Back to Journals » OncoTargets and Therapy » Volume 14
Expression and Prognosis of Sperm-Associated Antigen 1 in Human Breast Cancer
Authors Lin S, Lv Y, Zheng L, Mao G, Peng F
Received 13 November 2020
Accepted for publication 17 March 2021
Published 16 April 2021 Volume 2021:14 Pages 2689—2698
DOI https://doi.org/10.2147/OTT.S288484
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Shuangyan Lin,1 Yanbo Lv,1 Luoning Zheng,1 Genxiang Mao,2 Fang Peng1
1Department of Pathology, Zhejiang Hospital, Hangzhou, Zhejiang, People’s Republic of China; 2Department of Geriatrics, Zhejiang Provincial Key Laboratory of Geriatrics, Hangzhou, Zhejiang, People’s Republic of China
Correspondence: Fang Peng
Department of Pathology, Zhejiang Hospital, 12 Lingyin Road, Hangzhou, Zhejiang, People’s Republic of China
Email [email protected]
Background: Sperm-associated antigen 1 (SPAG1) has been identified as a marker of pancreatic cancer progression and promoter of cell motility; however, its role in breast cancer is not completely understood.
Methods: SPAG1 expression in breast cancer tissues and normal tissues was obtained from online databases. Knockdown function assays were designed and conducted to verify the functional role of SPAG1 in breast cancer cell lines. Cell counting and MTT assays were used to assess cell proliferation. Cell flow cytometry assay was used for cell cycle phase arrest, and fluorescence microscopy was used for colony formation assessment.
Results: Both the mRNA and protein levels of SPAG1 were significantly higher in the breast cancer tissues than in the normal tissues. In addition, SPAG1 is significantly related to many clinicopathological features of breast cancer, such as age (> 51 years), estrogen receptor (ER) (+), progesterone receptor (PR) (+), and nodal status (+), non-triple negative breast cancer (TNBC), not basal-like and not basal-like and not TNBC. Survival analysis indicates that breast cancer patients with low expression of SPAG1 had a significantly better prognosis with relapse-free survival (RFS). Functional experiment analysis revealed that knockdown of SPAG1 suppressed cell proliferation and colony-forming ability.
Conclusion: Our results suggested a possible role of SPAG1 in breast cancer pathogenesis.
Keywords: breast cancer, SPAG1, UALCAN, proliferation, colony formation
Introduction
Breast cancer has long been the greatest threat to women’s health. Its incidence and mortality rates are expected to increase significantly in the next 5 to 10 years,1 and it is increasingly detected in young women (<45 years).2 Based on the status of molecular markers for estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (Her-2), four major molecular subtypes of breast cancer include luminal A, luminal B, Her-2 positive, and triple-negative breast cancer (TNBC).3 Although great progress has been made in the diagnosis and treatment of breast cancer, the current survival period of breast cancer patients is not satisfactory, especially in patients with TNBC.4 Although many research efforts have been concentrated on detecting suitable molecular targets, breast cancer prognosis has not sufficiently improved.5–8 More studies on how to minimize side effects and improve the quality of life in breast cancer patients are needed.
Sperm-associated antigen 1 (SPAG1), also known as SP75, TPIS, CT140, CILD28, HSD-3.8, HEL-S-268, is located at chromosome 8q22.2. The transcript of SPAG1 consists of 3760 bp (RefSeq NM_172218.2) and encodes a protein with 926 amino acid residues.9 SPAG1 expression was found in many normal tissues such as gastrointestinal tract, pancreas, tonsils, lung, skin, liver, kidney, and tracheal, bronchial, and nasal epithelium.10–13 SPAG1 was found to be upregulated in pancreatic cancer,11 seminoma,14 kidney,14 colon,15 and other cancers.15 However, the significance and role of the SPAG1 in breast cancer are poorly understood. In the present study, we first analyzed clinical significance of SPAG1 expression for breast cancer patients by bioinformatics in the public databases. Then, cell counting and MTT assays were used to assess cell proliferation, cell flow cytometry assay for cell cycle phase arrest, and fluorescence microscopy for colony formation assessment.
