (25% vs 18%), KRAS (28% vs 22%), KEAP1 (20% vs 14%), LRP1B (31% vs 26%), and TP53 (46% vs.41%). ZFHX4 is a member of the zinc-finger homeobox transcription factor protein family, which encompasses a total of 42 members. The suppression of this regulator, which controls the commencement phase of glioblastoma tumor-forming cells and oversees the activity of SOX2 and CHD4, not only hampers cancer progression but also results in prolonged survival for glioblastoma patients.42 Elevated ZFHX4 levels were observed in LUAD tissue samples, which were associated with aggressive characteristics of immunotherapy outcomes LUAD and an unfavorable prognosis.43 XIRP2 safeguards against the breakdown of actin filaments, a process that is particularly altered in metastatic breast cancer and gastric cancer.44–46 LRP1B is commonly mutated in NSCLC, and its mutations are correlated with high TMB. Frequent inactivated mutations of LRP1B have been reported in many cancers. Furthermore, the association between LRP1B genetic alterations and TMB remained unaffected by TP53 mutations.47 KRAS mutations are present in about 30% of NSCLCs.48 In KRAS-mutant LUAD, ICI efficacies are consistently high.49 KRAS-mutant patients who had high KEAP1 mutation frequencies exhibited limited expression of immune biomarkers, including PD-L1. Mutations in KEAP1 are linked to reduced survival in patients with LUAD who undergo chemotherapy or receive single-agent ICI.50,51 Furthermore, individuals with KEAP1 gene alterations exhibited an elevated tumor mutational burden compared to those with the wild-type gene.50 Mutations in TP53 serve as prognostic indicators and are linked to elevated mutation load in NSCLC. This could be utilized to forecast clinical reactions for combined therapies targeting CTLA-4 and PDL1 in NSCLC.52 According to our IRGPI model, we hypothesized that specific genetic mutations in tumors impact the tumor microenvironment (TME). This impact includes increased expression of immune checkpoints, secretion of pro-inflammatory factors, and recruitment of immune cells. Consequently, these mutations affect the effectiveness of treatments and the outcomes of immunotheThereforeents.53,54 Consequently, subgroups at greater risk with an increased number of genetic mutations experience poorer treatment results, aligning with our observations on survival.
Next, we investigate the correlation between IRGPI and predictive indicators for immunotherapy, such as PD-L1 and TMB. In general, neoplasms displaying PD-L1 expression typically demonstrate a more favorable reaction to treatments directed at PD-1/PD-L1 than neoplasms lacking PD-L1 expression, particularly those lacking PD-L1 expression. The IRGPI supported the differentiation in PD-L1 expression between the two groups. Furthermore, we believed that the localization and strength of PD-L1 expression hold greater significance in immunohistochemistry compared to transcriptome data. Hence, additional investigation is required to elucidate the connection between PD-L1 and IRGPI. Moreover, TMB has the potential to serve as an indicator for predicting the efficacy of ICI treatment in LUAD. We found, however, that the IRGPI risk does not differ from the risk associated with a single TMB. Additionally, our IRGPI includes the CTLA-4 gene, which has been utilized in ICI therapy. Furthermore, individuals who were administered nivolumab (an anti-PD1 drug) in combination with ipilimumab (an anti-CTLA-4 drug) experienced a positive impact on their survival, irrespective of their TMB level.35,55 Therefore, there were alternative mechanisms at play.