Materials and Methods
The Expression of SPAG1 in Human Protein Atlas (HPA) Database and UALCAN Database
The HPA database is dedicated to providing the information on tissue and cell distribution for all 24,000 human proteins.16 The expression and distribution of each protein in 48 normal human tissues, 20 tumor tissues, 47 cell lines, and 12 blood cells were examined by immunohistochemistry. We determined the SPAG1 mRNA expression in different normal and cancer tissues using the HPA database. SPAG1 expression in breast cancer and normal breast tissues was analyzed by using UALCAN database.17
Clinical and pathohistological features of SPAG1 in breast cancer
We further studied the differences in SPAG1 expression depending on different clinical and pathological parameters in breast cancer using bc-GenExMiner database,18 which is a database of breast cancer expression. SPAG1 expression was examined by age, estrogen receptor (ER) status, progesterone receptor (PR) status, human epidermal growth factor receptor 2 (HER2) status, lymph node status, triple negative status, basal-like status, scarff-bloom-richardson (SBR) grade, nottingham prognostic index (NPI) score, RSCMC subtypes and HU’s subtypes.
Kaplan–Meier Plotter Analysis
Kaplan–Meier plotter database has been established using gene survival information of cancer patients.19 We explored the correlations between SPAG1 expression and overall survival, relapse-free survival, distant metastases-free survival, or post-progression survival in breast cancer patients using Kaplan-Meier plotter database.
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
The normal breast cell line (MCF-10A) was obtained from the cell bank of Guan Dao Biotechnology (Shanghai, China), and the breast cancer cell lines (T-47D and MDA-MB-231) were purchased from American Type Culture Collection (ATCC, USA). RNA of all three cell lines was isolated using TRIzol kit (Shanghai Pufei Biotechnology, China). SPAG1 primer sequences were (forward) 5′-TTTAATGGAGCTGGATGGACC-3′ and (reverse) 5′-GCTGTATTTACTGAGGGCGTCT-3′. GAPDH primer sequences were (forward) 5′-TGACTTCAACAGCGACACCCA-3′ and (reverse) 5′-CACCCTGTTGCTGTAGCCAAA-3′. All reaction mixtures contained 0.6 μL of the RT-qPCR cDNA product, 6.0 μL SYBR premix ex taq (DRR041B, Takara, Japan), 0.3 μL upstream primer (5 μM), 0.3 μL downstream primer (5 μM), and 5.1 μL RNase-free H2O. PCR reaction was conducted with an initial denaturation step of 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. After all the cycles, 95°C denaturation was performed for 15 s, followed by cooling at 60°C. From 60°C, the temperature was increased until achieving the final temperature of 95°C for 15 s. The relative mRNA expression was calculated with the 2−ΔΔCt method, using GAPDH mRNA expression level for normalization.
shRNA Expression Lentiviral Vector Construction and Cell Transfection
A short hairpin RNA (shRNA) targeting SPAG1 sequences (CCCTGAGAAACTTCCGATA) and a control with scrambled non-silencing RNA (TTCTCCGAACGTGTCACGT) were designed and synthesized to form the recombinant lentiviral SPAG1 shRNA expression vector and its control vector. The accuracy of the constructed vectors was verified via DNA sequencing. The verified lentiviral vectors carrying SPAG1 shRNA and non-silencing RNA were packaged by co-transfecting with Helper 1.0 and Helper 2.0 plasmids in 293T cells. The resulting recombinant lentiviruses were then harvested by centrifugation and purification after 48-h cell culture and named as shSPAG1 and shCtrl. For lentivirus transfection, the cells were cultured in 6-well plates before being transfected with either shSPAG1 or shCtrl according to the multiplicity of infection (MOI) protocol.