Various alterations in the TME can significantly impact and dictate diverse immunotherapeutic reactions. Gaining insight into the terrain of the tumor microenvironment (TME) might present a novel approach to managing LUAD.56 Nevertheless, the immune cell compositions varied among the subgroups of IRGPI. Furthermore, the existing computational methods for estimating immune infiltration can be classified into two primary groups: gene signature-based techniques (xCell, MCP-counter) and deconvolution-based methods (CIBERSORT, TIMER, EPIC, quanTIseq). The group organized as high risk exhibited a notable presence of Monocytes, M0 and M2 macrophages, activated dendritic cells, neutrophils, and activated mast cells as determined by the application of CIBERSORT. On the other hand, the low-risk subgroup exhibited an abundance of naïve B cells, plasma cells, CD8 T cells, resting memory CD4 T cells, activated memory CD4 T cells, Tregs, and M1 macrophages. Additionally, the high-risk group showed a higher presence of immune cells on various platforms displayed in the immune cell bubble chart, including common lymphoid progenitor, neutrophil at xCELL, and CD4+ T cell at QUANTISEQ (all P <0.05). Utilizing various algorithms offers enhancements to previous publications that relied solely on the CIBERSORT algorithm. Furthermore, as the accessibility of tumor profile data obtained through single-cell techniques continues to grow, there will be advancements in characterizing the patterns and computational estimation approaches for immune cells that infiltrate tumors. Recent studies have shown that in immune cells with differential expression, a favorable prognosis is suggested by the presence of high levels of T cell infiltrations, specifically cytotoxic CD8 T cells.57–59 M1 Macrophages in the majority of cancer types primarily generate proinflammatory substances, which could be associated with acute inflammation and strongly linked to improved prognostic outcomes in non-small cell lung cancer (NSCLC), hepatocellular carcinoma (HCC), ovarian cancer, and gastric cancer. On the other hand, in general, M2 macrophages promote the proliferation and formation of invasive characteristics. The densities of M2 macrophages are associated with chronic inflammation and the emergence of an unfavorable prognosis in breast, ovarian, bladder, gastric, and prostate cancers.57,58 Our study findings substantiate these conclusions. Furthermore, it was discovered that the group at a greater risk exhibited a higher presence of cells that suppress the immune system, signals related to metabolism, and a more active advancement of tumors. Conversely, the group at a lower risk demonstrated a heightened capacity for repairing damage and signs associated with the immune system. The suggestion was that the subgroup with low risk experienced a favorable outcome in terms of survival.
By integrating with other clinicopathological features, as well as molecular and cellular characteristics, the IRGPI grouping can be identified and validated as a distinct immune subtype of LUAD. Regarding the classification of LUAD immune subtypes, the high-risk subgroup had a higher number of patients classified as the C3 subtype and a lower number of patients classified as the C4 subtype. According to Song’s research, the C3 group, also known as the immune-cold subtype, exhibited the least amount of B cell infiltrations and T cell receptor (TCR) repertoire diversity. Additionally, it had higher levels of neoantigens and mutation rates, leading to poorer prognostic outcomes in patients with LUAD. The C4, also known as the immune-hot subtype, exhibited significant infiltration of immune cells, a wide range of leukocyte fractions, and diverse TCR and B cell receptor (BCR) repertoires.20 Consequently, individuals in the low-risk category of IRGPI experience more significant advantages from ICI therapy due to their heightened immune response towards the development and advancement of tumors, in contrast to those with a high risk of IRGPI. Moreover, Stemming from a consistent and replicable immune subtype, the low-risk subgroup exhibited a higher proportion of C2 (IFN-dominant) and C3 (inflammatory). In contrast, the high-risk subgroup had a greater prevalence of C1 (wound healing) and C4 (lymphocyte depleted).21 C2 displayed elevated levels of M1/M2 macrophage polarizations and robust CD8 signals. Moreover, it revealed high proliferative rates with overriding progressing type I immune responses. C3 was characterized by elevated expression of Th17 and Th1 genes, moderate to low proliferation of tumor cells, and reduced levels of aneuploidy. Tumors categorized under C2 and C3 have the potential for a favorable prognosis. On the other hand, C1 demonstrated the expression of genes related to angiogenesis, showed high proliferation rates, and preferred Th2 cells in adaptive immune infiltrations, resulting in poorer outcomes. Furthermore, C4 demonstrated a plentiful macrophage pattern characterized by diminished Th1 and an increased M2 reaction.21 The results suggested which was in agreement with our present study. In order to integrate the classification of lung adenocarcinoma based on anatomic, histopathological, and mutational characteristics, specifically TRU, PI, and PP,22 the naming of transcription subtypes is combined. The PP subtype was enriched with KRAS mutations, along with the inactivated STK11 tumor suppressor gene. In comparison, the PI subtype was characterized by dense histopathology and concurrent mutations of TP53 and NF1. Moreover, the TRU subtype exhibited a higher prevalence of EGFR-mutated tumors and tumors expressing kinase fusion, associated with a favorable prognosis. The results show that the three different subtypes (n = 221) are evenly distributed among the two subgroups (P = 0.804, chi-square test). One explanation for our results may be that the IRGPI can predict the immune risk not approved by this classification. Another theory suggests that the TCGA database does not primarily consist of most LUAD patients, with a total of 221 patients (n = 221).
Hence, variations in two IRGPI within the TME could indicate distinct immune advantages derived from ICI treatment (specifically anti-PD1 and anti-CTLA-4) as determined by TIDE. TIDE showed a correlation between the onset of T cell dysfunctions in tumors with high levels of cytotoxic T lymphocyte (CTL) infiltration and the absence of T cell infiltrations in cancers with low CTL levels, which can be used to predict immune-checkpoint blockade (ICB) responses. This method involves computational modeling of the dual processes responsible for cancer’s evasion of the immune system. Our research revealed that the high-risk group demonstrated elevated T cell exclusion and MSI scores while exhibiting lower scores for TIDE and T cell dysfunction. The group with minimal risk experienced the contrary result. The lower T cell exclusion and MSI score were the primary factors contributing to the favorable outcomes observed in low-risk patients. However, MSI-high status in LUAD is used as a predictive biomarker for ICI efficacy with positive correlation. Additionally, it is crucial to prioritize the impact of T cell exclusion in LUAD and evaluate the effectiveness of TIDE within our IRGPI. Survival analysis was conducted on a UC cohort that received anti-PD-L1 treatment to confirm the prognostic significance of IRGPI further. Our findings indicate that IRGPI can differentiate various patient outcomes following anti-PD-L1 therapy.
According to a recent investigation, specific indicators, like TIDE and TIS (Tumor Inflammation Signature), can potentially anticipate how a patient will react to immunotherapy. In patients with melanoma treated with first-line anti-PD1 or anti-CTLA-4 antibodies, the TIDE score has shown superior accuracy in predicting prognosis compared to other biomarkers, such as PD-L1 levels and mutational loads. Furthermore, the TIS, which is currently being investigated, utilizes an 18-gene pattern to detect adaptive immune reactions in tumors through the assessment of gene expressions associated with presenting antigens, cytotoxic cells, and interferon-gamma (IFNγ) functions.60 Developed to forecast responses to anti-PD-1, this method has been validated in a limited NSCLC group from nivolumab clinical trials, demonstrating a favorable association with survival.61 Nevertheless, TIDE and TIS primarily concentrated on the role and condition of T lymphocytes, failing to capture the intricacies of the tumor microenvironment implicated in immunotherapeutic reactions. Additionally, both markers concentrated on the patient’s reaction to immunotherapy, which was significant in making clinical treatment choices. In our research, the prognostic significance of IRGPI may be a superior indicator of OS compared to TIDE and TIS during extended monitoring periods. Furthermore, IRGPI consisted of merely four genes, making it more detectable than TIDE and TIS.
Above all, our IRGPI offers broad prospects for clinical applications. It’s meaningful to provide how to use the model briefly. Initially, physicians must undergo an ethical evaluation to obtain subsequent patient data over time and acquire their preserved tumor samples. Next, RNA-seq analysis is required for every LUAD patient to assess the expression of the four genes and integrate them into the model for risk score calculation. The median risk score, as indicated by the risk scores, can be utilized for further analysis, including immune checkpoint assessment, TMB analysis, and evaluation of immune cell microenvironment. Nevertheless, our ongoing study had several constraints. Initially, the expenses for conducting genome sequencing on a large scale with multiple samples are exorbitant. As a result of social ethics and other factors, relatively few cancer samples are available. Moreover, the accuracy of this model needs to be further expanded to include patients for processing. Furthermore, this study necessitates additional verification of biological tests.