Western Blot
The extracted proteins from harvested cells were separated on 10% SDS-PAGE gel and electrotransferred onto the PVDF (polyvinylidene fluoride) membrane (Millipore, USA) following the standard SOP (standard operating procedure).20–22 The membranes were then blocked with 5% non-fat dry milk in TBST (Tris buffered saline Tween) buffer for 1 h at room temperature and incubated with primary rabbit anti-human SPAG1 polyclonal antibody at a concentration of 1:2000 (Sigma, f1804) for 2 h at room temperature. GAPDH antibody (mouse anti-GAPDH monoclonal antibody, 1:2000, sc-32,233) from Santa Cruz was used as the loading control. After washing, the membranes were incubated with the secondary antibody (1:2000, sc-2005, Santa Cruz, USA) for 1.5 h at room temperature, and visualized using the Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific, USA).
Cell Proliferation Assay
The transfected shSPAG1 and shCtrl cells were adjusted to 2000 cells/100 μL per well and cultured at 5% CO2 and 37°C for 24 h. The plates were then analyzed for GFP expression in each well using the Celigo Imaging Cytometer (Nexcelom, USA) over a 5-day period. Subsequently, MTT assay was conducted. Briefly, the shSPAG1 and shCtrl transfected cells were collected and adjusted to 2000 cells/well for continuous detection over a 5-day period. Then, 20 μL MTT (5 mg/mL) was added to each well for 4-h before the termination of the culture. The optical densities of each well were measured at 490 nm by using a M2009PR microplate reader (Tecan infinite, Swiss).
Cell Cycle and Colony Formation
The shSPAG1 and shCtrl cell cultures were treated with trypsin and resuspended in the standard medium after achieving 80% confluence. After washing, the cells were collected, stained with 0.8 mL propidium iodide (PI), and then filtered through a 300-mesh nylon mesh and analyzed by flow cytometry using a FACS Calibur instrument (Millipore, USA) at a flow rate of 300 ~ 800cells/s. For colony formation assays, a total of 500 cells/well were seeded onto six-well plates, cultured at 37°C and 5% CO2 over a period of 10 days. Then, 4% paraformaldehyde was added to fix the cells before staining with purple crystal (Shanghai Sangon, China) 1000 μL/well for 20 min. After that, colonies were counted manually.
Statistical Analysis
Statistical analysis was performed by using SPSS software (version 18; IBM Corp., Armonk, NY, USA). All experiments were repeated three times. The results were shown as mean ± standard deviation (SD). An unpaired two-tailed Student’s t-test was used to analyze the differences between two groups. P < 0.05 was considered statistically significant.
Results
Expression Profile of SPAG1 in Human Normal and Cancer Tissues
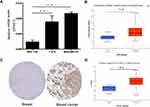
A high and detectable expression level of a gene is one of the most important traits needed for being considered a promising diagnostic or prognostic biomarker.23 Therefore, in the first place, we determined the expression of SPAG1 in different cancer tissues using the Human Protein Atlas (HPA) database. The results demonstrated that the top 10 cancer tissues according to the expression level of SPAG1 protein were the liver cancer, thyroid cancer, pancreatic cancer, head and neck cancer, lung cancer, renal cancer, urothelial cancer, endometrial cancer, breast cancer, and glioma (Figure 1A). The top 10 cancer tissues sorted by mRNA levels of SPAG1 were colorectal cancer, stomach cancer, urothelial cancer, lung cancer, breast cancer, pancreatic cancer, head and neck cancer, cervical cancer, endometrial cancer, and glioma (Figure 1B). The expression of SPAG1 mRNA and protein in different types of normal tissues was relatively low compared with the expression in cancer tissues, especially in the breast tissues, which was also analyzed using the HPA database (Figure 1C and D). Taken together, among all types of cancer, breast cancer presented the highest expression of SPAG1 in both mRNA and protein levels.
Next, we determined the expression of SPAG1 in breast cancer cell lines compared with normal breast cell line. Figure 2A shows that SPAG1 expression in two breast cancer cell lines (MDA-MB-231 and T-47D) was significantly higher than in the normal cell line (MCF-10A). We further analyzed the SPAG1 expression in breast cancer expression data from TCGA samples based on UALCAN database (Figure 2B). The mRNA of level of SPAG1 was higher in breast cancer tissues compared with normal breast tissues. In addition, the protein level of SPAG1 was also upregulated in breast cancer compared with normal breast tissue from HPA database and UALCAN database (Figure 2C and D). These results revealed that upregulation of SPAG1 may be correlated with the progression of breast cancer.