Conclusions
To summarize, a strong immune-related prognostic biomarker that indicated the immune microenvironment was created and verified. It can predict patient outcomes and reflect immunotherapeutic efficacies for LUAD patients. Our research presents a unique IRGPI that can forecast prognostic results for patients with LUAD and offers a valuable resource to fulfill clinical needs. However, additional investigations are necessary to elucidate this aspect.
Data Sharing Statement
The information backing the results of this research can be obtained by contacting the author. Ethical approval has been obtained for the patients included in the TCGA and GEO databases. Users can freely download pertinent data for research purposes and publish relevant articles.
Author Contributions
Jiaming He and Tiankuo Luan were responsible for the conception and design. Jiaming He and Gang Zhao were responsible for modifying part of the R language scripts. Yingxue Yang was responsible for data collection and screening. All authors took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This study was not supported by the foundation.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. André T, Shiu KK, Kim TW, et al. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N Engl J Med. 2020;383(23):2207–2218. doi:10.1056/NEJMoa2017699
2. Bellmunt J, de Wit R, Vaughn DJ, et al. Pembrolizumab as Second-Line Therapy for Advanced Urothelial Carcinoma. N Engl J Med. 2017;376(11):1015–1026. doi:10.1056/NEJMoa1613683
3. Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015;373(1):23–34. doi:10.1056/NEJMoa1504030
4. Hodi FS, O’Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi:10.1056/NEJMoa1003466
5. Socinski MA, Jotte RM, Cappuzzo F, et al. Atezolizumab for First-Line Treatment of Metastatic Nonsquamous NSCLC. N Engl J Med. 2018;378(24):2288–2301. doi:10.1056/NEJMoa1716948
6. Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373(2):123–135. doi:10.1056/NEJMoa1504627
7. Garon EB, Hellmann MD, Rizvi NA, et al. Five-Year Overall Survival for Patients With Advanced Non‒Small-Cell Lung Cancer Treated With Pembrolizumab: results From the Phase I KEYNOTE-001 Study. J Clin Oncol. 2019;37(28):2518–2527. doi:10.1200/JCO.19.00934
8. Borghaei H, Brahmer J. Nivolumab in Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med. 2016;374(5):493–494. doi:10.1056/NEJMc1514790
9. Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373(17):1627–1639. doi:10.1056/NEJMoa1507643
10. Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387(10027):1540–1550. doi:10.1016/S0140-6736(15)01281-7
11. Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol. 2019;30(8):1321–1328. doi:10.1093/annonc/mdz167
12. Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14(11):655–668. doi:10.1038/nrclinonc.2017.88
13. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi:10.1186/1471-2105-9-559
14. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. doi:10.1101/gr.239244.118
15. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
16. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi:10.1093/nar/gkaa407
17. Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi:10.1038/s41591-018-0136-1
18. Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi:10.1172/JCI91190
19. Tian Y, Yang J, Lan M, Zou T. Construction and analysis of a joint diagnosis model of random forest and artificial neural network for heart failure. Aging. 2020;12(24):26221–26235. doi:10.18632/aging.202405
20. Song Y, Yan S, Fan W, et al. Identification and Validation of the Immune Subtypes of Lung Adenocarcinoma: implications for Immunotherapy. Front Cell Dev Biol. 2020;8:550. doi:10.3389/fcell.2020.00550
21. Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity. 2018;48(4):812–830.e14. doi:10.1016/j.immuni.2018.03.023
22. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi:10.1038/nature13385
23. Snyder A, Nathanson T, Funt SA, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: an exploratory multi-omic analysis. PLoS Med. 2017;14(5):e1002309. doi:10.1371/journal.pmed.1002309
24. Sholl LM, Hirsch FR, Hwang D, et al. The Promises and Challenges of Tumor Mutation Burden as an Immunotherapy Biomarker: a Perspective from the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol. 2020;15(9):1409–1424. doi:10.1016/j.jtho.2020.05.019
25. Dubuisson A, Fahrner JE, Goubet AG, et al. Immunodynamics of explanted human tumors for immuno-oncology. EMBO Mol Med. 2021;13(1):e12850. doi:10.15252/emmm.202012850
26. Li S, Borowitz MJ. CD79a(+) T-cell lymphoblastic lymphoma with coexisting Langerhans cell histiocytosis. Arch Pathol Lab Med. 2001;125(7):958–960. doi:10.5858/2001-125-0958-CTCLLW
27. Mangogna A, Cox MC, Ruco L, et al. Rituximab Plus Chemotherapy Provides No Clinical Benefit in a Peripheral T-Cell Lymphoma Not Otherwise Specified with Aberrant Expression of CD20 and CD79a: a Case Report and Review of the Literature. Diagnostics. 2020;10(6):341. doi:10.3390/diagnostics10060341
28. Hof J, Visser L, Höppener DJ, et al. B Cells as Prognostic Biomarker After Surgery for Colorectal Liver Metastases. Front Oncol. 2020;10:249. doi:10.3389/fonc.2020.00249