Clinical and Pathological Features of SPAG1 in Breast Cancer
To further study the differential expression of SPAG1 in breast cancer, we analyzed the bc-GenExMiner database based on different clinicopathological parameters in breast cancer. As shown in Figure 3, SPAG1 was markedly upregulated in breast cancer with age (>51 years) (Figure 3A), ER (+) (Figure 3B), PR (+) (Figure 3C), nodal status (+) (Figure 3E), not TNBC (Figure 3F), not basal-like (Figure 3G), and not basal-like and not TNBC (Figure 3H). Furthermore, SPAG1 expression was significantly associated with scarff-bloom-richardson (SBR) grade (Figure 3I), RSCMC subtypes (Figure 3K) and Hu’s subtypes (Figure 3L). However, no significant difference in SPAG1 expression was observed between Her-2 (+)/Her-2 (-) (Figure 3D) and NPI score (Figure 3J). Taken together, all these results indicated that SPAG1 was an important molecular indicator of breast cancer.
Kaplan–Meier Plotter Analysis
Then, we further explored the prognostic values of SPAG1 in breast cancer by using the Kaplan–Meier plotter database. As shown in Figure 4, no statistical significance was observed between the SPAG1 high-expression group and SPAG1 low-expression group for predicting overall survival (Figure 4A), distant metastasis-free survival (Figure 4C) and post-progression survival (Figure 4D). However, breast cancer patients with low expression of SPAG1 had a significantly more favorable prognosis with relapse-free survival (RFS) (Figure 4B). The results demonstrated that high expression of SPAG1 was associated with poor prognosis of breast cancer.
Downregulation of SPAG1 Inhibited Cell Proliferation
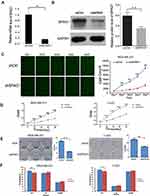
To further explain the molecular mechanism of SPAG1 in breast cancer, we conducted downregulation of SPAG1 experiments in MDA-MB-231 cells and T-47D cells. Post-transfection, the expression of SPAG1 was analyzed via RT-qPCR for mRNA and Western blot for protein. As shown in Figure 5, both the mRNA of SPAG1 (Figure 5A) and the protein expression of SPAG1 (Figure 5B) were significantly inhibited in shSPAG1 cells compared to that of the control (P<0.01), indicating successful down regulation of gene expression by shSPAG1. Then, we monitored the cell growth of the transfected cells with the shSPAG1 or shCtrl lentiviruses by measuring the GFP expression in MDA-MB-231 cells (Figure 5C) and MTT assay over a 5-day period in MDA-MB-231 cells and T-47D cells (Figure 5D). The proliferation was significantly lower in the shSPAG1 cells than in the control cells (P < 0.01). The difference in cell proliferation between the shSPAG1 and shCtrl cells was observed on the second day of the analysis. The difference became more notable in a time-dependent manner, indicating inhibitory effect of SPAG1 knockdown on cell proliferation. These data indicated that SPAG1 contributed to cell proliferation in breast cancer.
Downregulation of SPAG1 Decreased Cell Colony Formation
In order to further study whether SPAG1 has an effect on cell cloning ability, we investigated colony formation by light microscopy. Our data showed that downregulation of SPAG1 gene in MDA-MB-231 and T-47D cells significantly decreased the number of colonies compared with the control cells (P < 0.05, Figure 5E). The results showed that downregulation of SPAG1 inhibited cell colony formation in breast cancer.
Effects of Downregulation of SPAG1 on the Cell Cycle Progression
To further understand the role of SPAG1 in breast cancer cells, we further examined the effect of SPAG1 downregulation on cell cycle progression in breast cancer cells by fluorescence activated cell sorting (FACS). Our result revealed that the shSPAG1 group had fewer cells in the G1 phase (P<0.05) compared with the control group, but there was no significant change in the cells of S, G2/M phase (P>0.05, Figure 5F).
Discussion
Sperm-associated antigen 1 (SPAG1), localized primarily in the neck and midpiece of pachytene primary spermatocytes, has been implicated in infertility and tumorigenesis.24 Here we showed that although SPAG1 was expressed in many tumor tissues, breast cancer presented the highest expression of SPAG1 in both mRNA and protein levels. A previous study showed that SPAG1 was an important regulator of mammalian oocyte meiotic progression and was associated with meiotic spindles; its depletion severely compromised M-phase entry (germinal vesicle breakdown [GVBD]) and polar body extrusion via its involvement in AMPK and MAPK signaling pathways.24 SPAG1 was also reported to play a role in the cytoplasmic assembly and/or trafficking of the axonemal dynein arms.25
Silina et al showed that SPAG1 was immunogenic and presented high expression in normal colon, but upregulated prominently in lung and breast cancers,15 which is consistent with our results. In our study, we found the expression of SPAG1 was significantly upregulated in breast cancer compared with the normal breast tissue. It was also showed that SPAG1 was colocalized with tubulin, and has been shown as a progression marker of pancreatic cancer and promoter of cell motility.11 In our study, we demonstrated that SPAG1 was related to the development of breast cancer. Although SPAG1 was highly expressed in breast cancer tissue and was related to many clinicopathological features, SPAG1 did not have significantly related to TNBC or basal-like breast cancer. As is well known, TNBC is a more aggressive subtype of breast cancer with the absence of the expressions of ER, PR, and Her-2, and has the poorest prognosis, due to the lack of molecular targeted treatment. TNBC sometimes called as triple negative basal-like breast cancer because of expressing basal marker proteins.26 Furthermore, TNBC is heterogeneous by showing several different histopathological and molecular subtypes with different prognosis.27,28 Therefore, the high expression of SPAG1 in breast cancer is not related to TNBC or basal like breast cancer, it needs more study for the potential mechanism. Additionally, we found that breast cancer patients with low expression of SPAG1 had a significantly more favorable prognosis only with RFS, based on the Kaplan–Meier plotter database. In summary, SPAG1 might be a promising biomarker and therapeutic target for breast cancer.
At present, the cellular mechanism of SPAG1 in breast cancer is poorly understood. Yet, here we downregulated the SPAG1 and examined its effects on the proliferation, colony formation, and cell cycle progression in the MDA-MB-231 cells and T47D cells. Our results demonstrated that the proliferation activity and the number of colonies were significantly suppressed in SPAG1 knockdown cells relative to the control. Cell cycle analysis showed the ratio of G1 phase was significantly reduced, while the ratio of G2/M and S phase increased slightly with no significant difference. Further studies are needed to clarify the mechanism.
Conclusion
In our current study, both the mRNA and protein levels of SPAG1 were highly expressed in breast cancer cells. SPAG1 expression was significantly related to many clinicopathological features, and low expression was related to RFS. Furthermore, SPAG1 suppressed cell proliferation and colony formation. Taken together, these results implied that SPAG1 may play an essential role in breast cancer pathogenesis and development. However, further studies are required to precisely characterize the molecular mechanism of SPAG1.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
This work is supported by the Zhejiang Provincial Public Welfare Technology Research Program (LGF18H160028), National Natural Science Foundation (81771520), Zhejiang Provincial Health Bureau Foundation (2019KY257)
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Greaney ML, Sprunck-Harrild K, Ruddy KJ, et al. Study protocol for young & strong: a cluster randomized design to increase attention to unique issues faced by young women with newly diagnosed breast cancer. BMC Public Health. 2015;15(1):37. doi:10.1186/s12889-015-1346-9
2. Anastasiadi Z, Lianos GD, Ignatiadou E, Harissis HV, Mitsis M. Breast cancer in young women: an overview. Updates Surg. 2017;69(3):313–317. doi:10.1007/s13304-017-0424-1
3. Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321(3):288–300. doi:10.1001/jama.2018.19323
4. Tan R, Li H, Huang Z, et al. Neural functions play different roles in Triple Negative Breast Cancer (TNBC) and non-TNBC. Sci Rep. 2020;10(1):3065. doi:10.1038/s41598-020-60030-5
5. Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi:10.1038/35021093
6. Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi:10.1038/nature10983
7. Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12(5):323–334. doi:10.1038/nrc3261
8. Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi:10.1038/nature11412
9. Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21(11):932–939. doi:10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N
10. Zhang ML, Wang LF, Miao SY, Koide SS. Isolation and sequencing of the cDNA encoding the 75-kD human sperm protein related to infertility. Chin Med J. 1992;105(12):998–1003.