29. Kishimoto T, Akira S, Narazaki M, Taga T. Interleukin-6 family of cytokines and gp130. Blood. 1995;86(4):1243–1254.
30. Garbers C, Hermanns HM, Schaper F, et al. Plasticity and cross-talk of interleukin 6-type cytokines. Cytokine Growth Factor Rev. 2012;23(3):85–97. doi:10.1016/j.cytogfr.2012.04.001
31. Schwertschlag US, Trepicchio WL, Dykstra KH, et al. Hematopoietic, immunomodulatory and epithelial effects of interleukin-11. Leukemia. 1999;13(9):1307–1315. doi:10.1038/sj.leu.2401514
32. Huang G, Yu L, Cooper LJ, et al. Genetically modified T cells targeting interleukin-11 receptor α-chain kill human osteosarcoma cells and induce the regression of established osteosarcoma lung metastases. Cancer Res. 2012;72(1):271–281. doi:10.1158/0008-5472.CAN-11-2778
33. Taniguchi K, Karin M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin Immunol. 2014;26(1):54–74. doi:10.1016/j.smim.2014.01.001
34. Pastor MD, Nogal A, Molina-Pinelo S, et al. IL-11 and CCL-1: novel Protein Diagnostic Biomarkers of Lung Adenocarcinoma in Bronchoalveolar Lavage Fluid (BALF). J Thorac Oncol. 2016;11(12):2183–2192. doi:10.1016/j.jtho.2016.07.026
35. Hellmann MD, Paz-Ares L, Bernabe Caro R, et al. Nivolumab plus Ipilimumab in Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2019;381(21):2020–2031. doi:10.1056/NEJMoa1910231
36. Collins AV, Brodie DW, Gilbert RJ, et al. The interaction properties of costimulatory molecules revisited. Immunity. 2002;17(2):201–210. doi:10.1016/s1074-7613(02)00362-x
37. Chambers CA, Kuhns MS, Egen JG, Allison JP. CTLA-4-mediated inhibition in regulation of T cell responses: mechanisms and manipulation in tumor immunotherapy. Annu Rev Immunol. 2001;19:565–594. doi:10.1146/annurev.immunol.19.1.565
38. Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi:10.1146/annurev.immunol.26.021607.090331
39. Linsley PS, Bradshaw J, Greene J, et al. Intracellular trafficking of CTLA-4 and focal localization towards sites of TCR engagement. Immunity. 1996;4(6):535–543. doi:10.1016/s1074-7613(00)80480-x
40. Chae YK, Arya A, Iams W, et al. Current landscape and future of dual anti-CTLA4 and PD-1/PD-L1 blockade immunotherapy in cancer; lessons learned from clinical trials with melanoma and non-small cell lung cancer (NSCLC). J Immunother Cancer. 2018;6(1):39. doi:10.1186/s40425-018-0349-3
41. Burris HA, Infante JR, Ansell SM, et al. Safety and Activity of Varlilumab, a Novel and First-in-Class Agonist Anti-CD27 Antibody, in Patients With Advanced Solid Tumors. J Clin Oncol. 2017;35(18):2028–2036. doi:10.1200/JCO.2016.70.1508
42. Chudnovsky Y, Kim D, Zheng S, et al. ZFHX4 interacts with the NuRD core member CHD4 and regulates the glioblastoma tumor-initiating cell state. Cell Rep. 2014;6(2):313–324. doi:10.1016/j.celrep.2013.12.032
43. Xia W, Mao Q, Chen B, et al. The TWIST1-centered competing endogenous RNA network promotes proliferation, invasion, and migration of lung adenocarcinoma. Oncogenesis. 2019;8(11):62. doi:10.1038/s41389-019-0167-6