11. Neesse A, Gangeswaran R, Luettges J, et al. Sperm-associated antigen 1 is expressed early in pancreatic tumorigenesis and promotes motility of cancer cells. Oncogene. 2007;26(11):1533–1545. doi:10.1038/sj.onc.1209961
12. Lin W, Zhou X, Zhang M, et al. Expression and function of the HSD-3.8 gene encoding a testis-specific protein. Mol Hum Reprod. 2001;7(9):811–818. doi:10.1093/molehr/7.9.811
13. Takaishi M, Huh N. A tetratricopeptide repeat-containing protein gene, tpis, whose expression is induced with differentiation of spermatogenic cells. Biochem Biophys Res Commun. 1999;264(1):81–85. doi:10.1006/bbrc.1999.1477
14. Biermann K, Heukamp LC, Steger K, et al. Genome-wide expression profiling reveals new insights into pathogenesis and progression of testicular germ cell tumors. Cancer Genomics Proteomics. 2007;4(5):359–367.
15. Silina K, Zayakin P, Kalnina Z, et al. Sperm-associated antigens as targets for cancer immunotherapy: expression pattern and humoral immune response in cancer patients. J Immunother. 2011;34(1):28–44. doi:10.1097/CJI.0b013e3181fb64fa
16. Ponten F, Schwenk JM, Asplund A, Edqvist PH. The human protein atlas as a proteomic resource for biomarker discovery. J Intern Med. 2011;270(5):428–446. doi:10.1111/j.1365-2796.2011.02427.x
17. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
18. Jezequel P, Campone M, Gouraud W, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131(3):765–775. doi:10.1007/s10549-011-1457-7
19. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi:10.1007/s10549-009-0674-9
20. Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76(9):4350–4354. doi:10.1073/pnas.76.9.4350
21. Burnette WN. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate–polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112(2):195–203. doi:10.1016/0003-2697(81)90281-5
22. Fuentes JM, Lompre AM, Moller JV, Falson P, le Maire M. Clean Western blots of membrane proteins after yeast heterologous expression following a shortened version of the method of Perini et al. Anal Biochem. 2000;285(2):276–278. doi:10.1006/abio.2000.4784
23. Lou W, Liu J, Ding B, et al. Five miRNAs-mediated PIEZO2 downregulation, accompanied with activation of hedgehog signaling pathway, predicts poor prognosis of breast cancer. Aging. 2019;11(9):2628–2652. doi:10.18632/aging.101934
24. Huang C, Wu D, Khan FA, Jiao X, Guan K, Huo L. The GTPase SPAG-1 orchestrates meiotic program by dictating meiotic resumption and cytoskeleton architecture in mouse oocytes. Mol Biol Cell. 2016;27(11):1776–1785. doi:10.1091/mbc.e16-02-0132
25. Knowles MR, Ostrowski LE, Loges NT, et al. Mutations in SPAG1 cause primary ciliary dyskinesia associated with defective outer and inner dynein arms. Am J Hum Genet. 2013;93(4):711–720. doi:10.1016/j.ajhg.2013.07.025
26. Bao B, Prasad A. Targeting CSC in a most aggressive subtype of breast cancer TNBC. Adv Exp Med Biol. 2019;1152:311–334.
27. Liao HY, Zhang WW, Sun JY, Li FY, He ZY, Wu SG. The clinicopathological features and survival outcomes of different histological subtypes in triple-negative breast cancer. J Cancer. 2018;9(2):296–303. doi:10.7150/jca.22280
28. Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13(11):674–690. doi:10.1038/nrclinonc.2016.66
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.