44. Pacholsky D, Vakeel P, Himmel M, et al. Xin repeats define a novel actin-binding motif. J Cell Sci. 2004;117(Pt 22):5257–5268. doi:10.1242/jcs.01406
45. Paul MR, Pan TC, Pant DK, et al. Genomic landscape of metastatic breast cancer identifies preferentially dysregulated pathways and targets. J Clin Invest. 2020;130(8):4252–4265. doi:10.1172/JCI129941
46. Li X, Wu WK, Xing R, et al. Distinct Subtypes of Gastric Cancer Defined by Molecular Characterization Include Novel Mutational Signatures with Prognostic Capability. Cancer Res. 2016;76(7):1724–1732. doi:10.1158/0008-5472.CAN-15-2443
47. Chen H, Chong W, Wu Q, et al. Association of LRP1B Mutation With Tumor Mutation Burden and Outcomes in Melanoma and Non-small Cell Lung Cancer Patients Treated With Immune Check-Point Blockades. Front Immunol. 2019;10:1113. doi:10.3389/fimmu.2019.01113
48. Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet. 2016;387(10026):1415–1426. doi:10.1016/S0140-6736(16)00004-0
49. Jeanson A, Tomasini P, Souquet-Bressand M, et al. Efficacy of Immune Checkpoint Inhibitors in KRAS-Mutant Non-Small Cell Lung Cancer (NSCLC). J Thorac Oncol. 2019;14(6):1095–1101. doi:10.1016/j.jtho.2019.01.011
50. Marinelli D, Mazzotta M, Scalera S, et al. KEAP1-driven co-mutations in lung adenocarcinoma unresponsive to immunotherapy despite high tumor mutational burden. Ann Oncol. 2020;31(12):1746–1754. doi:10.1016/j.annonc.2020.08.2105
51. Frank R, Scheffler M, Merkelbach-Bruse S, et al. Clinical and Pathological Characteristics of KEAP1- and NFE2L2-Mutated Non-Small Cell Lung Carcinoma (NSCLC). Clin Cancer Res. 2018;24(13):3087–3096. doi:10.1158/1078-0432.CCR-17-3416
52. Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. 2018;33(5):843–852.e4. doi:10.1016/j.ccell.2018.03.018
53. Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275–287. doi:10.1038/nrc.2016.36
54. Dong ZY, Zhong WZ, Zhang XC, et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin Cancer Res. 2017;23(12):3012–3024. doi:10.1158/1078-0432.CCR-16-2554
55. Ma W, Gilligan BM, Yuan J, Li T. Current status and perspectives in translational biomarker research for PD-1/PD-L1 immune checkpoint blockade therapy. J Hematol Oncol. 2016;9(1):47. doi:10.1186/s13045-016-0277-y
56. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi:10.1038/s41591-018-0014-x
57. Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi:10.1016/j.immuni
58. Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27(4):462–472. doi:10.1016/j.ccell.2015.02.015
59. Fridman WH, Zitvogel L, Sautès-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. doi:10.1038/nrclinonc.2017.101
60. Danaher P, Warren S, Lu R, et al. Pan-cancer adaptive immune resistance as defined by the Tumor Inflammation Signature (TIS): results from The Cancer Genome Atlas (TCGA). J Immunother Cancer. 2018;6(1):63. doi:10.1186/s40425-018-0367-1
61. Damotte D, Warren S, Arrondeau J, et al. The tumor inflammation signature (TIS) is associated with anti-PD-1 treatment benefits in the CERTIM pan-cancer cohort. J Transl Med. 2019;17(1):357. doi:10.1186/s12967-019-2100-3
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.